Abstract
Thirty-one urease-positive thermophilic Campylobacter (UPTC) isolates, including three reference strains (NCTC12892, NCTC12895 and NCTC12896), and three Campylobacter lari isolates, which were isolated from several countries and sources, were compared genotypically by using multilocus enzyme electrophoresis (MLEE). We examined allelic variation around seven enzyme loci, including the adenylate kinase, alkaline phosphatase, catalase, fumarase, malic enzyme, malate dehydrogenase, and l-phenylalanyl-l-leucine peptidase loci. MLEE typing revealed the presence of 23 different electrophoretic types (ETs) among the 31 UPTC isolates, and 14 isolates shared six electrophoretic profiles. Three different ETs were identified for the three C. lari isolates examined, and no ETs were shared by UPTC and C. lari isolates. Quantitative analyses were subsequently performed by using allelic variation data, and the results demonstrated that the mean genetic diversity was 0.655. In conclusion, MLEE demonstrated that the UPTC isolates examined are genetically hypervariable and form a cluster separate from the C. lari cluster.
Urease-positive thermophilic Campylobacter (UPTC), a microaerophilic, gram-negative bacterium, is an organism that was identified only relatively recently in England (2). After the original descriptions of UPTC appeared (2), isolates of UPTC were found in France (1, 11), Northern Ireland (15), and The Netherlands (5), and UPTC strains have also recently been found in Japan (10). The UPTC group was first described by Bolton et al. (2), and strains were isolated from river water, seawater, mussels, and cockles. In 1988, Mégraud et al. reported the first isolation of UPTC from human clinical infections, and organisms were isolated from an appendix, as well as from human feces (11). This group of organisms has frequently been found to comprise the most common Campylobacter strains in shellfish and water (15).
Taxonomically, UPTC isolates have been classified as biochemical variants of Campylobacter lari, as demonstrated by Owen et al. (13), although there have been few reports of characterization of subspecies of these organisms. Recently, the usefulness of multilocus enzyme electrophoresis (MLEE) typing, which has been employed for eukaryotic genetics, was demonstrated for analysis of bacterial populations (14), including campylobacters (12). In MLEE analysis, variations in alleles of housekeeping genes encoding enzymes can be detected by estimating variations in the net electrophoretic charges of the polypeptides. Until now, MLEE analysis of the genetic relatedness of UPTC isolates has not been performed; it was the aim of this study to examine the relatedness of a small population of UPTC isolates from several countries that originated from shellfish, marine water, freshwater, seagulls (Larus sp.), and humans.
MATERIALS AND METHODS
Origins of UPTC and C. lari isolates.
We used 31 UPTC isolates, including 3 reference strains (NCTC12892, NCTC12895, and NCTC12896), and three C. lari isolates; these organisms were isolated in several countries from different sources, as shown in Table 1.
TABLE 1.
UPTC and C. lari isolates examined by MLEE typing
| Isolate | Taxon | MLEE ET | Source | Country |
|---|---|---|---|---|
| 2 | UPTC | A | Oysters | Northern Ireland |
| 11 | UPTC | B | Mussels | Northern Ireland |
| 142 | UPTC | C | Oysters | Northern Ireland |
| 145 | UPTC | D | Mussels | Northern Ireland |
| 163 | UPTC | E | Mussels | Northern Ireland |
| 182 | UPTC | F | Water | Northern Ireland |
| 412 | UPTC | G | Mussels | Northern Ireland |
| 467 | UPTC | H | Mussels | Northern Ireland |
| 472 | UPTC | I | Mussels | Northern Ireland |
| 475 | UPTC | J | Mussels | Northern Ireland |
| 476 | UPTC | K | Mussels | Northern Ireland |
| 480 | UPTC | L | Quality control | Northern Ireland |
| 484 | UPTC | M | Mussels | Northern Ireland |
| 485 | UPTC | N | Mussels | Northern Ireland |
| 487 | UPTC | I | Mussels | Northern Ireland |
| 494 | UPTC | O | Unknown | Northern Ireland |
| 497 | UPTC | P | Unknown | Northern Ireland |
| 504 | UPTC | Q | Mussels | Northern Ireland |
| 505 | UPTC | K | Mussels | Northern Ireland |
| NCTC12892 | UPTC | D | River water | England |
| NCTC12895 | UPTC | K | Mussels | England |
| NCTC12896 | UPTC | R | Mussels | England |
| 3980 | UPTC | S | Seawater | England |
| 10231 | UPTC | T | River water | England |
| CF89-12 | UPTC | U | River water | Japan |
| CF89-14 | UPTC | U | River water | Japan |
| 89049 | UPTC | C | Human | France |
| 92251 | UPTC | V | Human | France |
| C6 | UPTC | W | Seagull | Northern Ireland |
| C25 | UPTC | W | Seagull | Northern Ireland |
| A3 | UPTC | W | Seagull | Northern Ireland |
| 448 | C. lari | X | Mussels | Northern Ireland |
| 477 | C. lari | Y | Mussels | Northern Ireland |
| JCM2530 | C. lari | Z | Seagull | Japan |
Preparation of lysates for MLEE.
Each isolate was grown on 16 Mueller-Hinton agar plates supplemented with 5% (vol/vol) defibrinated sheep blood at 42°C for 48 h under microaerophilic conditions. Cells (ca. 1011 CFU) were harvested by scraping the surfaces of the plates with a sterile loop and suspending the cells in 2 ml of buffer (10 mM Tris, 1 mM EDTA, 0.5 mM NADP; pH 6.8). Lysates were prepared by subjecting the suspended cells to two cycles of sonication (30 s) in a MSE 150-W ultrasonic disintegrator (model Mk2; Sanyo Inc., Tokyo, Japan) at minimal power and cooled in an ice bath. The cells and particles remaining after lysis were removed by centrifugation (30,000 × g, 15 s, 4°C), and the straw-colored lysates were stored in 200-μl portions at −70°C.
Specific enzymes used.
All the isolates were assayed for the following enzymes: adenylate kinase, alkaline phosphatase, catalase, fumarase, malic enzyme, malate dehydrogenase, and l-phenylalanyl-l-leucine peptidase.
Electrophoresis and staining of enzymes.
The methods described previously by Selander and colleagues (14) were employed for electrophoresis and staining.
Calculation of genetic parameters.
The mobilities of the enzymes from the different isolates on the same gel slice were compared visually by using an illuminated light box. Replicate control strains were analyzed on each gel slice, which facilitated easy visual comparison of the gels. For each enzyme, distinct electromorphs were numbered in order of decreasing anodal migration. The repeated absence of enzymatic activity was scored as a null character. Each isolate was characterized on the basis of its combination of electromorphs for the seven enzymatic loci assayed, and distinct profiles of electromorphs corresponding to unique multilocus genotypes (electrophoretic types [ETs]) were designated arbitrarily with letters (ETs A to Z).
Genetic diversity (h) was calculated for each locus as follows: h = (1 − Σxi2)[n/(n − 1)], where xi is the frequency of the ith allele at the locus, n is the number of ETs, and n/(n − 1) is a correction factor for the bias in small samples. Pairwise comparisons between ETs were analyzed statistically by calculating the Euclidean distance between clustered pairs of strains, which gave an unweighted matrix of coefficients of genetic distance for the enzymatic loci.
RESULTS
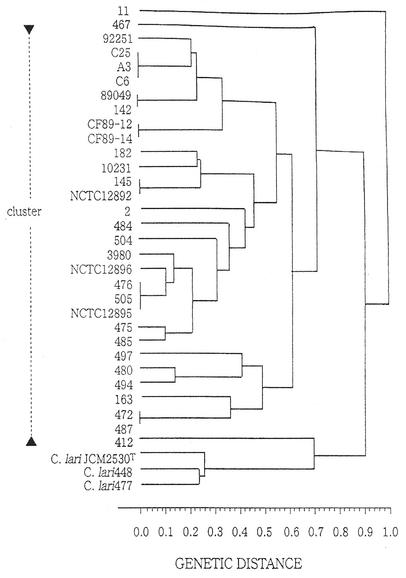
All 31 UPTC isolates, as well as the three C. lari isolates employed in this study, were gram negative, catalase and oxidase positive, negative for hydrolysis of hippurate, and resistant to cephalothin. All organisms grew microaerophilically at 37 and 43°C but not at 25°C. A positive urease test was recorded for all UPTC isolates but not for the C. lari isolates. Electrophoresis of a control strain with a known isoenzyme type was performed in both the left and right outer lanes, as well as in the central lane, and this allowed strict control of assignment of allelic types both within the same gel and among multiple gels. MLEE typing with seven enzyme loci revealed the presence of 23 different ETs for the 31 UPTC isolates; isolates C6, C25, and A3 had the same ET profile, as did isolates 89049 and 142, isolates CF89-12 and CF89-14, isolates 145 and NCTC12892, isolates 476, 505, and NCTC12895, and isolates 472 and 487. Three different ETs were obtained for the three C. lari isolates examined, and no ETs were shared by UPTC isolates and C. lari isolates. Quantitative analyses were subsequently performed by using allelic variation, and the results demonstrated that the mean genetic diversity was 0.655. In relation to allelic variation, l-phenylalanyl-l-leucine peptidase was the most diverse and malic enzyme was the least diverse in terms of the number of different alleles found. The genetic relatedness of the 31 UPTC isolates and three C. lari isolates is shown in the dendrogram in Fig. 1; in this dendrogram the C. lari isolates formed a cluster distinct from the UPTC cluster.
FIG. 1.
Dendrogram showing the genetic relationships of 31 isolates of UPTC and three isolates of C. lari, as determined by MLEE analysis of seven enzyme loci.
DISCUSSION
The aims of this study were to use MLEE typing to demonstrate the genetic relatedness of UPTC isolates in a diverse population of organisms collected in several countries and to demonstrate the relationship of these organisms to C. lari. Using MLEE, we were able to assign an ET to all of the isolates of both UPTC and C. lari examined, and thus this study did not suffer from the typeability problems associated with serology and phage-typing schemes. There was a high degree of hypervariability within the UPTC isolates, and 23 ETs were obtained for a population consisting of 31 UPTC isolates. Thus, the UPTC group of organisms behaved like their close phylogenetic neighbors, Campylobacter jejuni and C. lari (12), exhibiting a high degree of variability when MLEE was used as the typing technique. However, in the present study, the MLEE analysis clustered several isolates into similar types. For example, the cluster containing isolates C6, C25, and A3 was composed of isolates obtained from seagulls in Northern Ireland, which were isolated from coastal regions approximately 50 miles apart, probably reflecting the movement of seagulls around local coastal regions. It is also interesting that isolate 89049, a human clinical isolate from France, clustered with isolate 142, which was isolated from oysters in Northern Ireland approximately 4 years after the French report. In addition, the clustering of isolates 476 and 505 and the clustering of isolates 472 and 487 demonstrate that similar types are distributed in shellfish growing in in-shore marine waters around Northern Ireland.
Historically, the UPTC group of organisms has never been formally recognized in the Approved Lists of Bacterial Names; in this list UPTC isolates have been regarded as members of a biovar of C. lari. Some of the early work of Owen et al. (13), in which one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cellular protein was used, showed that the eight UPTC isolates in their study clustered into five profile types, which were suggested to be biovars of C. lari. With the development of discriminatory molecular techniques, such as 16S ribosomal DNA sequence analysis and sequence analysis of other conserved gene loci, including the recA locus (8), it has been possible to ascertain the relationship between UPTC isolates and C. lari. Recently, On et al. [S. L. W. On, P. I. Fields, T. Broman, L. O. Helsei, C. Fitzgerald, C. S. Harrington, S. Laevens, A. G. Steigerwalt, B. Olsen, and P. A. R. Vandamme, Int. J. Med. Microbiol. 291(Suppl. 31):144, 2001] demonstrated in a comprehensive polyphasic study, in which they used phenotypic characterization, whole-cell protein profiling, amplified fragment length polymorphism, 16S ribosomal DNA sequencing, and DNA-DNA hybridization, that the species C. lari comprises at least three taxa, including “C. lari subsp. lari,” “C. lari subsp. subantarcticus,” and “C. lari subsp. ureasum.” In addition, these workers noted that UPTC isolates were more heterogeneous than “C. lari subsp. lari,” which is in agreement with our findings. Likewise, recently, Duim et al. [B. Duim, J. Wagenaar, H. P. Endtz, J. R. Dijkstra, and P. A. R. Vandamme, Int. J. Med. Microbiol. 291(Suppl. 31):143-144, 2001] used a polyphasic approach similar to that of On et al. [Int. J. Med. Microbiol. 291(Suppl. 31):144, 2001] and showed that UPTC isolates form a group that is distinct from C. lari.
More recently, multilocus sequence typing (MLST) has emerged as an novel typing tool for bacterial population genetic studies (for a review see reference 6), and it has been used successfully to subtype a variety of bacterial and fungal pathogens, including Enterococcus faecium (7), Neisseria meningitidis (9), and Candida albicans (3). The increased use of MLST is largely a result of the widespread adoption and availability of automated DNA sequencing in many microbiology laboratories. MLST molecularly examines nucleotide differences in several gene loci, whereas MLEE relies on cellular detection of variation at gene loci by starch gel electrophoresis under nondenaturing conditions. Although MLST has become the method of choice for population genetic studies, the validity of and data obtained from MLEE analysis are comparable and equally valid, if workers are prepared to expend the time and labor necessary to perform the latter technique. Table 2 shows the differences in the MLST and MLEE typing techniques. To date, there have not been any reports of using MLST for characterization of UPTC subspecies, although the technique has been successfully employed to examine the clonality of C. jejuni populations (4).
TABLE 2.
Comparison of MLEE and MLST
| Technique | Main laboratory technique involved | Subjectivity | Ability for interlaboratory comparison | Requirement for complex equipment | Simplicity | Cost | Repeat- ability |
|---|---|---|---|---|---|---|---|
| MLEE | Cellular (protein analysis) | Low to moderate | Low | Low | Moderate | Moderate (mainly labor) | Moderate |
| MLST | Molecular (PCR plus DNA sequencing) | Low | High | High | High | Moderate (mainly con- sumables) | High |
In conclusion, in this study using MLEE, we demonstrated that the UPTC isolates examined are genetically hypervariable and form a cluster separate from the C. lari cluster.
Acknowledgments
We thank Francis Mégraud, Bordeaux, France, for providing certain UPTC isolates.
This work was supported in part by the Department of Health & Social Services (Northern Ireland) and by the British Council (Tokyo).
REFERENCES
- 1.Bezian, M. C., G. Ribou, C. Barberis-Giletti, and F. Megraud. 1990. Isolation of a urease-positive thermophilic variant of Campylobacter lari from a patient with urinary tract infection. Eur. J. Microbiol. Infect. Dis. 9:895-897. [DOI] [PubMed] [Google Scholar]
- 2.Bolton, F. J., A. V. Holt, and D. N. Hutchinson. 1985. Urease-positive thermophilic campylobacters. Lancet i:1217-1218. [DOI] [PubMed]
- 3.Bougnoux, M. E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endtz, H. P., J. S. Vliegenthart, P. Vandamme, H. W. Weverink, N. P. van den Braak, H. A. Verbrugh, and A. van Verkum. 1997. Genotypic diversity of Campylobacter lari isolated from mussels and oysters in The Netherlands. Int. J. Food Microbiol. 34:79-88. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 7.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honda, M., S. Matsushita, O. Murayama, B. C. Millar, J. E. Moore, and M. Matsuda. 2002. Cloning and sequence analysis of the recA gene in urease-positive thermophilic Campylobacter (UPTC). Br. J. Biomed. Sci. 59:166-169. [DOI] [PubMed] [Google Scholar]
- 9.Kriz, P., J. Kalmusova, and J. Felsberg. 2002. Multilocus sequence typing of Neisseria meningitidis directly from cerebrospinal fluid. Epidemiol. Infect. 128:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda, M., A. Kaneko, M. Fukuyama, T. Itoh, M. Shingaki, M. Inoue, and J. E. Moore. 1996. First finding of urease-positive thermophilic strains of campylobacter in river water in the Far East, namely, in Japan, and their phenotypic and genotypic characterization. J. Appl. Bacteriol. 81:608-612. [Google Scholar]
- 11.Megraud, F., D. Chevrier, N. Desplaces, A. Sedallian, and I. L. Guesdon. 1988. Urease-positive thermophiic campylobacter (Campylobacter lari variant) isolated from an appendix and from human feces. J. Clin. Microbiol. 26:1050-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore, J. E., M. M. Garcia, and R. H. Madden. 2002. Subspecies characterization of porcine Campylobacter coli and Campylobacter jejuni by multilocus enzyme electrophoresis typing. Vet. Res. Commun. 26:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Owen, R. J., M. Costas, L. Sloss, and F. J. Bolton. 1988. Numerical analysis of electrophoretic protein patterns of Campylobacter laridis and allied thermophilic campylobacters from the natural environment. J. Appl. Bacteriol. 65:69-78. [DOI] [PubMed] [Google Scholar]
- 14.Selander, R. K., D. A. Caugant, H. Ochinan, J. M. Musser, M. N. Gilmour, and T. S. Vhittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson, I. G., and J. E. Moore. 1996. Presence of Salmonella spp. and Campylobacter spp. in shellfish. Epidemiol. Infect. 116:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]