Abstract
We investigated the microbial diversity of biofilms found in dental unit water systems (DUWS) by three methods. The first was microscopic examination by scanning electron microscopy (SEM), acridine orange staining, and fluorescent in situ hybridization (FISH). Most bacteria present in the biofilm were viable. FISH detected the β and γ, but not the α, subclasses of Proteobacteria. In the second method, 55 cultivated biofilm isolates were identified with the Biolog system, fatty acid analysis, and 16S ribosomal DNA (rDNA) sequencing. Only 16S identified all 55 isolates, which represented 13 genera. The most common organisms, as shown by analyses of 16S rDNA, belonged to the genera Afipia (28%) and Sphingomonas (16%). The third method was a culture-independent direct amplification and sequencing of 165 subclones from community biofilm 16S rDNA. This method revealed 40 genera: the most common ones included Leptospira (20%), Sphingomonas (14%), Bacillus (7%), Escherichia (6%), Geobacter (5%), and Pseudomonas (5%). Some of these organisms may be opportunistic pathogens. Our results have demonstrated that a biofilm in a health care setting may harbor a vast diversity of organisms. The results also reflect the limitations of culture-based techniques to detect and identify bacteria. Although this is the greatest diversity reported in DUWS biofilms, other genera may have been missed. Using a technique based on jackknife subsampling, we projected that a 25-fold increase in the number of subclones sequenced would approximately double the number of genera observed, reflecting the richness and high diversity of microbial communities in these biofilms.
Biofilms have been implicated as major causes of infection and in the pathogenesis of several diseases (6). During the past 2 decades, it has been established that water used in dental treatment has high microbial counts, typically ranging from 104 to >106 CFU/ml (2). Previous studies addressing dental unit (DU) water supply (DUWS) contamination have confirmed that the high bacterial count is due to the shedding of biofilm bacteria from the lumen surface of dental waterline tubing into treatment water (2, 27, 31, 32). The relatively high surface area/volume ratio associated with the DU waterline (DUWL) tubing, periods of stagnation (when the DU is not in use) of water in the lines, and laminar flow conditions with low shear forces near the lumen wall of the waterlines provide the opportunity for the development of bacterial biofilms. The main mass of the biofilm, the exopolysaccharide, provides added protection to the biofilm organisms by limiting the diffusion of antimicrobial agents as well as by acting as a nutrient source for the bacterial community (7). The presence of bacterial biofilms also increases resistance to flow in the DUWL and can act as a reservoir for potential pathogens. For example, in biofilms, Legionella pneumophila can survive within amoeba, which can protect the bacteria from chlorination (18). L. pneumophila has been shown to occur in DUWL biofilms (4), and biocides have been shown to be less effective against the organism in biofilms than against those in the planktonic phase. The introduction into patients of such high levels of microorganisms is of concern particularly for immunocompromised individuals (22).
Although studies have assessed numbers of bacteria in the bulk water delivered from dental units (2, 3, 17, 30), few have identified the organisms. Even less attention has been given to the types of organisms present in the biofilm (27), the primary source of bacteria in the DUWS. Up to now, the microbial community in the bulk water and in the DUWL biofilm has only been assessed by cultivation methods. However, conventional culture methods do not provide a representative profile of the true composition of microbial communities in nature (20). When microorganisms in oligotrophic aquatic systems such as DUWS are examined in situ by using fluorescent oligonucleotide probes in combination with epifluorescence microscopy, most of the bacteria appear to be metabolically active (16). A second culture-independent technique involves amplification and analyses of 16S rRNA gene sequences. This method is becoming increasingly popular to characterize novel bacterial communities in the environment. Thus, culture-independent techniques detect a more complete representation of the microbial community (29).
The aims of this study were to examine and characterize the DUWL biofilm community by using a combination of both culture-dependent and -independent methodologies. The three-dimensional structure of the biofilm was examined by scanning electron microscopy (SEM), nucleic acid stains, and fluorescence in situ hybridization (FISH). Bacterial groups were distinguished by morphotypes by confocal scanning laser microscopy (CSLM). Cultivated bacteria were isolated on a standard oligotrophic medium, R2A, developed specifically for quantification of bacteria from potable water. (26). Bacterial isolates were identified by three commercially available methods: Biolog (Biolog, Inc., Haywood, Calif.), gas-liquid chromatography (GLC) of fatty acid methyl esters (FAMEs), and 16S ribosomal DNA (rDNA) sequencing. Culture-independent methods were used to characterize the bacterial community by sequencing subcloned PCR-amplified 16S rDNAs. This study provides the first in situ examination of DUWL biofilms and the use of rDNA sequencing methods to characterize these biofilms.
MATERIALS AND METHODS
DUs.
The tubing samples were obtained from a total of 12 DUs in Baltimore, Md. The water from these units was used routinely during patient treatment. The tubing was obtained from DUs that were in use 5 to 7 years. There was no particular problem with the units or tubing. The portion of the DUWL closest to the patient, the air/water syringe tube, which is approximately 3 ft long, was selected. After removal from the DU, the tubing was transferred to the laboratory in less than 5 min for analysis.
SEM analysis.
For SEM analysis, DUWL tubing was taken from two DUs. From each 3-ft section of tubing, 1-in. sections were cut from each end and the middle of the tube. Sections were fixed and stored in 0.1% glutaraldehyde in 0.2 M cacodylate buffer (Electron Microscopy Sciences, Fort Washington, Pa.) at 4°C. The fixative was flushed by being washed twice in 0.2 M cacodylate buffer, followed by increasing concentrations of ethanol (30 to 100%). The sections were then dried overnight in hexamethyldisilazane (HMDS) (Electron Microscopy Sciences). The next day, specimens were mounted onto aluminum studs, sputter coated with gold-palladium, and examined via SEM (JEOL model 4000).
Embedding of DUWL biofilm.
For biofilm embedding, tubing sections were processed from two DUs. A modification of the method (5) previously used for embedding flow-cell biofilm was developed. Biofilm in the tubing was fixed by introducing 5 ml of 4% paraformaldehyde into the lumen with a 14-gauge syringe. The tubes were clamped at both ends for 1 h at room temperature. The fixative was flushed gently (so as not to disturb the biofilm structure) from the tubing five times with phosphate-buffered saline (PBS). The biofilm was then embedded by gently introducing 20% acrylamide into the tubing with a syringe. The acrylamide was allowed to polymerize at room temperature for 1 h. The polyvinyl chloride (PVC) tubes were cut into 1-in. sections and then cut longitudinally with a sterile razor blade to expose the polymerized acrylamide. The solidified cylinder of acrylamide with embedded biofilm was carefully lifted from the lumen.
Fluorescence microscopy with general nucleic acid stain.
A 10-mm-long cylindrical section of the embedded biofilm was stained with 0.1% acridine orange (Sigma, St. Louis, Mo.), washed in double-distilled H2O, and then mounted onto a hanging drop microscope slide. The biofilm community structure was examined with a fluorescence microscope (Nikon E-600). CSLM (Zeiss-LSM 510) was also used to image a similarly stained piece of polyacrylamide-embedded biofilm.
FISH.
Universal oligonucleotide probes specific for the 16S rRNA regions of Proteobacteria were used for in situ hybridization of the embedded biofilm (21). The α subclass of Proteobacteria was detected with the ALF1b probe labeled with Cy5, the β subclass was detected by the BET42a probe labeled with fluorescein, and the γ subclass was detected by the GAM42a probe labeled with rhodamine. The permeable acrylamide block was prepared for hybridization as described by Christensen et al. (5). Briefly, an acrylamide-embedded biofilm section was incubated in prehybridization solution (30% formamide, 0.9 M NaCl, 0.1 M Tris [pH 7.5], 0.1% sodium dodecyl sulfate [SDS]) for 30 min. The block was then incubated overnight in hybridization solution (30% formamide, 0.9 M NaCl, 0.1 M Tris [pH 7.5]) containing all three probes. Each probe was added to a final concentration of approximately 2.5 μg/ml in the hybridization solution. On day 2, the acrylamide block was incubated with prewarmed prehybridization solution for 30 min and rinsed with washing solution (0.1 M Tris [pH 7.5], 0.9 M NaCl) for 30 min. The block was then rinsed with double-deionized H2O, mounted on a hanging drop microscope slide, and examined with a standard fluorescence microscope (Nikon E-600) and CSLM (Zeiss LSM-510).
DUWL biofilm extraction.
Biofilm was extracted from 3-ft-long sections of DUWL tubing obtained from five DUs. After the water was drained, a 4-ft-long piece of stainless steel wire with a sterile brush at one end was inserted into each tube to scrape the biofilm from the luminal surface. After each scraping, the brush was rinsed with sterile distilled water. The brushing was repeated until no visible biofilm was removed. The collected biofilm samples from each tube were centrifuged (Microfuge 18; Beckman, Palo Alto, Calif.) briefly at 3,000 rpm to pellet the cells.
DNA extraction from DUWL biofilm.
Community nucleic acids were extracted from the biofilm by physical disruption of cells by bead beating as described by Hugenholtz et al. (12) with some modifications. Approximately 200 μl of biofilm sample was suspended in 1 ml of lysis buffer (200 mM Tris [pH 8.0], 50 mM EDTA, 200 mM NaCl, 2 mM sodium citrate, 10 mM CaCl2) containing 20 mg of lysozyme per ml (Sigma, St. Louis, Mo.). The sample was incubated at 37°C for 1 h with occasional vortexing. Proteinase K (to 1 mg/ml [Sigma, St. Louis, Mo.]) and SDS (0.3% [Sigma]) were then added, and the mixture was incubated further at 70°C for 30 min. Samples were reciprocated on a Mini-Beadbeater (Biospec Products, Inc., Bartlesville, Okla.) at low speed for 2 min in the presence of 3% (wt/vol) SDS and approximately 0.5 g of 0.1-mm-diameter zirconium beads (Biospec Products, Inc.). The lysate was then treated with RNase (Sigma) for 20 min at 37°C. Proteins were precipitated on ice with 200 μl of protein precipitation solution (ABI Systems, Norwalk, Conn.). The sample was centrifuged for 5 min, and nucleic acids were precipitated from the aqueous phase with 1 volume of isopropanol. The DNA pellet was then washed with 70% ethanol and suspended in 50 μl of 10 mM Tris-1 mM EDTA (pH 8.0).
16S rDNA clone library construction.
Community rDNAs were PCR amplified from 30 ng of bulk DNA from each of the five biofilm samples. Primers 533F (5′-AGAGTTTGATC/TA/CTGGCTCAG-3′) and 1492R (5′-CGGC/TTACCTTGTTACGAC-3′) were used to amplify a 950-bp region of the 16S rDNA gene (30). The reaction mixtures contained 1× PCR buffer; 2.5 mM MgCl2; 200 μM each dATP, dCTP, dTTP, and dGTP; 300 nM each forward and reverse primer; and 0.025 U of Taq polymerase (Invitrogen, Carlsbad, Calif.). Reaction mixtures were incubated for an initial denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 10 s. PCR products were resolved on a 1.7% low-melting-point agarose gel (Invitrogen). The amplified PCR product was excised from the gel and purified according to the manufacturer's instructions with the Wizard purification kit (Promega, Madison, Wis.). Clone libraries of PCR product were generated with a TOPO TA cloning kit in accordance with the manufacturer's instructions (Invitrogen). Recombinants were selected with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-IPTG (isopropyl-β-d-thiogalactopyranoside) indicator Luria-Bertani agar plates supplemented with 100 μg of ampicillin per ml. A clone library was created for each of the biofilm samples processed, and the results were pooled.
Determination of nucleotide sequences.
M13F and M13R primers corresponding to vector DNA flanking the cloned insert (Invitrogen) were used in an amplification reaction to screen 480 clones. PCR was initiated as described above, except the annealing temperature was 65°C and 30 cycles were performed. The PCR products were resolved on a 1.7% agarose gel, and 170 clones were picked that contained the 1-kb insert. The PCR products were purified by filtration (PCR purification kit; Millipore, Bedford, Mass.) and sequenced bidirectionally with the universal oligonucleotide primers 533F and 1492R. Products were separated in the 3700 DNA capillary sequencer (ABI Systems, Foster City, Calif.). Both strands of template DNA were sequenced, and 165 contigs were generated using Phred and Phrap (8, 9). Low-quality sequence was removed from the ends.
Isolation of DUWL biofilm microorganisms.
For the isolation of biofilm microorganisms, tubing was obtained from three DUs. Each tube was aseptically cut into 2-cm lengths. Four sections were randomly selected and cut longitudinally to expose the inner surface. All sections had an apparently uniform layer of biofilm. The biofilm was removed by scraping the wall with a sterile forceps or scalpel, and the scrapings were suspended in PBS. Each suspension was 10-fold serially diluted in PBS, spread onto R2A (26) agar plates in duplicate, and incubated at 28°C for 1 week. To obtain pure cultures, morphologically different CFU were selected for subculture on R2A.
Identification by Biolog.
The Microlog 2 Biolog (Biolog, Inc.) system assays 95 different carbon sources. Cultures to be tested were grown for 48 h on tryptic soy agar (TSA; BBL Microbiology Systems, Cockeysville, Md.). The cultures were harvested and suspended at the manufacturer's recommended density for gram-negative or gram-positive organisms in 18 to 20 ml of 0.85% saline. One hundred fifty microliters of each of the respective suspensions was added to each well of the plate, followed by incubation at 30°C for 48 h. Each plate was read automatically, and a similarity index was calculated for the 10 strains in the database most closely related to the test strain. A similarity index of >0.50 reports a genus and species name.
Identification by GLC.
For GLC analysis, both known and unknown organisms were cultivated, harvested, derivatized, and extracted according to the procedure described by the manufacturer (Microbial ID, Inc., Newark, Del.). Samples were analyzed with the Microbial Identification System on a Hewlett-Packard 5898A gas chromatograph (Palo Alto, Calif.). Cellular fatty acids were identified as their FAMEs on the basis of equivalent chain length data. The value is a representation of a fatty acid's retention time as it relates to a series of straight-chain saturated fatty acid methylated esters in the calibration mixture. Identification of an unknown organism is based on computer comparisons of its FAME profile with a library of previously determined profiles. The correlation is expressed as a similarity index (SI) on a numeric scale of 0 to 1.0. SI values of ≥0.6 are considered excellent matches (23).
Genomic DNA extraction and sequencing of 16S rDNA genes of cultured isolates.
Genomic DNA was extracted from each bacterial morphotype on R2A by using the QIAamp tissue kit (Qiagen, Hilden, Germany). The 16S rDNA gene was amplified by a PCR initiated with primers 533F and 1492R. The same primers were used for sequencing by the methodology described above for the subclones from the biofilm community DNA.
Phylogenetic analysis of sequence data.
All sequences were analyzed with BLAST (1) searches to determine the closest match from available database sequences. Alignment of the final data set was accomplished with ClustalX (15). Neighbor-joining distance matrix analysis and maximum-parsimony analysis were performed with PAUP computer programs to generate dendrograms (D. Swofford, PAUP∗ Beta, Sinauer Associates, Sunderland, Mass., 2000).
Jackknife statistical analysis.
We developed a jackknife-like subsampling method to quantify the relationship between the number of isolates examined, N, and the number of genera counted, or generic richness, R. We have used the distribution of abundances in the sample to relate the number of isolates examined in a subsample to the probability of finding a new genus, p(x), where x = 1, 2, 3…N. We then make a projection, S, for the number of new genera that we expect to find if the number of isolates is increased to N + M; the probability of finding a new species is a decreasing function of the number of new isolates counted, q(M), and it is fitted to p(x). We point out that the function p(x) is based only on the sampling distribution; that it is defined only at the counting numbers x = 1, 2, 3…N; and that R ≈ Σp(N). In contrast, q(M) is a general, flexible function written in closed form chosen to have the same shape as p(N), near N.
To compute p(N), we note that the probability of discovering a new genus by adding one isolate is approximately equal to the probability of removing an observed genus when an isolate is deleted at random, or in other words, the number of genera that were observed exactly once (called the singletons) divided by the total number of isolates examined. The value of function at p(x) is estimated by randomly removing N − x isolates and then counting the number of singletons in the subsample and dividing by x. The function p(x) was generated by repeating this 1,000 times for each x = 1, 2, 3…165. To generate the function q(M), we fitted a general three-parameter (μ, λ, θ) function, q(M) = μ exp (−λ Mθ), to the curve p(N) by a nonlinear fitting procedure. The expected number of genera that would be found after collecting M additional isolates is S (M) = R + ΣM q(M). We simulated sampling using the function q(M) for values of M ranging up to 4,000.
RESULTS
SEM analysis.
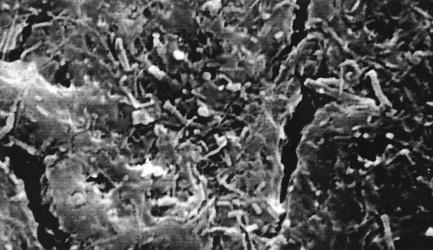
SEM analysis of tubing obtained from DU revealed a dense biofilm matrix in every field of the luminal surface of the tubing (Fig. 1). Bacteria of distinct morphotypes (cocci, rods, and filamentous bacteria) could be observed in the matrix, suggesting that DU biofilms are complex. The cracks or crevices seen in the figure are partially due to the stress on the sample during preparation for SEM analysis. However, these crevices reveal the thickness of the biofilm and confirm that this is a mature microbial community.
FIG. 1.
SEM of DT lumen surface shown at ×2,500. Cocci and rods are shown in a dense biofilm matrix.
Examination of embedded biofilm.
A dense matrix of cocci, rods, and filamentous bacteria stained with acridine orange was observed in every field of the embedded biofilm. In addition, larger, apparently unicellular organisms presumed to be eukaryotes were seen in some, but not all, fields. In contrast, no bacteria were visible on the inner surface of the tubing, implying that the entire biofilm had been removed and embedded. Only the β and γ rDNA probes hybridized with cells in the scanned fields and planes. No cells were hybridized with the α probe.
16S rDNA analysis of total biofilm community DNA.
Forty bacterial genera were represented among the sequences of 165 subclones of 16S rDNA genes amplified from genomic DNA from DUWL biofilm (Fig. 2). The six most common genera and their frequencies were as follows: Leptospira, 20%; Sphingomonas, 14%; Bacillus, 7%; Escherichia, 6%; Geobacter, 5%; and Pseudomonas, 5%. The major (∼55%) group of organisms belonged to the Proteobacteria. The α subdivision comprised ∼23%, including Sphingomonas (14%), Afipia (3%), Bradyrhizobium (3%), Blastobacter (1%), and, at <1%, Hyphomicrobium and Ketogulonigenium. The γ subdivision comprised ∼15%, including Escherichia (6%), Pseudomonas (5%), Legionella (3%), and Aeromonas (<1%). The β subdivision comprised ∼11%, including Leptothrix (4%), Roseateles (3%), Aquabacterium (1%), Herbaspirillum (1%), and, at <1%, Chlamydia, Azoarcus, and Aminomonas. The δ subdivision comprised ∼7% and included Geobacter (5%) and Bdellovibrio (2%).
FIG. 2.
Phylogram of 16S rDNA gene sequenced from 165 subcloned 16S fragments from community DNA without cultivation.
Bacteria comprising the other 45% of biofilm bacteria included Leptospira (20%), Bacillus (7%), Planctomyces (2%), Prosthecobacter (1%), and, at <1%, Pirellula, Flexibacter, Cytophaga, Rhodothermus, Geodermatophilus, Agrobacterium, Chlamydophila, Geodermatophilus, Matsuebacter, Mesorhizobium, Oceanospirillum, Rhizobium, Xanthomonas, and Micrococcus. Microorganisms other than bacteria were identified, including Odontella sinensis (5 of 165), Polydora ciliata (1 of 165), and endosymbionts of Acanthamoeba (5 of 165).
Identification of 55 cultured isolates.
The brushing technique was efficient at removing biofilm from the DUWL, because no bacteria were seen on the inner surface after the biofilm was removed for both culture-dependent and -independent analyses. Fifty-five morphologically distinct colonies were recognized on R2A medium from the DUWL biofilm samples (Fig. 3). The colonies were subcultured onto TSA as the required medium for Biolog and GLC identification. Of the 55 isolates, 6 were gram positive and 49 were gram negative, including 1 that did not grow on TSA (isolate 37) and therefore could not be tested by either Biolog or GLC.
FIG. 3.
Phylogram of 16S rDNA gene sequenced from 55 cultured isolates.
For Biolog analysis, only 47 of 55 (85%) of the isolates were tested. The other eight (15%) isolates did not grow sufficiently on TSA. Fourteen of 47 isolates (30%) were identified to the genus and species level (Table 1). Six genera were identified, including Pseudomonas (n = 4), Citrobacter (n = 3), Staphylococcus (n = 4), Bacillus (n = 1), Corynebacterium (n = 1), and Micrococcus (n = 1). Thirty-three (70%) of the 47 isolates that grew sufficiently were not identified by the Biolog system. For GLC analysis, 48 of 55 (87%) isolates were tested. The other six gram-positive isolates could not be tested by GLC. Seventeen of 48 (35%) isolates were identified to the species level. Species belonging to the following five genera were identified: Pseudomonas (n = 4), Escherichia (n = 3), Paracoccus (n = 2), Sphingomonas (n = 7), and Cornyebacterium (n = 1). Thirty-one (65%) of the 48 isolates that grew were not identified by the GLC method.
TABLE 1.
Identification of 55 cultured isolates by Biolog, GLC, and 16S rDNA sequencing
| Isolate | Identification bya:
|
||
|---|---|---|---|
| 16S rDNA | Biolog | GLC | |
| 1 | Pseudomonas stutzeri | P. stutzeri | P. stutzeri |
| 2 | Escherichia coli | Citrobacter braaki | E. coli |
| 3 | Afipia genospecies | Inconclusive | Inconclusive |
| 4 | E. coli | C. braaki | E. coli |
| 5 | E. coli | C. braaki | E. coli |
| 6 | Methanotroph, Afipia | Inconclusive | Inconclusive |
| 7 | Paracoccus solventivorans | Inconclusive | P. solventivorans |
| 8 | Staphylococcus epidermidis | S. epidermidis | Did not test |
| 9 | P. stutzeri | P. stutzeri | P. stutzeri |
| 10 | P. stutzeri | P. stutzeri | P. stutzeri |
| 11 | P. solventivorans | Inconclusive | P. solventivorans |
| 12 | Benzene-mineralizing consortium bacteria, Bacillus sp. | Inconclusive | Inconclusive |
| 13 | Paracraurococcus ruber | Inconclusive | Inconclusive |
| 14 | Acinetobacter radioresistens | Inconclusive | Inconclusive |
| 15 | Staphylococcus warneri | S. warneri | Could not identify; gram positive |
| 16 | Afipia genospecies | Inconclusive | Inconclusive |
| 17 | Afipia genospecies | Inconclusive | Inconclusive |
| 18 | Afipia genospecies | Inconclusive | Inconclusive |
| 19 | Afipia genospecies | Inconclusive | Inconclusive |
| 20 | Afipia genospecies | Inconclusive | Inconclusive |
| 21 | Afipia genospecies | Inconclusive | Inconclusive |
| 22 | Paracoccus thiocyanatus | Inconclusive | Inconclusive |
| 23 | P. thiocyanatus | Inconclusive | Inconclusive |
| 24 | Benzene-mineralizing consortium bacteria, Bacillus sp. | Inconclusive | Inconclusive |
| 25 | E. coli | Control (ML35) | Control (ML35) |
| 26 | Bosea thiooxidans | Inconclusive | Inconclusive |
| 27 | Sphingomonas alaskaensis | Could not test; insufficient growth | Sphingomonas adhaesiva |
| 28 | Bacillus sp. | Bacillus halodurans | Could not identify; gram positive |
| 29 | Human oral bacterium, Staphylococcus | S. warneri | Could not identify; gram positive |
| 30 | Sphingomonas sp. | Could not test; insufficient growth | S. paucimobilis |
| 31 | Afipia genospecies | Inconclusive | Inconclusive |
| 32 | Methanotroph, Afipia | Inconclusive | Inconclusive |
| 33 | α-Proteobacteria, Bosea | Inconclusive | Inconclusive |
| 34 | Afipia genospecies | Inconclusive | Inconclusive |
| 35 | Afipia genospecies | Inconclusive | Inconclusive |
| 36 | Afipia genospecies | Inconclusive | Inconclusive |
| 37 | Uncultured rape rhizospecies, Paracraurococcus | Could not test, grew on R2A medium only | Could not test; grew on R2A medium only |
| 38 | Afipia genospecies | Inconclusive | Inconclusive |
| 39 | α-Proteobacteria, Bosea | Inconclusive | Inconclusive |
| 40 | Sphingomonas sp. | Could not test; insufficient growth | S. paucimobilis |
| 41 | Sphingomonas asaccharolytica | Could not test; insufficient growth | S. paucimobilis |
| 42 | Afipia genospecies | Inconclusive | Inconclusive |
| 43 | Sphingomonas sp. | Could not test; insufficient growth | S. paucimobilis |
| 44 | Staphylococcus pasteuri | S. pasteuri | Could not identify; gram positive |
| 45 | Sphingomonas yanoikuyae | Could not test; insufficient growth | S. paucimobilis |
| 46 | Corynebacterium tuberculostearicum | Cornyebacterium singulare | Corynebacterium glucuronolyticum |
| 47 | Methanotroph B2, Afipia | Inconclusive | Inconclusive |
| 48 | P. stutzeri | P. stutzeria | P. stutzeri |
| 49 | Micrococcus luteus | M. luteus | Could not identify; gram positive |
| 50 | Afipia genospecies | Inconclusive | Inconclusive |
| 51 | Afipia genospecies | Inconclusive | Inconclusive |
| 52 | Benzene-mineralizing consortium bacteria, Bacillus sp. | Inconclusive | Inconclusive |
| 53 | Afipia genospecies | Inconclusive | Inconclusive |
| 54 | Caulobacter leidyi | Inconclusive | Inconclusive |
| 55 | Spingomonas sp. | Could not test; insufficient growth | S. paucimobilis |
Isolates in boldface were not identified by Biolog or GLC. Isolate 25 (E. coli ML35) was used as the positive control.
For 16S rDNA, all 55 (100%) cultured isolates were identified. They represented 13 genera and 17 bacterial species (Table 1). Forty-five of 55 (82%) isolates gave high-similarity matches (>95%) to the genus level. The most common organisms, Afipia (28%) and Sphingomonas (16%), belong to the α subdivision of Proteobacteria, whereas Pseudomonas (8%) and Escherichia (8%) belong to the γ subdivision. The following organisms were identified: Afipia (n = 15), Sphingomonas (n = 7), Paracoccus (n = 5), Pseudomonas (n = 4), Escherichia (n = 4), Staphylococcus (n = 3), Paracraurococcus (n = 1), Caulobacter (n = 1), Corynebacterium (n = 1), Bosea (n = 1), Acinetobacter (n = 1), Bacillus (n = 1), and Micrococcus (n = 1).
The other 10 sequences were identified to the genus level based on clustering in a phylogenetic tree (Fig. 3). With BLAST (1), these 10 sequences were most closely related to a sequence from a bacterium of unknown genus. However, in a neighbor-joining tree, the sequence most similar to “uncultured rape rhizospecies” by BLAST (1) was most closely related to the genus Paracraurococcus; the two sequences identified as α-Proteobacteria were most closely related to Bosea (α subdivision of Proteobacteria); the three isolates identified as methanotrophs were most closely related to Afipia (α subdivision of Proteobacteria); the three benzene-mineralizing consortia bacteria were most closely related to the Bacillus group; and the one human oral bacterium was most closely related to Staphylococcus.
Five isolates (no. 1, 9, 10, 46, and 48) were reported identically by all three methods: four belong to the genus Pseudomonas, and one belongs to the genus Corynebacterium. Nine isolates were identified consistently by 16S and GLC but not by Biolog. Six isolates were identified consistently by both 16S and Biolog but not by GLC. The only inconsistent identifications occurred for isolates 2, 4, and 5, which were identified as Escherichia coli with GLC and 16S rDNA analysis but as Citrobacter braaki with Biolog, despite the use of E. coli ML35 as a control.
Jackknife statistical analysis.
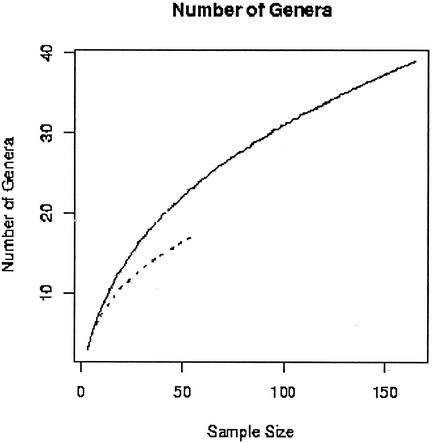
We used the jackknife procedure to estimate the rate at which new genera were observed from both sets of isolates. Based on this analysis, the 16S rDNA method found new genera more efficiently than culture methods (Fig. 4). Using the sample to estimate the rate at which new genera were being identified when the study was stopped, we projected that 80 genera would have been found after cloning a total of 4,000 subclones, and the curve does not have an asymptote. Therefore, the actual number of genera present in the DUWS biofilm is unknown.
FIG. 4.
Distribution of the number of genera detected as a function of the number of sequences analyzed. The dotted line represents the cultivated isolates, showing 55 sequences representing 13 genera, and the solid line represents the uncultured community, in which 165 sequences revealed 40 genera. Culture-independent methods found new genera more efficiently than culture-dependent methods.
DISCUSSION
The results of this study have revealed a complex, diverse microbial community in DUWL biofilms as determined by both culture and cultivation-independent methods. Analysis of 16S rDNA genes from the total biofilm community revealed a much more diverse community of organisms than culture methods. It was also a more efficient technique for analyzing the DUWL biofilm communities than either FISH or culture methods. The 16S rDNA technique detected all three classes of the Proteobacteria division, as well as other organisms. On the contrary, FISH failed to detect organisms belonging to the subdivision α-Proteobacteria, the dominant group revealed by both culture and subcloned 16S rDNA methodologies. This could be due to the hybridization stringency of the α probe not being compatible with the conditions required by the β or γ oligonucleotide probes or possible photo-bleaching of the Cy5 dye. Moreover, even though R2A is a nonselective medium used for heterotrophic plate counts of water samples (26), it failed to detect organisms of the β-Proteobacteria division that were detected by other methods. Our results complement a report by Wagner et al. (28) that R2A preferentially supports growth of the α- and γ-Proteobacteria.
For the identification of cultured isolates, the 16S rDNA sequence analysis was the superior method, yielding 100% identification to the genus level. The Biolog and GLC methods identified 25 and 31%, respectively. The most common cultured organism, Afipia, was not identified by either the Biolog or GLC method, because it was not in either database.
The genera identified by the culture-dependent and -independent methods were not identical either quantitatively or qualitatively. Quantitatively, Afipia was the most common isolate in the culture-dependent analysis, and Leptospira was the common isolate in the culture-independent analysis. However, Leptospira was not cultivated on R2A, indicative of a potential bias of the medium (28). In contrast, Sphingomonas was the second-most-common isolate by both methods. The amplification methods may also produce a biased estimate of the frequency, since the rates of amplification of different 16S rDNAs may be different (25).
The total diversity in a community is a difficult quantity to estimate, but several methods have been proposed (13). Obviously, the number of unique genera identified in a study increases with effort (number of isolates). The relationship between the number of unique entities identified and the number of isolates examined depends critically on the distribution of abundances—common entities are more likely to be found than rare ones, and they are likely to be counted several times. Since the total diversity and the distribution of abundances are not known, a priori, they must be estimated from the sample distribution. Collecting and identifying new isolates requires additional time and effort, but it is a simple matter to randomly eliminate samples as if they had never been counted. Based on the rate with which we found new genera in the biofilm, we projected that 80 genera instead of 40 would have been found had we collected 4,000 clones instead of 165. Clearly, genera that were present at low densities in the biofilm would be less likely to be counted. The projection is confirmed, to some extent, by comparing the genera discovered by the different methods; only 54% (7 of 13) of the genera counted by culture methods were discovered by the culture-independent methods.
Some organisms found in the DUWS may pose a health risk to some patients. Leptospira, the most common organism detected, can invade any susceptible mucosal membrane and cause leptospirosis. Sphingomonas and Legionella, two organisms found in the DUWL, are easily spread via aerosols. Sphingomonas sp. strains secrete viscous polysaccharides (24), which aid in their ability to adhere to and corrode surfaces of pipelines. Legionella species are known to cause respiratory infections (2). Both species have been found in hospital environments, including such devices as mechanical ventilators, catheters, and bronchofiberoscopes (11, 19). Their presence in DUWL is a concern, because studies have shown that aerosols generated from dental handpieces during treatment are sufficient to expose patients and dental personnel to the microorganisms in the DUWS (10).
The 40 genera identified by culture-independent methods represent a wide range of functional groups from symbionts to predators. The close proximity of organisms in biofilms permits interactions between distinct species based on their metabolic diversity. For example, Geobacter, a strict anaerobe involved in the reduction of Fe(III), may benefit from close association with Leptothrix, which is common in water distribution systems and is involved in the oxidation and chelation of iron. Another key metabolic reaction involves the oxidation of anaerobic ammonia (14). Planctomyces and Paracoccus remove toxic ammonia nitrogen and are used commercially in wastewater plants. Some organisms may utilize components in the DUWL tubing as a nutrient source. Roseateles depolymerans is involved in the degradation of polyhexamethylene carbonate and could be using the surface of the PVC tubing as a nutrient source. A predatory organism, Bdellovibrio, was also found in the biofilm. Bdellovibrio organisms are known to prey upon a wide range of gram-negative organisms.
In addition to bacteria, three eukaryotic organisms were found in the DUWL biofilm. Acanthamoeba typically inhabits domestic and industrial water supplies. Its presence is significant, since it is known to enhance the reproduction, environmental survival capacity, and pathogenicity of Legionella (4). The other organisms included Odentella, a species of photosynthetic phytoplanktons, and Polydora ciliata. The latter is a bristled worm, less than 1 mm in diameter and about 10 mm long, typically found attached to surfaces rich in calcium carbonate. Its role in this biofilm is unknown. The finding of eukaryotic organisms confirmed microscopic observations of nonbacterial morphotypes in the embedded biofilm.
The results of this study have provided a more complete representation of the types of organisms in the DUWL microbial community and the potential functional relationships between its members. The biodiversity revealed by 16S was much greater than that found by traditional culture-dependent approaches and is much greater than previously reported. Our sample probably found less than half of the actual number of genera present in these DUWL biofilms. These results should lead to a better appreciation of the metabolic and genetic diversity in biofilms. Continued investigations regarding DUWLs and the potential risk to patients are therefore warranted to safeguard patient health.
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) grant N44-DE-92628 and by the Dental Research Initiative Fund, University of Maryland, Baltimore.
We thank Yuansha Chen (University of Maryland Genomics Core Laboratory), Gerard J. Osterhaut (MIDI Corp.), and James Dick (Johns Hopkins University Department of Pathology) for technical guidance with sequencing and fatty acid analysis.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbeau, J., C. Gauthier, and P. Payment. 1998. Biofilms, infectious agents, and dental unit waterlines: a review. Can. J. Microbiol. 44:1019-1028. [DOI] [PubMed] [Google Scholar]
- 3.Barbeau, J., R. Tanguay, E. Faucher, C. Avezard, L. Trudel, L. Côté, and A. P. Prevost. 1996. Multiparametric analysis of waterline contamination in dental units. Appl. Environ. Microbiol. 62:3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cargill, K. L., B. H. Pyle, R. L. Sauer, and G. A. McFeters. 1992. Effects of culture conditions and biofilm formation on the iodine susceptibility of Legionella pneumophila. Can. J. Microbiol. 38:423-429. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, B. B., C. Sternberg, J. B. Anderson, R. J. Palmer, A. T. Nielsen, M. Givskow, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., K.-J. Cheng, G. G. Geesey, T. L. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 8.Ewing, B., and P. Greene. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 9.Ewing, B., L. Hillier, M. C. Wendl, and P. Greene. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 10.Fotos, P. G., H. N. Westfall, I. S. Snyder, R. W. Miller, and B. M. Mutchler. 1985. Prevalence of Legionella-specific IgG and IgM antibody in a dental clinic population. J. Dent. Res. 64:1382-1385. [DOI] [PubMed] [Google Scholar]
- 11.Hsueh, P.-R., L.-J. Teng, P.-C. Yang, Y.-C. Chen, H.-J. Pan, S.-W. Ho, and K.-T. Luh. 1998. Nosocomial infection caused by Sphingomonas paucimobilis: clinical features and microbiological characteristics. Clin. Infect. Dis. 26:676-681. [DOI] [PubMed] [Google Scholar]
- 12.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson, E. N., and L. Young. 2001. Ecology and industrial microbiology: learning and earning from diversity. Curr. Opin. Microbiol. 4:281-284. [Google Scholar]
- 15.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 16.Kalmbach, S., W. Manz, and U. Szewczyk. 1997. Dynamics of biofilm formation in drinking water: phylogenetic affiliation and metabolic potential of single cells assessed by formazan reduction and in situ hybridization. FEMS Microbiol. Ecol. 22:265-279. [Google Scholar]
- 17.Karpay, R. I., T. J. Plamondon, and S. E. Mills. 1999. Comparison of methods to enumerate bacteria in dental unit water lines. Curr. Microbiol. 38:132-134. [DOI] [PubMed] [Google Scholar]
- 18.Kuchta, J. M., J. S. Navratil, M. E. Shepherd, R. M. Wadowsky, J. N. Dowling, S. J. States, and R. B. Yee. 1993. Impact of chlorine and heat on the survival of Hartmannella vermiformis and subsequent growth of Legionella pneumophila. Appl. Environ. Microbiol 59:4096-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaitre, D., A. Elaichouni, M. Hundhausen, G. Claeys, P. Vanhaesebrouck, M. Vaneechoutte, and G. Verschraegen. 1996. Tracheal colonization with Sphingomonas paucimobilis in mechanically ventilated neonates due to contaminated ventilator temperature probes. J. Hosp. Infect. 32:199-206. [DOI] [PubMed] [Google Scholar]
- 20.Manz, W. 1999. In situ analysis of microbial biofilms by rRNA-targeted oligonucleotide probing. Methods Enzymol. 310:79-91. [DOI] [PubMed] [Google Scholar]
- 21.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 22.Martin, M. V. 1987. The significance of the bacterial contamination of dental unit water systems. Br. Dent. J. 163:152-153. [DOI] [PubMed] [Google Scholar]
- 23.Osterhaut, G. J., V. H. Shull, and J. D. Dick. 1991. Identification of clinical isolates of gram-negative nonfermentative bacteria by an automated cellular fatty acid identification system. J. Clin. Microbiol. 29:1822-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollock, T. J., and R. W. Armentrout. 1999. Planktanic/sessile dimorphism of polysaccharide-encapsulated sphingomonads. J. Ind. Microbiol. Biotechnol. 23:436-441. [DOI] [PubMed] [Google Scholar]
- 25.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reasoner, D. J., and E. E. Geldreich. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tall, B. D., H. N. Williams, K. S. George, R. T. Gray, and M. Walch. 1995. Bacterial succession within a biofilm in water supply lines of dental air-water syringes. Can. J. Microbiol. 41:647-654. [DOI] [PubMed] [Google Scholar]
- 28.Wagner, M., R. Amann, H. Lemmer, and K. H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams, H. N., A. Johnson, J. I. Kelley, M. L. Baer, T. S. King, B. Mitchell, and J. F. Hasler. 1995. Bacterial contamination of the water supply in newly installed dental units. Quintessence Int. 26:331-337. [PubMed] [Google Scholar]
- 31.Williams, H. N., H. Quinby, and E. Romberg. 1994. Evaluation and use of a low nutrient medium and reduced incubation temperature to study bacterial contamination in the water supply of dental units. Can. J. Microbiol. 40:127-131. [DOI] [PubMed] [Google Scholar]
- 32.Williams, J. F., A. M. Johnston, B. Johnson, M. K. Huntington, and C. D. Mackenzie. 1993. Microbial contamination of dental unit waterlines: prevalence, intensity and microbiological characteristics. J. Am. Dent. Assoc. 124:59-65. [DOI] [PubMed] [Google Scholar]