Abstract
Sole-carbon-source tests (Biolog), designed to identify bacteria, have become very popular for metabolically fingerprinting soil microbial communities, despite disadvantages associated with the use of carbon source profiles that primarily select for fast-growing bacteria. In this paper we describe the use of an alternative method that combines the advantages of the Biolog community-level physiological profile (CLPP) method, in which microtiter-based detection plates are used, with the ability to measure carbon dioxide evolution from whole soil. This method facilitates measurement over short periods of time (4 to 6 h) and does not require the extraction and culturing of organisms. Deep-well microtiter plates are used as test wells into which soil is placed. The apparatus to fill the deep-well plates and interface it with a second removable detection plate is described. Two detection systems, a simple colorimetric reaction in absorbent alkali and scintillation counting with radioactive carbon sources, are described. The methods were compared to the Biolog-CLPP system by using soils under different vegetation types and soil treated with wastewater sludge. We aimed to test the hypothesis that using whole soil would have specific advantages over using extracts in that more immediate responses to substrates could be obtained that would reflect activity rather than growth. The whole-soil method was more rapid and gave earlier detection of C source use. Also, the metabolic fingerprints obtained could discriminate between sludge treatments.
When the health and activity of a soil are assessed, a basic method is to measure the amount of carbon dioxide (CO2) that is being respired by soil microorganisms that are decomposing organic substrates within the soil (1). It is also possible to measure the substrate-induced respiration (SIR) by measuring the amounts of CO2 before and after addition of a substrate, such as glucose (2). This provides additional information about the size of the microbial biomass and how different soils might respond to stress, such as the addition of pollutants or organic matter. The individual species that comprise a soil microbial community have different abilities to respire different substrates, so that by adding different substrates it is possible to obtain a catabolic fingerprint of the community (12) or a community-level physiological profile (CLPP) (20). Measuring the respiration of a large number of carbon sources (e.g., 16 to 95 carbon sources) can be laborious and time-consuming as most methods require between 5 and 150 g of soil in 100-ml or 2.5-liter glass jars. Consequently, the methods used for measurement can be difficult to automate when large numbers of samples are processed.
Another method of testing multiple C sources involves extraction of the community from the soil into an aqueous suspension before inoculation into a microtiter test plate and measurement of the subsequent growth (16), usually by using commercially available test plates (Biolog). Use of sole-carbon-source tests in this way has become a popular way to measure changes in community structure and functional diversity (16, 27) and has been applied to a wide range of soil habitats undergoing changes in land use or disturbance due to pollution (3, 6, 8, 18, 26). This method can be used to simultaneously test the utilization of 95 different substrates as sole carbon sources. Carbon source utilization is indicated by the color development of a redox indicator dye, and changes in the overall patterns of carbon source utilization rates indicate differences in community composition. The technique is widely used in part because it is simple, because an automated measuring apparatus is used, and because the procedure yields a great deal of information about an important functional attribute of microbial communities. However, the fundamental basis of using such an approach for ecological studies has been questioned due to its dependence on inefficient extraction, the physiological status of inoculated cells, the subsequent growth of cells, and the relevance and concentrations of the C substrates used (19). Furthermore, analysis and interpretation of the data are often complicated (14), and the degree to which the results reflect function rather than structure has also been questioned (15). Nevertheless, such methods have been shown in some cases to be as sensitive as or more sensitive than measuring microbial biomass and respiration (18), and they have been used to detect the effects of important environmental changes on sensitive ecosystems (17).
Although it is possible to make several modifications to the original approach by selecting more ecologically relevant carbon substrates (8, 9) and using modified cultural conditions for assessing acid soils (26), the Biolog approach remains a method that relies on extraction of a soil suspension and subsequent growth of organisms. The results are biased towards organisms that are (i) readily extractable and (ii) able to develop rapidly within the pH-neutral, nutrient-rich, aqueous environment of the test well (23). These conditions tend to discriminate against fungi and other slowly growing organisms that have filamentous habits and a preference for more acidic conditions. The method also relies on substrates that are soluble, readily metabolized, and present at high concentrations (3 mg well−1). Most importantly, the method relies on growth of organisms in the wells, even though the original concept for identifying pure cultures did not (5), so that selection by the growth conditions and C substrates determines the CLPP. The rate of color development is dependent on the inoculum density, but in extracts of soil, which are diluted, it is difficult to get cell densities equivalent to those recommended by the manufacturers. Consequently, long lag phases in color development with the Biolog-CLPP technique are often reported. Most applications require an incubation time of between 72 and 168 h in order to collect kinetic data that allow numerical solutions to the bias attributed to differences in inoculum density (14).
There is, therefore, a need to develop methods that can be used to assess the whole soil population in a system and that rely more on indigenous microbial activity than on microbial growth. A whole-soil method based on the SIR response of slurried soil in glass jars has been developed (12) and applied (10, 11) in a way similar to the way in which Biolog-based techniques have been applied, but compared with Biolog-based techniques, it is more laborious and still requires high substrate concentrations. These two approaches have not previously been compared in the same study, and it is difficult to be equivocal about the merits of the different approaches. Here we describe a rapid microrespirometry method that can be performed in a 96-well microtiter plate, which has the advantage of being compact and holding many samples and/or replicates and can be read with existing plate readers. We used this method to compare the Biolog and whole-soil approaches. We hypothesized that use of whole soil would have specific advantages compared to methods in which extracts are used because more immediate responses to substrates could be obtained. This should reflect activity rather than growth and open up new avenues for CLPP approaches to assess microbial community structure.
MATERIALS AND METHODS
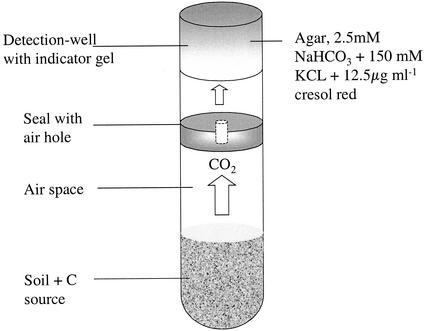
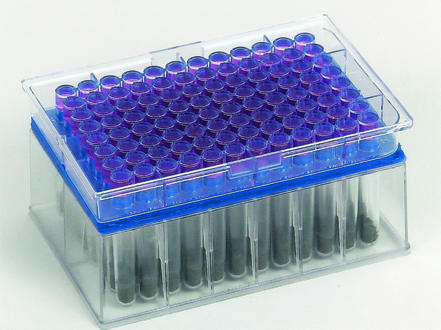
The microrespirometry system described here, referred to below as MicroResp (United Kingdom and United States patents have been applied for), consists of two microtiter plates placed face to face; one of these plates, a deep-well plate with a capacity of 1.2 ml (Thermo LifeSciences, Basingstoke, United Kingdom), holds soil samples with added carbon sources, and the other plate is a system for detecting the evolved carbon dioxide (Fig. 1). The two plates are sealed together with a silicone rubber gasket with interconnecting holes between the corresponding wells (Fig. 2). The assembly is held together firmly with a purpose-designed clamp (data not shown). The moisture content of sieved soil is preadjusted to the required level (40% of the water-holding capacity) so that after the carbon source in solution is added, the moisture content is 60% of the soil's water-holding capacity.
FIG. 1.
Schematic diagram of a deep well connected to a detection well, showing the position and composition of the dye detection system.
FIG. 2.
Assembled MicroResp system comprising a deep-well microtiter plate to hold soil, an interconnecting gasket, and a top plate with a detection gel.
We used two methods to detect the evolved carbon dioxide for assaying substrate utilization. The first method is a colorimetric method that relies on the change in the pH of a solution of bicarbonate in quasi-equilibrium with the well headspace (Fig. 2). CO2 absorption in the alkali, which is set in a gel containing a pH indicator dye (21), causes a color change. The indicator dye, cresol red (12.5 ppm, wt/wt), potassium chloride (150 mM), and sodium bicarbonate (2.5 mM) are set in 150 μl of Noble agar (1%) in each well of the detection plate. After they set, the plates are stored in plastic bags with wet paper towels and soda lime to ensure that they do not desiccate or react with atmospheric CO2. A calibration curve for absorbance versus headspace equilibrium CO2 concentration was determined by equilibrating dye solutions at different CO2 concentrations prepared with standard gas mixtures (21) or by incubating four very different soils in soil jars, with or without glucose (30 mg of soil ml of water−1), for 6 h and measuring the headspace CO2 concentration every 2 h. Each jar also contained four microtiter wells detached from breakable CombiStrips (Thermo LifeSciences) and prepared in the manner described above for the detection plate. CombiStrips comprise strips of eight microtiter wells, each of which is detachable; however, the wells can be reassembled in a plate carriage to be read as a 96-well plate when required. At the end of 6 h these strips were put in a plate carriage and read with the plate reader.
In the second detection system (the radioactive test) radioactive substrates are used, and the radioactive carbon dioxide that evolves is detected by trapping it in 40 μl of alkali (2 M NaOH). In this case the alkali solution is not set in gel so the system is assembled upside down. The moist soil placed in the deep well remains in place and does not fall out when the apparatus is turned upside down. After incubation, the lower detection well microtiter plate is removed, 200 μl of scintillant (Wallac Optiphase Supermix) is added to the wells, and the radioactivity is determined with a microplate liquid scintillation counter (Wallac MicroBeta TriLux counter; Wallac Oy, Turku, Finland). Incubation is usually for 24 to 48 h.
The carbon sources were selected from the previously described carbon sources (9) based on carbon sources that are ecologically relevant to soil (specifically as plant root exudates) and that can be dissolved in water (Table 1). Each carbon source was dissolved in deionized water and prepared as a stock solution at a concentration designed to deliver 30 mg of C g of soil water−1 (25) in each deep well when 25 μl was dispensed. Some less soluble substrates were prepared as stock solutions to deliver 7.5 mg of C ml−1 (Table 1). The C source was dispensed before the soil was added to ensure that the soil contacted the C source at the same time for all wells. The position of each C source was recorded on a template but the configuration was the reverse of the desired configuration so that the configuration on the detection plate, which was inverted on the deep-well plate, was in the desired order.
TABLE 1.
Concentrations of different carbon sources added to soil in MicroResp CLPP tests when the colorimetric and radioactive (14C) detection systems were used
| Carbon source | Concn (mg g of soil water−1)
|
|
|---|---|---|
| Colorimetric test | Radioactive test | |
| None | 0 | |
| l-Alanine | 7.5 | |
| l-Arabinose | 30 | 0.77 |
| l-Arginine | 30 | |
| dl-Aspartic acid | 7.5 | 1.14 |
| γ-Aminobutyric acid | 30 | |
| Protocatechuic acid | 30 | |
| Citric acid | 30 | |
| d-Fructose | 30 | |
| d-Galactose | 30 | 0.77 |
| d-Glucosamine hydrochloride | 0.92 | |
| N-Acetylglucosamine | 7.5 | |
| d-Glucose | 30 | 0.77 |
| Glycine | 1.29 | |
| l-Leucine | 0.45 | |
| l-Lysine hydrochloride | 30 | 0.67 |
| l-Malic acid | 30 | |
| oxalic acid | 30 | |
| d-(+)-Trehalose | 30 | |
For the 14C method, radiolabeled substrate was mixed with cold substrate to give 200 Bq well−1 and approximately 1 mg ml of soil water−1, respectively. The actual quantities of substrate (Table 1) were calculated so that the substrate consumed approximately 100 and 70 μl of oxygen well−1, assuming complete oxidation, during the 6 and 144 h of incubation for the colorimetric and 14C methods, respectively. This ensured that the wells did not become anaerobic during extended incubation.
Soil was placed into the deep-well plate by using a third device made from a 300-μl well microtiter plate from which the bottom had been removed and replaced with a Perspex sliding base. Three hundred microliters (total volume) of soil was placed into this plate, and the plate was tapped gently to ensure consistent packing. The weight of soil was recorded, and the packing density was calculated. The plate was positioned over the deep-well plate, and the false bottom was removed, which allowed the soil to fall into the deep wells and make contact with the C source solution. The deep-well plate was then immediately sealed with the gasket and detection plate.
For colorimetric detection the detection plate was read immediately before and after 6 h of incubation at 25°C with a microtiter plate reader (VMAX; Molecular Devices, Wokingham, United Kingdom) at 590 nm. The absorbance after 6 h was normalized for any differences recorded at zero time before exposure and then converted to the headspace CO2 concentration by using the calibration curve. The average amount of CO2 that evolved per sample was calculated and used to normalize individual C source concentrations before the multivariate analysis.
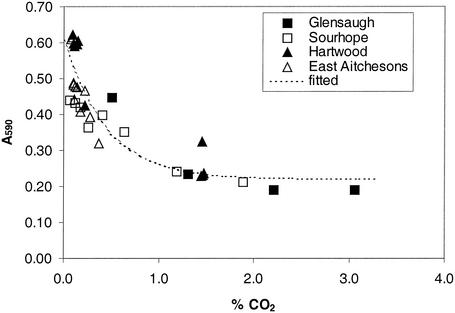
The calibration of dye color with CO2 concentration by using equilibrated solutions (data not shown) gave calibration data similar to previously published data (21). Using the respiration of different soils to calibrate the dye color set in agar in microtiter strips also revealed an exponential relationship between absorbance and the percentage of CO2, but the parameters were different from those of previously described relationships (21) (Fig. 3). The best fit for the calibration curve was as follows: percentage of CO2 = A + B × e−kx, where k is −ln(R), R is 0.106, B is 0.384, A is 0.222, and x is A590 (Fig. 3). This curve was based on four soils measured over 6 h and was highly significant (P < 0.001; r2= 0.79). The calibration curve represented the dynamic equilibration that occurs over short incubation times, reflected the operating conditions more accurately, and was therefore used in all subsequent calculations of respired CO2.
FIG. 3.
Calibration curve for absorbance (A590) versus percentage of CO2 for four different soils.
Biolog CLPPs were determined for the same soils by direct incubation of soil suspensions in a combination of GN2 and MT2 plates (Biolog Inc., Hayward, Calif.) containing 125 different C sources in individual wells to determine changes in the relative and absolute rates of utilization of individual substrates. Soil samples (10 g) in 100 ml of sterile deionized water were shaken on a wrist action shaker at full speed for 10 min. After centrifugation at 500 × g for 10 min to separate the soil, 5 ml of each suspension was used to make a 10-fold dilution series up to 10−3, from which 150 μl of supernatant was inoculated into each well of a GN2 plate (Biolog Inc.) and an MT plate prepared with 30 additional, ecologically relevant carbon sources (9). The plates were incubated in the dark at 25°C in a sealed polyethylene bag and read at a wavelength of 590 nm at zero time and every 12 h for 7 days by using an automated plate reader (VMAX; Molecular Devices) and Microlog Rel 3.7 software (Biolog Inc.).
Two experiments were used to test the different systems. In the first experiment, the colorimetric method was used with soils that had been treated with sewage sludge (4). Six treatments were used, including an unamended control, a digested sludge low in metals, an undigested sludge low in metals, a Zn-rich digested sludge, a Cd-rich digested sludge, and a Cu-rich undigested sludge. All the sludges were obtained from sewage works in the United Kingdom. The sludges rich in metals were selected to be predominantly enriched in Zn, Cu, or Cd compared to other metals and had been used to obtain target concentrations of Zn, Cu, and Cd of 300, 135, and 3 mg/kg, respectively, in the soil (4). Because the type of sewage treatment is likely to affect the availability of readily available organic matter in a sludge, both undigested and digested sludges low in metals were included to separate effects due to metals and effects due to organic input. The treatments were replicated in three fully randomized blocks in individual plots (6 by 8 m). The experiment was performed at the Hartwood Research Station in Lanarkshire, Scotland (National Grid Reference NS855602) with soil that is a medium-texture sandy clay loam (pH 5.8, 21% clay, 4.7% organic carbon). Soil samples were taken with an auger to a depth of 25 cm in 2001, 3 years after the last application of sludge. Twenty-five cores were taken in each plot, and the soil was pooled to obtain a composite sample for each plot. The soil samples were sieved (mesh size, <2 mm) and sorted to remove soil animals and plant debris. The moisture content was then adjusted to 40% of the water-holding capacity, and the samples were incubated for 2 to 4 days in polyethylene bags at 25°C to condition them after the disturbance due to sieving.
In the second experiment, the radioactive carbon source method was used to measure rates of C source utilization in soil from a coniferous forest at Fetteresso, Aberdeenshire, Scotland (National Grid Reference NO819869). Soil samples (five replicates) down to a depth of 15 cm were obtained from a 50-year-old plantation forest of Sitka spruce (Picea sitchensis) on a common major soil subgroup (brown forest soil with gleying). After the litter layer was removed, the soil FH horizon was pretreated as described above. The soil had total C and N contents of 10.7 and 0.52%, respectively, and a pH in water of 3.16. Only the time course of C source utilization is described below to illustrate the utility of the method; the complete results of the study will be presented elsewhere.
Statistical analysis.
The average C source utilization results (i.e., the average well color development [AWCD] for the Biolog test or the amount of CO2 respired for the MicroResp test) for each sample were compared, as were the individual C source responses. The soil respiration values (colorimetric method) and absorbance data (Biolog test) for the various carbon sources and treatments were subjected to multivariate analysis initially standardized by subtracting the no-carbon control value and dividing by the average value for each sample. C source data for equivalent times in the mid-exponential phase were obtained by fitting a logistic curve to the growth expressed in AWCD for each sample and finding the time of the central point of inflexion. A weighted average was determined from the individual C source data obtained at times on either side of the point of inflexion, with weights calculated from the fitted lines around the point of inflexion. This procedure simulated the approach of Garland (15) in that kinetic data were used to correct for differences in the initial inoculum density. Principal-component analysis was then performed on the correlation matrix, followed by canonical variate (CV) analysis of the first 10 principal components, which explained >95% of the variation (Table 2). The aim of the principal-component analysis was to reduce the dimensionality enough to allow CV analysis to be carried out and ensure that similar dimensionality was being compared between methods. The mean Mahalanobis distance for all groups was used to measure the overall separation of the groups. The significance of the mean distance was tested by using a randomization test. The samples were randomly reassigned to treatments, the CV analysis was performed, and the mean distance was noted. This was repeated 1,000 times, and the number of times that the mean distance exceeded the distance for the real data was recorded and used to calculate a P value. All computations were carried out by using Genstat 5, release 4.22 (VSN International Ltd., Oxford, United Kingdom).
TABLE 2.
Variation explained in the first 10 principal components, mean Mahalanobis distances between treatment groups, and significance (P values) from CV analysis of sole-carbon-source tests when MicroResp and Biolog CLPP methods were used with different numbers of C sources
| Test | No. of C sources | % Variation | Mean group distance | P value |
|---|---|---|---|---|
| MicroResp colorimetric | 15 | 97.3 | 18.07 | 0.021 |
| Biolog | 125 | 98 | 4.49 | 0.801 |
| Biolog GN | 95 | 93.5 | 5.63 | 0.584 |
| Biolog | 15 | 99.3 | 4.57 | 0.806 |
RESULTS AND DISCUSSION
MicroResp colorimetric detection.
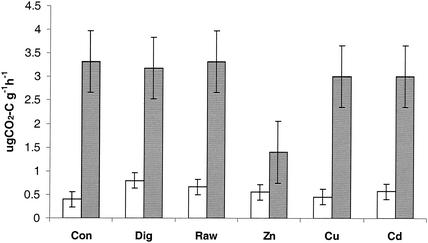
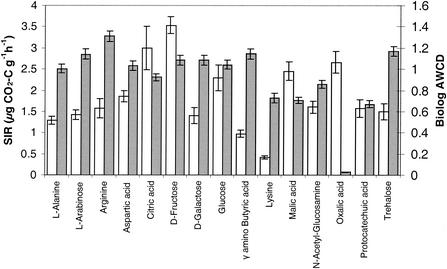
The average basal respiration (with no carbon source) for each treatment showed that the digested blank sludge had significantly greater soil respiration than the unamended control soils, but no other significant differences between different types of sludge-amended soil were found (Fig. 4). In contrast to the basal respiration, the average SIR data showed that the Zn-rich sludge treatment resulted in significantly lower SIR than any other treatment (Fig. 4). Addition of all the C sources resulted in detectable SIR in all soil samples (Fig. 5). The highest level of SIR was observed with d-fructose, and the lowest level of SIR was observed with l-lysine. Addition of oxalic acid resulted in no detectable SIR in the Biolog plates, in contrast to the relatively large response in the MicroResp system (Fig. 5). The overall evenness of the SIR values for the different C sources was greater in the Biolog-CLPP system than in the MicroResp system. Consequently, the whole-soil method may provide a greater opportunity for discriminating different samples. The general findings for stimulation of the basal respiration by sludge addition are consistent with trends observed in the same experiment in which basal respiration was measured by analysis of headspace gas over a 6-h period (4). The effect of Zn-rich sludge on SIR also suggests that there was an effect on the overall microbial biomass that has not been detected previously (4).
FIG. 4.
Basal respiration (open bars) and SIR for glucose (gray bars) in a pasture soil treated with either a no-sludge control (Con), digested blank sludge (Dig), blank undigested sludge (Raw), digested sludge rich in either zinc (Zn) or cadmium (Cd), or undigested sludge rich in copper (Cu). The error bars indicate a least significant difference at 0.05.
FIG. 5.
Substrate-induced responses for 15 carbon sources in a pasture soil as determined by using the MicroResp colorimetric detection system (open bars) and Biolog AWCD (gray bars). The error bars indicate ± standard error.
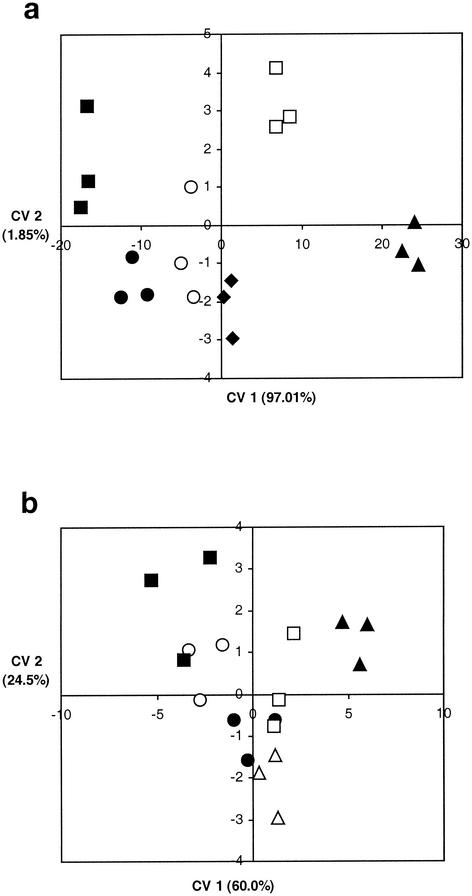
The multivariate analysis of the MicroResp data obtained by the colorimetric method for the 15 C sources showed that there were significant differences between the sludges (Table 2). The greatest difference was between the Zn-rich sludge and the other sludges (Fig. 6a) and was primarily on CV 1 with high ordination coordinates. CV 1 explained 97.05% of the variation. The two undigested sludges were also slightly discriminated on CV 2, which explained 1.85% of the variation, although the differences were not significant overall. The effects of the Zn-rich sludge on the community structure are in contrast to the results of other studies of metal-rich sludges, in which Biolog CLLPs were found to be most affected by Cu-rich sludges (4). The soil used in this experiment contained high levels of ammonium nitrate-extractable Zn, possibly due to a lower soil pH (pH 5.8) than the pH of arable agricultural fields (4).
FIG. 6.
Plot of ordination of first and second CVs for MicroResp CLPPs with 15 C sources (a) and Biolog CLPPs with 95 C sources (b) generated by CV analysis of sole-carbon-source tests for soil that received either no sludge (○), digested sludge (•), undigested sludge (▪), undigested sludge rich in Cu (□), digested sludge rich in Zn (▴), or digested sludge rich in Cd (⧫).
Biolog CLPPs.
The highest Biolog AWCD value for the same 15 C sources that were used for the MicroResp method was the AWCD value for arginine, and the lowest was the AWCD value for oxalic acid. Lower utilization was also found for l-lysine, malic acid, and protocatechuic acid (Fig. 5). However, there was not a strong correlation between the two methods for these C sources.
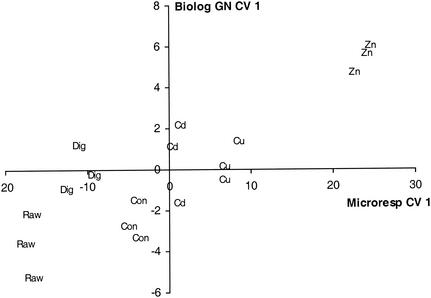
The multivariate analysis of Biolog CLPPs when either all 125 C sources, 95 GN C sources, or just the 15 C sources were used did not result in any overall significant separation between treatments (Table 2). The cumulative percentages of variation explained in the first 10 principal components were similar for all methods (Table 2). The greatest distance between treatments was found when the 95 Biolog GN C sources were used, and an ordination pattern similar to that obtained with the MicroResp system was found (Fig. 6), although it was not statistically significant for the Biolog data. The similarity of the pattern was strongly influenced by the effect of the Zn-rich sludge ordinates on CV 1, and there was a significant correlation (P < 0.05; r2= 0.72) between the first CVs of the two techniques (Fig. 7), confirming that they produced similar ordination patterns. There was no significant correlation, however, when the Zn treatments were omitted (r2= 0.34) or between the second CVs of the two techniques (data not shown).
FIG. 7.
Correlation between first CVs of the MicroResp and Biolog GN CLPPs for the following six sludge treatments: no sludge (Con), digested sludge (Dig), undigested sludge (Raw), undigested sludge rich in Cu (Cu), digested sludge rich in Zn (Zn), and digested sludge rich in Cd (Cd).
MicroResp radioactive detection system.
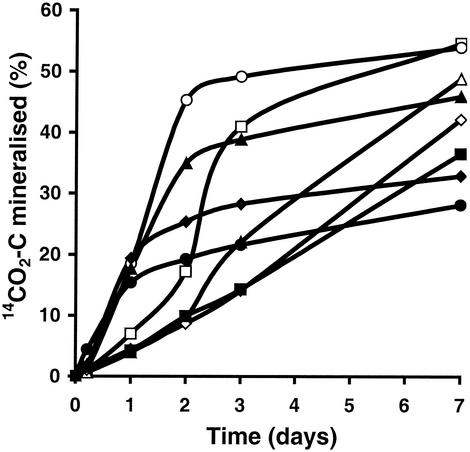
With the 14C MicroResp method the greatest utilization over 7 days was found for aspartic acid, followed by glycine, but different C sources had different patterns of utilization (Fig. 8). The time course of 14CO2 release from eight different substrates added to the forest soil (Fig. 8) showed that the response curves were generally sigmoidal for both amino acids and sugars except for d-glucosamine and l-lysine, whose response curves were more linear, over the time measured. The sigmoidal pattern is similar to previously published kinetic data for C source utilization in Biolog wells and reflects the normal kinetics of cell growth with lag, exponential, and stationary phases. However, the lag phase with the whole-soil method is much shorter than that observed with the Biolog method, in which detection of C source utilization occurs after 24 h or even 48 h (9). In contrast to Biolog plates (9), in the 14C MicroResp method C utilization could be detected after 5 h. The use of whole soil in the MicroResp system, therefore, does appear to reflect immediate activity within a 5-h period. Prolonged incubation can reveal further information about the kinetic response of organisms growing on the substrate. Thus, physiological information equivalent to that obtained from Biolog plates can be obtained without a prolonged lag phase when whole soil is used, and by using more C sources a CLPP could be constructed from the data.
FIG. 8.
Percentages of 14CO2 C mineralized over 7 days for eight substrates, glycine (□), l-aspartate (○), l-leucine (▵), l-lysine (⋄), galactose (▴), glucosamine (▪), glucose (⧫), and arabinose (•), added to Fetteresso forest soil.
It was not possible to test all of the C substrates used in Biolog GN plates as many were not readily available as 14C-labeled compounds. The advantage of the radioisotope method over the dye detection systems is that it is possible to categorically attribute the CO2 evolved to the degradation of the C substrate. Consequently, this approach would be very useful in biodegradation studies. In addition, the use of whole-soil and radioactive substrates means that C source concentrations lower than those currently used in SIR methods and Biolog approaches should be possible. Also, insoluble and/or more recalcitrant types of C sources can be tested over longer periods of time. This should overcome two of the main limitations of the existing Biolog approach, in which high substrate concentrations can cause toxicity problems (13), and it also opens the way to testing the less soluble and more recalcitrant substrates that are characteristic of soils, provided that 14C-labeled compounds are available. The cost and practicality of testing 14C-labeled compounds may, however, constrain extensive use of this approach.
The MicroResp system could be used in other applications. The system essentially allows gaseous transfer between two plates so it could be used for testing other gaseous products, including NH3, volatile metals (Hg, Se), or volatile organic compounds, provided that a suitable detection system can be found. The system could be used for testing either the inhibitory effects of volatile compounds or utilization of these compounds as C substrates without the problems associated with cross-contamination (24), provided that the volatile compounds do not interfere with the detection system. Similarly, it might be useful for other environmental samples, including waste, water, or other biologically active materials, such as cultured cells or biological tissues.
The main novelty of this approach is that we have miniaturized the process of measuring soil respiration and SIR in a microtiter plate system so that reading of the CO2 reactions can be carried out by using conventional automated plate readers. This was achieved by designing a new device in which two plates are joined together such that each well is sealed from the corresponding well but is interconnected to allow free gas exchange between the deep well containing soil and the detection well containing the detection system (either liquid alkali or gel with alkali and indicator dye) (Fig. 1 and 2). Gels with indicators for detection of gaseous products are already available (7), but they have not previously been included in a microtiter plate system to allow rapid, automated, quantitative measurement of absorbance (color). The scaling down of the method inevitably creates problems given the small amount of soil that is used. It is particularly important to prepare the soil carefully by sieving and mixing it to obtain a representative sample and by adjusting the moisture content to ensure homogeneous distribution in the deep wells. However, deep-well plates having larger volumes (5 ml) can be used.
The 96 individual wells of a microtiter plate can be filled with soil and various combinations of different substrates and controls set up with high levels of replication and numbers of samples, which can then be tested quickly with low consumable costs. The applications of the method include basal respiration tests, substrate-induced respiration tests (with different substrates), toxicity tests (e.g., tests for pollution-induced community tolerance to heavy metals or other chemical stresses), and pollutant degradation tests, as well as general assessment of the CLPP. For toxicity studies (e.g., pollution-induced community tolerance assays) (22), our method has the major advantage over Biolog of allowing toxicant availability to be determined in situ by soil properties rather than by the composition of the growth media. The ability to test large numbers of samples also means that the method could be used to rapidly identify constraints to bioremediation of contaminated soils by employing factorial combinations of substrates, acidity, N, and P to determine optimal levels that stimulate degradation.
In conclusion, our results do prove for the first time that the use of whole soil does have advantages (12) over the use of soil extract for determining CLPPs and that the microrespirometry method can provide meaningful and comparable data for substrate-induced responses of soil microorganisms to construct such CLPPs.
Acknowledgments
We are grateful for the technical assistance provided by Donna Heslin, Angela Norrie, Severine Montez, and David Riley.
This work was funded by the Scottish Executive, Environment and Rural Affairs Department.
REFERENCES
- 1.Anderson, J. P. E. 1982. Soil respiration, p. 833-871. In A. L. Page, R. H. Miller, and D. R. Keeney (ed.), Methods of soil analysis, part 2: chemical and microbiological properties. Madison, New York, N.Y.
- 2.Anderson, J. P. E., and K. H. Domsch. 1978. A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol. Biochem. 10:215-221. [Google Scholar]
- 3.Bååth, E., M. Diazravina, A. Frostegard, and C. D. Campbell. 1998. Effect of metal-rich sludge amendments on the soil microbial community. Appl. Environ. Microbiol. 64:238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacon, J. R., C. D. Campbell, M. C. Coull, B. J. Chambers, P. A. Gibbs, A. M. Chaudri, S. P. McGrath, C. H. Carlton-Smith, and M. N. Aitken. 2001. A long-term study of the effects of heavy metals in sewage sludge on soil fertility and soil microbial activity, p. 87-96. In R. K. Dhir, M. C. Limbachiya, and M. J. McCarthy (ed.), Recycling and reuse of sewage sludge. Thomas Telford, London, United Kingdom.
- 5.Bochner, B. R. 1989. Sleuthing out bacterial identities. Nature 339:157-158. [DOI] [PubMed] [Google Scholar]
- 6.Bossio, D. A., and K. M. Scow. 1995. Impact of carbon and flooding on the metabolic diversity of microbial communities in soils. Appl. Environ. Microbiol. 61:4043-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinton, W. F., and M. D. Droffner. April 1994. Test kits for determining the chemical stability of a compost sample. U.S. patent 5,320,807.
- 8.Bundy, J. G., G. I. Paton, and C. D. Campbell. 2001. Microbial communities in different soil types do not converge after diesel contamination. J. Appl. Microbiol. 91:1-13. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, C. D., S. J. Grayston, and D. J. Hirst. 1997. Use of rhizosphere carbon sources in sole carbon source tests to discriminate soil microbial communities. J. Microbiol. Methods 30:33-41. [Google Scholar]
- 10.Degens, B. P. 1998. Microbial functional diversity can be influenced by the addition simple organic substrates to soil. Soil Biol. Biochem. 30:1981-1988. [Google Scholar]
- 11.Degens, B. P. 1999. Catabolic response profiles differ between microorganisms grown in soils. Soil Biol. Biochem. 31:475-477. [Google Scholar]
- 12.Degens, B. P., and J. A. Harris. 1997. Development of a physiological approach to measuring the catabolic diversity of soil microbial communities. Soil Biol. Biochem. 29:1309-1320. [Google Scholar]
- 13.Fulthorpe, R. R., and D. G. Allen. 1994. Evaluation of Biolog MT plates for aromatic and chloroaromatic substrate utilization tests. Can. J. Microbiol. 40:1067-1071. [Google Scholar]
- 14.Garland, J. L. 1996. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 28:213-221. [Google Scholar]
- 15.Garland, J. L. 1997. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol. Ecol. 24:289-300. [Google Scholar]
- 16.Garland, J. L., and A. L. Mills. 1991. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 57:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, D., C. D. Campbell, J. A. Lee, T. V. Callaghan, and D. Gwynn-Jones. 2002. Arctic microorganisms respond more to elevated UV-B radiation than CO2. Nature 416:82-83. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, D., J. R. Leake, J. A. Lee, and C. D. Campbell. 1998. Changes in soil microbial biomass and microbial activities in response to 7 years simulated pollutant nitrogen deposition on a heathland and two grasslands. Environ. Pollut. 103:239-250. [Google Scholar]
- 19.Konopka, A., L. Oliver, and R. F. Turco. 1998. The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb. Ecol. 35:103-115. [DOI] [PubMed] [Google Scholar]
- 20.Lehman, R. M., F. S. Colwell, D. B. Ringelberg, and D. C. White. 1995. Combined microbial community-level analyses for quality assurance of terrestrial subsurface cores. J. Microbiol. Methods 22:263-281. [Google Scholar]
- 21.Rowell, M. J. 1995. Colorimetric method for CO2 measurement in soil. Soil Biol. Biochem. 27:373-375. [Google Scholar]
- 22.Rutgers, M., I. M. vantVerlaat, B. Wind, L. Posthuma, and A. M. Breure. 1998. Rapid method for assessing pollution-induced community tolerance in contaminated soil. Environ. Toxicol. Chem. 17:2210-2213. [Google Scholar]
- 23.Smalla, K., U. Wachtendorf, H. Heuer, W. T. Liu, and L. Forney. 1998. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl. Environ. Microbiol. 64:1220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stronggunderson, J. M., and A. V. Palumbo. 1994. Alternative method for rapidly screening microbial isolates for their potential to degrade volatile contaminants. J. Ind. Microbiol. 13:361-366. [DOI] [PubMed] [Google Scholar]
- 25.West, A. W., and G. P. Sparling. 1986. Modifications to the substrate-induced respiration method to permit measurement of microbial biomass in soils of differing water contents. J. Microbiol. Methods 5:177-189. [Google Scholar]
- 26.Yao, H., Z. He, M. J. Wilson, and C. D. Campbell. 2000. Microbial biomass and community structure in a sequence of soils with increasing fertility and changing land use. Microb. Ecol. 40:223-237. [DOI] [PubMed] [Google Scholar]
- 27.Zak, J. C., M. R. Willig, D. L. Moorhead, and H. G. Wildman. 1994. Functional diversity of microbial communities: a quantitative approach. Soil Biol. Biochem. 26:1101-1108. [Google Scholar]