Abstract
A partially annotated proteome reference map of the food pathogen Listeria monocytogenes was developed for exponentially growing cells under standardized, optimal conditions by using the sequenced strain EGDe (serotype 1/2a) as a model organism. The map was developed by using a reproducible total protein extraction and two-dimensional (2-D) polyacrylamide gel electrophoresis analysis procedure, and it contained 33 identified proteins representing the four main protein functional classes. In order to facilitate analysis of membrane proteins, a protein compartmentalization procedure was assessed. The method used provided partial fractionation of membrane and cytosolic proteins. The total protein 2-D profiles of three serotype 1/2a strains and one serotype 1/2b strain isolated from food were compared to the L. monocytogenes EGDe proteome. An average of 13% of the major protein spots in the food strain proteomes were not matched in the strain EGDe proteome. The variation was greater for the less intense spots, and on average 28% of these spots were not matched. Two of the proteins identified in L. monocytogenes EGDe were missing in one or more of the food isolates. These two proteins were proteins involved in the main glycolytic pathway and in metabolism of coenzymes and prosthetic groups. The two corresponding genes were found by PCR amplification to be present in the four food isolates. Our results show that the L. monocytogenes EGDe reference map is a valuable starting point for analyses of strains having various origins and could be useful for analyzing the proteomes of different isolates of this pathogen.
Listeria monocytogenes is a gram-positive facultatively intracellular pathogen that is mainly associated with infections in certain human risk groups, including pregnant woman, newborns, and immunocompromised patients (40). It is widespread in nature and may be transferred to humans by contaminated foods (12). This pathogen is able to survive food processing technologies, such as high concentrations of salt and relatively low pHs, and it is capable of multiplication at refrigeration temperatures (24). Many of the preservation methods and cleaning compounds used in the food industry target the bacterial cell membrane, and furthermore, many successful drugs act by modulating the activity of membrane proteins (37). Methods that monitor membrane proteins in particular are therefore of vital importance.
Two-dimensional (2-D) polyacrylamide gel electrophoresis of bacterial proteins was introduced more than 25 years ago (29). This technique is based on separation of proteins by isoelectric point (pI) in the first dimension and by molecular weight in the second dimension. Over the years, the technique has been improved, and it now has the potential to resolve thousands of proteins in a complex sample (15).
Previously, in 2-D analyses of L. monocytogenes proteins workers focused on responses to stress, including resistance to antimicrobial compounds (9, 17, 34), pH stress (7, 28, 32, 35), high salinity (11), or cold shock (2, 19, 41). We know of no studies in which 2-D analysis was used in which the workers focused on membrane proteins in L. monocytogenes. 2-D electrophoretic analysis has also been used for identification and classification of Listeria (16); Gormon and Phan-Thanh observed that the proteome similarity was greatest for strains belonging to the same serovar and that there was more variation between serovars.
Our general knowledge about the molecular constituents of L. monocytogenes has been greatly enhanced by the recent release of the genomic sequence of L. monocytogenes EGDe (14). The release of this sequence provided a resource for comparison of genomes and proteomes of strains of L. monocytogenes from various sources. Strain EGDe is an animal isolate belonging to serotype 1/2a, which is the only serotype that is prevalent in illness, as well as in foods and food processing facilities (see reference 22 for a recent review). This strain may therefore be expected to be a good reference organism for clinical isolates, as well as food isolates. There is, however, relatively high genetic diversity within serotype 1/2a, and there is pronounced diversity in food strains in general (22).
In this study, we constructed a total protein 2-D reference map of abundant proteins in exponentially dividing cells under standardized, optimal growth conditions using L. monocytogenes EGDe as the model organism. Furthermore, we evaluated a membrane protein extraction procedure for L. monocytogenes based on the method developed for Escherichia coli by Ames and Nikaido (1). In order to assess how well the reference map represents strains originating from food with a focus on serotype 1/2a, we compared the L. monocytogenes EGDe proteome reference map with the 2-D profiles of a serotype 1/2b isolate and several serotype 1/2a food isolates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. monocytogenes EGDe (animal isolate; serotype 1/2a) (14), B73 (meat isolate; serotype 1/2a) (10), 412 (isolated from raw salted pork; serotype 1/2a) (18), 386 (isolated from heat-treated pork; Danish Meat Research Institute, Roskilde, Denmark; serotype 1/2b), and O57 (isolated from lightly pickled salmon; serotype 1/2a) (3) were maintained on brain heart infusion broth or agar at 37°C.
Preparation of cells prior to protein isolation.
L. monocytogenes strains were grown without shaking until the mid-exponential phase (optical density at 600 nm, 0.45 to 0.5). Chloramphenicol (Sigma, St. Louis, Mo.) was added to a final concentration of 20 μg/ml to halt protein synthesis. Bacterial cells were harvested by centrifugation (8,000 × g, 15 min, 4°C). Each cell pellet was washed once with 10 mM phosphate-buffered saline (pH 7.0) and twice with 32 mM Trizma Pre-Set crystals (pH 7.5) (Sigma). The wash buffers were also supplemented with chloramphenicol at a final concentration of 20 μg/ml. Each washed cell pellet was resuspended in TE (10 mM Tris, 1 mM EDTA; pH 7.5) containing a Complete minitablet (protease inhibitors; one cocktail minitablet per 5 ml of TE; Roche, Mannheim, Germany) and stored at −80°C. Cell suspensions were thawed on ice and transferred to FastProtein Blue tubes (Bio 101, Carlsbad, Calif.). The cells were disrupted with a FastPrep Instrument FP 120 (Bio 101) at a maximum tube velocity of 6.5 m/s for 45 s and subsequently chilled on ice. This cycle was repeated five times. All chemicals and materials were obtained from Amersham Biosciences (Little Chalfont, Buckinghamshire, United Kingdom), unless indicated otherwise.
Fractionation of cellular proteins.
The method used for fractionation of cellular proteins was based on the method of Ames and Nikaido (1), modified as follows. Unbroken cells and cellular debris were sedimented by centrifugation (16,000 × g, 4°C, 25 min), and the supernatant was treated with DNase I at a concentration of 85 μg/ml and RNase I (Boehringer, Mannheim, Germany) at a concentration of 4.2 μg/ml and incubated at 37°C for 30 min. In order to separate the membrane fraction from the cytosolic fraction, the homogenate was centrifuged (100,000 × g, 4°C, 80 min). The clarified supernatant, which was considered the cytosolic fraction, was removed and stored at −20°C. The yellow pellet was resuspended in 200 μl of a 1% (wt/vol) sodium dodecyl sulfate (SDS) solution containing 100 mM dithiothreitol (DTT) (Sigma) and boiled for 5 min. To the boiled sample, 9.5 M urea, 100 mM DTT, 8% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 4% (vol/vol) Pharmalyte (pH 3 to 10) were added to the final concentrations indicated. The mixture was incubated at 30°C for 2 h. Insoluble material was removed by centrifugation (18,000 × g, 45 min, 28°C), and the resulting supernatant was considered the membrane fraction. The membrane fraction was stored at −80°C or used immediately for isoelectric focusing (IEF).
Total protein isolation.
Cell lysates were treated with nucleases as described above. To each homogenate, 9.5 M urea, 100 mM DTT, 4% (wt/vol) CHAPS, and 2% (vol/vol) Pharmalyte (pH 3 to 10) were added to the final concentrations indicated. The mixture was incubated at 30°C for 2 h. Insoluble material was removed by centrifugation (18,000 × g, 85 min, 28°C). The clarified supernatant was carefully removed and stored at −80°C or used immediately for IEF.
First-dimension electrophoresis.
IEF was carried out by using 11-cm precast Immobline DryStrips with a linear pH 4 to 7 gradient and a Multiphore II apparatus according to the manufacturer's instructions. For total protein gels, approximately 75 and 7.5 μg of protein were cup loaded at the anodic end for Coomassie blue- and silver-stained gels, respectively. Prior to loading of the cytosolic fraction, 15 μl of the protein preparation was added to 85 μl of solubilization solution (9.5 M urea, 100 mM DTT, 4% [wt/vol] CHAPS, 2% [vol/vol] Pharmalyte [pH 3 to 10]) and incubated at 30°C for 1 h. When we used a protein load for the membrane fraction gels that was similar to the protein load described above for the total protein samples, a low number of spots was visualized. In order to increase the number of spots visualized, approximately twice as much protein was utilized for compartmentalized gels (i.e., approximately 150 and 15 μg of protein for Coomassie blue- and silver-stained gels, respectively). Protein concentrations were determined with a PlusOne 2-D Quant kit (Amersham Biosciences). The following voltage gradient was applied: from 0 to 300 V in 0.01 h; 300 V for 6.5 h; from 300 to 3,500 V for 5 h; and 3,500 V for 8 h.
Second-dimension electrophoresis.
Electrophoresis in the second dimension was performed on precast ExcelGel XL SDS 12-14 gels by using a Multiphore II apparatus as described in the manufacturer's instructions. IEF strips were equilibrated in SDS equilibration buffer as recommended by the manufacturer, with the following modifications: (i) the concentration of SDS and iodoacetamide (Sigma) was increased from 2 to 4% (wt/vol); and (ii) each of the equilibration steps was carried out for 30 min instead of 15 min. MultiMark multicolored standards (Novex, San Diego, Calif.) were electrophoresed in the second dimension to determine the relative molecular masses of proteins. Gels were stained with either Coomassie blue R250 or silver; for silver staining an automated silver stainer was used. Gels that were prepared for mass spectrometry were stained with Coomassie colloidial blue G250 by using a previously described procedure (26).
Image analysis.
Coomassie blue-stained gels were scanned at a resolution of 200 dots per in. and were analyzed by using the Z3 2-D gel image analysis system, version 2.00 (Compugen Ltd., Jamesburg, N.J.). Total numbers of spots were determined by automated spot detection followed by manual editing. The major spots used for analysis were defined as the spots with a minimum spot area of 50 pixels and a minimum spot contrast of 25. For the comparison of total protein gels, a cutoff value of 200 predominant spots was used in order to standardize the number of spots compared. Spots that did not fit the criteria for major spots were considered minor spots. A minimum of three Coomassie blue-stained gels were used for each sample, and a typical gel was used for computer-aided analysis. The absence of spots was verified visually on all Coomassie blue-stained gels and also on silver-stained gels for each sample.
Protein identification.
For N-terminal sequencing, electroblotting, staining, and storage of the blot were done as described previously (36). N-terminal sequencing was performed with a 491 Procise automated sequencer (Perkin-Elmer, Wellesley, Mass.).
Sequence data for internal peptides were acquired with a quadropole time of flight mass spectrometer after electrospray ionization with tandem mass spectrometry. The membrane or cytosolic fraction gels of L. monocytogenes EGDe were destained and cut away from the backing, and selected spots were excised and dried at room temperature under reduced pressure. In-gel digestion was performed on the dried gel pieces by treatment with 30 to 60 μl of trypsin (2 μg/ml) in 50 mM NH4HCO3 (sequencing grade; Promega, Madison, Wis.) overnight at 37°C. The supernatant was removed and stored, and the gel pieces were incubated with 40 to 80 μl of 5% formic acid for 30 min. The same volume of acetonitrile was added, and incubation was continued for 15 min. The supernatant was removed and pooled with the first supernatant, and the volume was reduced with a Speed Vac to approximately 15 μl. The peptides generated were desalted by using C18-ZipTip (Millipore, Bedford, Mass.) as recommended by the manufacturer. Elution of the purified peptides was carried out with 5 μl of 65% methanol-0.5% formic acid. For electrospray analysis and subsequent peptide sequencing, 3-μl portions of a ZipTip-purified sample were placed into Au/Pd-coated nanospray glass capillaries (Protana, Odense, Denmark). The tip of each capillary was placed orthogonally in front of the entrance hole of a quadrupole time of flight mass spectrometry instrument (Q-TOF II; Micromass, Manchester, United Kingdom) equipped with a nanospray ion source. A capillary voltage between 750 and 1,000 V and a cone voltage of 35 V were applied. Doubly and triply charged peptides were chosen for collision-induced tandem mass spectrometry fragmentation experiments, and the corresponding parent ions were selectively transmitted from the quadrupole mass analyzer into the collision cell. Argon was used as the collision gas, and the kinetic energy was set between 20 and 35 eV. The resulting daughter ions were separated with an orthogonal time of flight mass analyzer. Peptide microsequencing and protein identification were carried out with the Peptide-Sequencing program in the BioLynx software (version 3.4; Micromass) and with the Sonar program (Proteometrics, New York, N.Y.), respectively. The trypsin fragment sequences obtained were compared to the proteins predicted from the L. monocytogenes EGDe genome sequence (http://genolist.pasteur.fr/ListiList/).
Bioinformatics.
The theoretical molecular weights and pIs of identified proteins were calculated from the predicted amino acid sequences by using the ProtParam tool at the Expasy website (http://www.expasy.ch/tools/protparam.html). The grand average hydropathy (GRAVY) values were calculated by the method of Kyte and Doolittle (23) by using ProtParam. Transmembrane domains (TMDs) were predicted by using TMpred with default settings (http://www.ch.embnet.org/software/TMPRED_form.html). Only TMD values greater than 500 were considered to be significant.
The L. monocytogenes EGDe total protein reference map is available at http://www.mli.kvl.dk/foodmicro/special/index.htm.
PCR analysis of the GAPDH and phosphomethylpyrimidine kinase genes.
The presence of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and phosphomethylpyrimidine kinase genes was investigated by performing colony PCR with primers designed by using the L. monocytogenes EGDe genome sequence (lmo2459 and lmo0662, respectively) to cover substantial parts of the corresponding reading frames. For lmo2459, primers gapF1 (5′-GTCTAGCATTCCGTCGTATTC-3′) and gapR1 (5′-AGCTCATTTCGTTATCGTACC-3′) were used, and these primers provided 915 bp of the 1,011-bp coding region (nucleotides 44 to 958). For lmo0662, primers thiDF1 (5′-CAATGGACCCAGACAACAAC-3′) and thiDR1 (5′-TGCGACAGCTTCTTCAAC-3′) were used, and they provided 584 bp of the 816-bp coding region (nucleotides 128 to 711). The PCR was performed for 30 cycles with annealing at 52°C for 1 min.
RESULTS
Total protein profile.
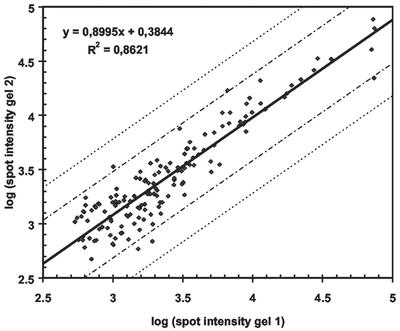
A total of 261 spots were detected in the L. monocytogenes EGDe total protein Coomassie blue-stained gel (Fig. 1a). The reproducibility of the total protein profiles was evaluated by using 2-D gels of proteins extracted from two independent cultures of L. monocytogenes EGDe. The logarithms of the spot intensities from the two gels are shown in Fig. 2. For most of the spots (97%) the difference in intensity was less than 2.5-fold (within the area delineated by the first set of dashed lines in Fig. 2). Moreover, no outliers were observed. The results indicate that when this procedure was used for proteome comparison, differences in spot intensity that were greater than fivefold (delineated by the second set of dashed lines in Fig. 2) were due to biological variation rather than experimental variation.
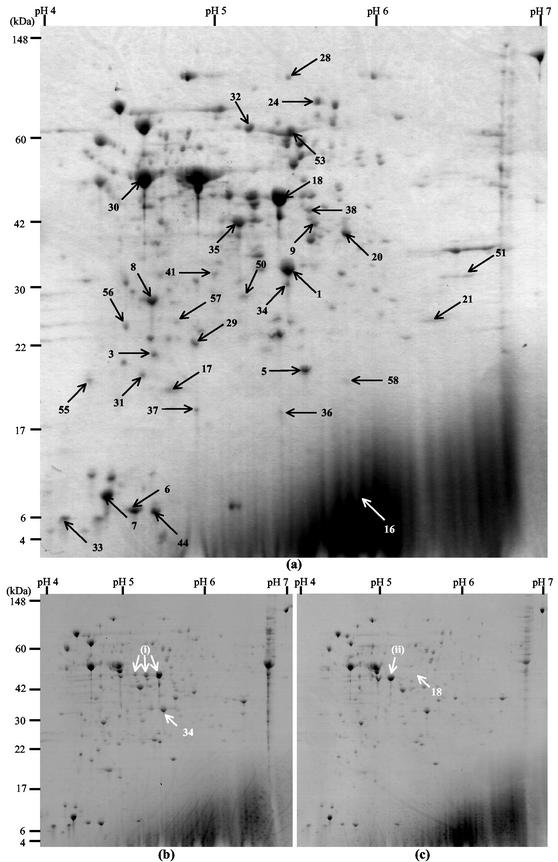
FIG. 1.
Coomassie blue R250-stained 2-D gels of total cellular proteins from L. monocytogenes EGDe (a), B73 (b), and 412 (c). The numbered spots represent proteins identified in the L. monocytogenes EGDe total protein reference map. The proteins identified are described in Table 1. Strains B73 and 412 lacked spots 34 and 18, respectively. Arrows (i) indicate spots in strain B73 corresponding to GAPDH identified by Michel Hebraud (personal communication), the leftmost of which is indicated by arrow (ii) in the strain 412 gel (see text for details).
FIG. 2.
Double-logarithmic plot showing the reproducibility of two independent protein extractions and 2-D analyses of L. monocytogenes EGDe total protein isolation procedures. The logarithms of the spot intensities are plotted, and the regression line and R2 value are shown. The two sets of dashed lines indicate 2.5- and 5-fold differences in intensity.
Compartmentalization of cellular proteins.
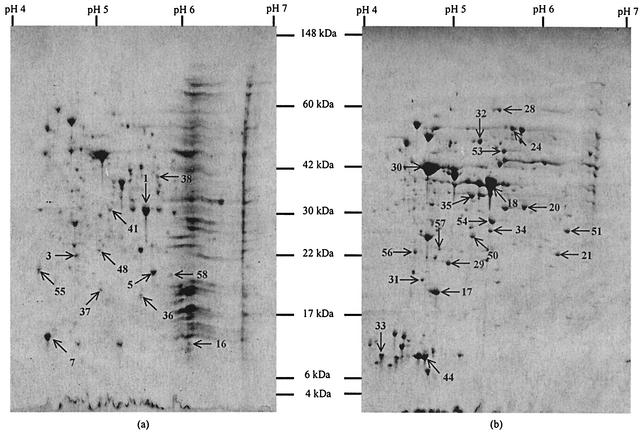
Visual comparisons of the 2-D profiles of the membrane and cytosolic fractions of L. monocytogenes EGDe suggested that there was a distinctive overall protein pattern for each of the fractions (Fig. 3). An image analysis of the Coomassie blue-stained membrane fraction gel revealed 208 spots, 106 of which were considered to be major (see above for definition of major spots). A similar analysis of the cytosolic fraction gel revealed 420 spots, 236 of which were major. A larger protein load was added to gels that contained the compartmentalized fractions than to the gels that contained the total protein samples. Computer-aided comparison of the membrane and cytosolic fraction gels showed that only 27 of the major spots in the membrane fraction were unique (i.e., were not discernible in the cytosolic fraction), whereas 161 of the major spots were unique in the cytosolic fraction. However, many of the common spots were enriched and had substantially greater intensity in one of the two fractions.
FIG. 3.
Coomassie blue R250-stained 2-D gels of the membrane fraction (a) and cytosolic fraction (b) of protein extracts of L. monocytogenes EGDe. Spots from the two fractions that were excised and identified are numbered. Proteins that were identified are described in Table 1.
Identification of proteins.
A total of 12 unique or enriched spots from the membrane fraction and 20 unique or enriched spots from the cytosolic fraction (Fig. 3) were excised and identified. Additionally, four spots from a total protein gel were analyzed. The proteins identified and their characteristics are shown in Table 1. All of the trypsin fragments sequenced from each of the spots analyzed exhibited 100% identity with the corresponding regions in the predicated amino acid sequence of the protein. The theoretical pI and molecular weight were generally in good agreement with the experimentally observed values. Clear deviations in theoretical and experimentally observed pIs could have been due to posttranslational modification.
TABLE 1.
Proteins identified from 2-D gels of L. monocytogenes EGDe
| Spota | Protein identification | Mol wt (103)
|
pl
|
Functional classb | TMD predictionc | GRAVY value | ||
|---|---|---|---|---|---|---|---|---|
| Observed | Predicted | Observed | Predicted | |||||
| Spots excised from membrane fraction gels | ||||||||
| 1 | FbaA (lmo2556), similar to fructose-1,6-bisphosphate aldolase | 23 | 30.0 | 5.47 | 5.20 | 2.1.1 | 1/0 | −0.034 |
| 3 | lmo2829, similar to yeast protein Frm2p in fatty acid signaling | 20 | 22.2 | 4.77 | 4.70 | 2.4 | 1/1 | −0.174 |
| 5 | RplJ (lmo0250), ribosomal protein L10 | 16 | 17.7 | 5.60 | 5.36 | 3.7.1 | 1/0 | −0.080 |
| 7 | RplL (lmo0251), ribosomal protein L12 | 10 | 12.5 | 4.54 | 4.54 | 3.7.1 | 1/1 | 0.143 |
| 16 | lmo2149, similar to proteins with no known function | 9.25 | 11.9 | 6.00 | 5.53 | 5.2 | 0/0 | −0.488 |
| 29 | ClpP (lmo2468), ATP-dependent Clp protease proteolytic subunit | 18.5 | 21.6 | 5.06 | 4.94 | 4.1 | 3/2 | −0.072 |
| 36 | TufA (lmo2653), very similar to translation elongation factor EF-Tu | 13.5 | 43.34 | 5.49 | 4.81 | 3.7.4 | 1/0 | −0.272 |
| 37 | lmo1580, similar to proteins with no known function | 14 | 16.9 | 5.05 | 4.98 | 5.2 | 1/0 | −0.192 |
| 38 | MreB (lmo1548), similar to cell shape-determining protein | 35 | 35.5 | 5.66 | 5.16 | 1.1 | 2/1 | 0.061 |
| 41 | lmo1011, similar to tetrahydrodipicolinate succinylase | 24 | 24.8 | 5.14 | 4.95 | 2.2 | 6/2 | 0.193 |
| 55 | LmaA (lmo0118), antigen A | 16.1 | 18.1 | 5.40 | 4.47 | 4.5 | 1/0 | −0.093 |
| 58 | lmo0273, similar to proteins with no known function | 16 | 18.8 | 5.80 | 5.45 | 5.2 | 0/0 | −0.387 |
| Spots excised from cytosolic fraction gels | ||||||||
| 1 | FbaA (lmo2556), similar to fructose-1,6-bisphosphate aldolase | 23 | 30.2 | 5.5 | 5.20 | 2.1.1 | 1/0 | −0.034 |
| 17 | Fri (lmo0943), nonheme iron-binding ferritin | 14.7 | 18.1 | 4.83 | 4.86 | 4.1 | 0/0 | −0.371 |
| 18 | GAPDH (lmo2459), similar to GAPDH | 38 | 36.3 | 5.3 | 5.20 | 2.1.2 | 1/0 | −0.115 |
| 20d | Pfk (lmo1571), similar to 6-phosphofructokinase | 30 | 34.4 | 5.79 | 5.46 | 2.1.1 | 4/1 | −0.104 |
| 21 | Upp (lmo2538), similar to uracil phosphoribosyltransferase | 30 | 22.9 | 5.77 | 5.70 | 2.3 | 1/1 | −0.035 |
| 24 | PykA (lmo1570), similar to pyruvate kinases | 64 | 62.6 | 5.64 | 5.39 | 2.1.1 | 3/1 | −0.034 |
| 28 | PnpA (lmo1331), polynucleotide phosphorylase | 85 | 79.6 | 5.49 | 5.23 | 2.3 | 2/1 | −0.308 |
| 29 | ClpP (lmo2468), ATP-dependent Clp protease proteolytic subunit | 18.5 | 21.6 | 4.96 | 4.94 | 4.1 | 3/0 | −0.072 |
| 30 | Eno (lmo2455), similar to enolase | 43 | 46.5 | 4.71 | 4.70 | 2.1.2 | 1/0 | −0.244 |
| 31 | lmo0796, similar to proteins with no known function | 16 | 19.5 | 4.71 | 4.69 | 5.2 | 0/0 | −0.460 |
| 32 | Pgm (lmo2456), similar to phosphoglycerate mutase | 53 | 56.1 | 5.29 | 5.10 | 2.1.2 | 0/0 | −0.342 |
| 33 | CspB (lmo2016), similar to major cold shock protein | 8.5 | 7.29 | 4.24 | 4.44 | 4.1 | 0/0 | −0.398 |
| 34 | ThiD (lmo0662), similar to phosphomethylpyrimidine kinase ThiD | 22 | 28.8 | 5.41 | 5.25 | 2.5 | 2/1 | −0.025 |
| 35 | Tsf (lmo1657), translation elongation factor | 32.5 | 32.6 | 5.20 | 5.11 | 3.5.3 | 0/0 | −0.430 |
| 44 | PtsH (lmo1002), phosphotransferase system phosphocarrier protein (Hpr) | 9 | 9.4 | 4.73 | 4.81 | 1.2 | 1/0 | −0.067 |
| 50 | Adk (lmo2611), similar to adenylate kinases | 22 | 24.2 | 5.23 | 5.08 | 2.3 | 0/0 | −0.586 |
| 51 | SerS (lmo2747), seryl-tRNA synthetase | 22 | 49.2 | 6.26 | 5.26 | 3.7.2 | 1/0 | −0.553 |
| 53 | lmo0355, similar to flavocytochrome c fumarate reductase chain A | 49 | 54.6 | 5.54 | 5.71 | 1.4 | 2/2 | −0.489 |
| 56 | lmo2376, similar to peptidyl-prolyl cis-trans isomerase | 19.5 | 21.5 | 4.57 | 4.59 | 3.9 | 0/0 | −0.327 |
| 57 | DeoD (lmo1856), purine nucleoside phosphorylase | 20 | 25.5 | 4.84 | 4.87 | 2.3 | 2/1 | 0.073 |
| Spots excised from total protein gels | ||||||||
| 6 | GroES (lmo2069), class I heat shock protein | 8.50 | 10.05 | 4.72 | 4.59 | 3.9 | 0/0 | −0.104 |
| 7 | RplL (lmo0251), ribosomal protein L12 | 10.85 | 12.44 | 4.57 | 4.54 | 3.7.1 | 1/1 | 0.143 |
| 8 | Tpi (lmo2457), triosephosphate isomerase | 27.42 | 26.86 | 4.77 | 4.78 | 2.1.2 | 2/1 | 0.096 |
| 9d | MptA (lmo0096), similar to mannose-specific phosphotransferase system enzyme IIAB | 35 | 34.99 | 5.35 | 5.32 | 1.2 | 1/1 | −0.123 |
| 9d | Pfk (lmo1571), similar to 6-phosphofructokinase | 35 | 34.42 | 5.35 | 5.46 | 2.1.1 | 1/1 | −0.104 |
Spots 6, 7, and 8 were identified by N-terminal sequencing, and all other spots were identified by time of flight mass spectrometry.
Functional class codes according to genolist (http://genolist.pasteur.fr/ListiList/help/function-codes.html): 1.1, cell wall; 1.2, transport/binding proteins and lipoproteins; 1.4, membrane bioenergetics; 2.1.1, metabolism of carbohydrate-specific pathways; 2.1.2, main glycolytic pathways; 2.2, metabolism of amino acids and related molecules; 2.3, metabolism of nucleotides and nucleic acids; 2.4, metabolism of lipids; 2.5, metabolism of coenzymes and prosthetic groups; 3.5.3, RNA synthesis—elongation; 3.7.1, protein synthesis—ribosomal proteins; 3.7.2, protein synthesis—aminoacyl-tRNA sysnthetases; 3.7.4., protein synthesis—elongation. 3.9, protein folding; 4.1, adaptation to atypical conditions; 4.5, miscellaneous; 5.2, from other organisms.
Detected TMDs/significant TMDs as calculated at http://www.ch.embnet.org/software/TMPREDform.html.
Spot identified previously (17).
From the 36 spots that were analyzed, 33 different proteins were identified, as 3 proteins were found in more than one fraction. Spots 1 (fructose-1,6-bisphosphate aldolase) and 29 (ClpP) were excised from both the membrane and cytosolic fraction gels, and spot 7 (ribosomal protein L12) was isolated from the membrane fraction and total protein gels. The proteins identified represented each of the four major functional classes. Four of the proteins were cell envelope and cellular process category proteins (class 1), 14 were intermediary metabolism class proteins (class 2), seven were information pathways class proteins (class 3), three were considered to be involved in adaptation to atypical conditions (subclass 4.1), and one was antigen A (miscellaneous function subclass 4.2). Four proteins had no known function in Listeria (Table 1).
Membrane proteins generally contain hydrophobic domains. One measure of average protein hydrophobicity is the GRAVY value, and hydrophobic and hydrophilic proteins have positive and negative GRAVY values, respectively. A total of five proteins had positive GRAVY values. These included the ribosomal protein L12, the cell shape-determining protein MreB, and tetrahydrodipicolinate succinylase (spots 7, 38, and 41, respectively) from the membrane fraction and purine nucleotide phosphorylase, DeoD (spot 57), from the cytosolic fraction. One protein from the total protein gels, spot 8 corresponding to triosephosphate isomerase (Tpi), also had a positive GRAVY value.
The TMpred program predicts the likelihood that a protein traverses a membrane, as well as the most likely orientation of the protein in the membrane. TMDs were detected in 21 of the 33 proteins identified. Of the TMDs detected, the only significant ones were those for four proteins excised from the membrane fraction (spots 3, 7, 38, and 41), for eight proteins from the cytosolic fraction (spots 20, 21, 24, 28, 29, 34, 53, and 57), and for two proteins from the total protein gel (spots 8 and 9). Spot 53 (a homologue of flavocytochrome c fumarate reductase chain A) from the cytosolic fraction had the highest predicted TMD score (2,305). All proteins with a positive GRAVY value also had at least one significant predicted TMD.
Five of the spots sequenced (spots 17, 18, 24, 34, and 50) were unique to the cytosolic fraction; two of these (spots 24 and 34) contained proteins with significant predicted TMDs. In the membrane fraction, spots 16, 36, 37, and 38 were unique, and MreB (spot 38) had a significant TMD and a positive GRAVY value. One of the five proteins with positive GRAVY values, DeoD (spot 57), was enriched in the cytosolic fraction. There was, therefore, no distinct correlation between the GRAVY value or predicted TMD and the fraction.
Proteome reference map of L. monocytogenes EGDe.
Comparison of the membrane and cytosolic fraction gels with total protein gels revealed that all spots found in the fraction gels were present in total protein gels stained with either Coomassie blue or silver. Subsequently, a total protein 2-D reference map of L. monocytogenes EGDe containing the 33 proteins identified was constructed (Fig. 1).
Comparison of food isolates with L. monocytogenes EGDe.
The reference map was used to assess the similarity between L. monocytogenes serotype 1/2a and 1/2b strains isolated from food and the serotype 1/2a animal strain EGDe. A visual comparison indicated that the total protein profiles of the strains isolated from food were very similar to the strain EGDe profile (Fig. 1). The numbers of spots that were not matched in L. monocytogenes EGDe after computer-aided analysis of gels containing total protein samples are shown in Table 2. From four to eight of the major spots in the food strains were not matched in the L. monocytogenes EGDe profile. L. monocytogenes 412 (serotype 1/2a) had the highest number of unmatched major proteins and also lacked one of the proteins identified in L. monocytogenes EGDe (see below). Comparisons of the minor spots resulted in an up-to-10-fold increase in the number of unmatched spots in the L. monocytogenes EGDe profile. Strain O57 (serotype 1/2a) had the highest number of unmatched minor spots. The proteome of strain 386 (serotype 1/2b) did not differ more from the reference strain EGDe proteome than the proteomes of the serotype 1/2a food isolates differed.
TABLE 2.
Comparison of 2-D total protein profiles of L. monocytogenes food isolates and L. monocytogenes EGDe: numbers of major and minor spots in the food isolates that were not matched in strain EGDe (serotype 1/2a)
| L. monocytogenes strain (serotype) | No. of unmatched major spots (%)a | No. of unmatched minor spots (%)b |
|---|---|---|
| B73 (1/2a) | 4 of 57 (7) | 43 of 143 (30) |
| 412 (1/2a) | 8 of 42 (19) | 40 of 158 (25) |
| O57 (1/2a) | 7 of 46 (15) | 52 of 154 (34) |
| 386 (1/2b) | 5 of 54 (9) | 32 of 146 (22) |
Only spots with a minimum spot area of 50 pixels and a minimum spot contrast of 25 were considered major spots.
The minor spots considered were the 200 most significant spots that did not fit the criteria for major spots.
Of the 33 spots identified in the L. monocytogenes EGDe profile, only 2 were not detected in the total protein profiles of all the food isolates analyzed. Spot 18 (GAPDH) was absent from the L. monocytogenes 412 profile, and spot 34 (phosphomethylpyrimidine kinase) was missing from the strain B73, O57, and 386 profiles. The presence of the two corresponding genes was examined by colony PCR by using internal primers covering a substantial part of the reading frame. For each gene, strains B73, 412, O57, and 386 gave PCR products that were the same size as strain EGDe products, showing that all four food strains contained the corresponding chromosomal regions without discernible insertions or deletions.
DISCUSSION
Proteome reference map of L. monocytogenes EGDe.
A partially annotated 2-D map of L. monocytogenes EGDe total protein was constructed from the proteins identified (Fig. 1a and Table 1). This map contains proteins belonging to each of the four major functional classes defined for L. monocytogenes EGDe (14). Only 6 of the 33 proteins identified, Fri (9, 17, 19, 31, 32), GAPDH (8), Pfk (17, 39), Pgm (8), TufA (8), and the mannose-specific phosphotransferase system enzyme IIAB (8, 17, 34), have been identified previously in L. monocytogenes by 2-D analysis.
Previously generated 2-D total protein maps of other microorganisms contained very few if any proteins having an overall hydrophobic amino acid composition (25). In the 2-D reference map presented here, five of the proteins identified had positive GRAVY values, and 42% of the proteins had significant predicted TMDs.
Evaluation of procedures.
Our data analysis showed that our procedure (i.e., total protein isolation, IEF, second-dimension electrophoresis, and spot quantification) had good reproducibility and that with confidence we can consider differences in spot intensity that are greater than fivefold meaningful variations in protein expression.
The L. monocytogenes genome contains 2,853 annotated open reading frames (14). It has been estimated that 30% of the open reading frames in previously sequenced organisms encode transmembrane proteins (30, 41), which corresponds to approximately 850 transmembrane proteins in the L. monocytogenes EGDe proteome. By using an experimental window consisting of molecular masses ranging from 4 to 148 kDa and pIs ranging from 4 to 7, we detected 261 spots in Coomassie blue-stained gels of total protein samples and 208 spots in gels of membrane fraction extracts from L. monocytogenes EGDe. The differences in coding capacities and visualized proteins could be explained by some of the following possibilities: (i) some proteins were not in the experimental window; (ii) certain proteins were insoluble in the IEF sample buffer (37); (iii) Coomassie blue staining was less sensitive; (iv) some proteins were not expressed under the growth conditions employed or at the growth phase at the time of cell harvesting; and (v) membrane proteins are generally low-copy-number proteins so they were not very abundant.
To our knowledge, this is the first report of the use of 2-D gel electrophoresis to specifically assess membrane protein profiles of L. monocytogenes. A modification of one of the first protocols used to visualize membrane proteins (1) was employed, since we performed the first-dimension electrophoresis with immobilized pH gradients, which have yielded highly reproducible protein profiles in different laboratories (5) and provide increased protein loading capacity (33). The compartmentalization procedure was assessed by analyzing the 33 proteins identified (Fig. 3 and Table 1). We compared the GRAVY values and predicted TMDs with the spot intensities in the two fractions; however, no direct correlation was observed. For example, only one of the five proteins with positive GRAVY values (spot 7, ribosomal protein L12) was substantially enriched in the membrane fraction, and ClpP (spot 29), which has two predicted TMDs, was distinctly more intense in the cytosolic fraction than in the membrane fraction (excised and identified from both fractions). However, three of the five proteins with positive GRAVY values were from the membrane fraction. Although our methods only gave an indication of cellular location, since proteins with negative GRAVY values may contain hydrophobic domains and the TMD prediction algorithm is not ideal for prokaryotes (27), the observations suggest that the procedure results in partial but incomplete fractionation that does not clearly reflect the cellular locations of the proteins.
Comparison of food isolates with strain EGDe.
L. monocytogenes EGDe has on several occasions been noticed to be less robust than a number of other strains when the organisms are subjected to stresses, including acid (6) or carbon dioxide (A.-M. Jydegaard-Axelsen and S. Knøchel, unpublished data). Nonetheless, our results indicate that L. monocytogenes EGDe should be a useful reference organism for the study of the proteomes of strains isolated from food. Our results do not, however, permit speculation concerning the cause of the different phenotypes or concerning adaptation to different environments.
We compared both major and minor proteins of three serotype 1/2a L. monocytogenes strains and one serotype 1/2b strain originating from food with the strain EGDe proteins (Table 2). An average of 13% of the major proteins of the food strains were not detected in strain EGDe. When the intensity of the spots being compared was decreased, the average percentage of unmatched spots in strain EGDe increased to 28%.
Of the 33 spots identified in strain EGDe, 2 were missing in one or more of the food isolates. The two proteins were GAPDH involved in the main glycolytic pathway and phosphomethylpyrimidine kinase involved in the metabolism of coenzymes and prosthetic groups. PCR analyses indicated that the two corresponding genes were present in all five strains. Thus, the absence of the spots could have been due to either an extremely low level of expression (below the detection limit of silver-stained gels) or a change in pI or molecular weight that gave rise to a different location on the gel. GAPDH is an essential enzyme in the glycolytic pathway and is expected to be highly expressed in all organisms, even though reduced expression may in some cases be sufficient to sustain normal growth (38). The L. monocytogenes EGDe genome sequence does not contain a reading frame corresponding to an auxiliary GAPDH protein, as is seen in some gram-positive bacteria (13, 42). The GAPDH protein has been reported to exist in several forms with a conserved molecular weight but different pIs (4, 42). Similarly, three forms of GAPDH have been identified by 2-D gel analysis of L. monocytogenes (M. Hebraud, personal communication). In Fig. 1b, arrows marked (i) indicate the locations of the three forms of GAPDH in strain B73, and the rightmost of these arrows indicates spot 18. A comparison of Fig. 1b and c shows that for strain 412 there was increased intensity of the leftmost of the putative GAPDH forms [Fig. 1c, arrow (ii)], corresponding to the missing spot 18. This indicates that strain 412 contains only one form of GAPDH, in contrast to the other strains tested, all of which contain three possible forms of the protein. Functional roles of the different forms of the GAPDH protein have not been determined yet (42).
In our study, we compared serotype 1/2a and 1/2b strains. The variations between serotypes were determined in a previous study (16) to be larger than the variations within serotypes. Only 46.7% of the spots were common among L. monocytogenes strains across serotypes, and serotypes 1/2a and 1/2b were in two different major clusters (16). In our hands, the difference between serotype 1/2b and 1/2a strains was not greater than the variation within serotype 1/2a. Genomic comparison by subtractive hybridization showed that 5% of the genome of a serotype 4b strain did not hybridize to the genome of L. monocytogenes EGD, a variant of strain EGDe and also a serotype 1/2a strain (20). The greater variability observed at the proteomic level can be attributed to the fact that while a single amino acid change in a protein can result in a shift in pI, resulting in a detectable modification in the 2-D pattern (21), the corresponding DNA change should not be registered by hybridization.
By constructing an L. monocytogenes EGDe total protein reference map based on a reproducible protein extraction and 2-D analysis procedure, we established a platform for further study of protein expression in this pathogen. The observed variations indicate that most of the predominant proteins in food isolates can be identified with a degree of confidence from the EGDe map, while identification of less intense spots requires greater caution.
Acknowledgments
We thank Carmen Buchrieser and Philippe Glaser for serotyping L. monocytogenes B73, 412, 386, and O57, Henrik Siegumfeldt for constructing the reference map website, and Lene Gertman for expert technical assistance.
This work was supported by the Food Biotechnology Program of the Danish Ministry for Food, Agriculture, and Fisheries (grant BIOT99-8) and by a grant from The Danish Rectors' Conference and the National Research Foundation (South Africa) to M.R. and J.W.H.
REFERENCES
- 1.Ames, G. F., and K. Nikaido. 1976. Two-dimensional gel electrophoresis of membrane proteins. Biochemistry 15:616-623. [DOI] [PubMed] [Google Scholar]
- 2.Bayles, D. O., B. A. Annous, and B. J. Wilkinson. 1996. Cold stress proteins induced in Listeria monocytogenes in response to temperature downshock and growth at low temperatures. Appl. Environ. Microbiol. 62:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Embarek, P. K., and H. H. Huss. 1993. Heat resistance of Listeria monocytogenes in vacuum packaged pasteurized fish fillets. Int. J. Food Microbiol. 20:85-95. [DOI] [PubMed] [Google Scholar]
- 4.Cash, P., E. Argo, L. Ford, L. Lawrie, and H. McKenzie. 1999. A proteomic analysis of erythromycin resistance in Streptococcus pneumoniae. Electrophoresis 20:2259-2268. [DOI] [PubMed] [Google Scholar]
- 5.Corbett, J. M., M. J. Dunn, A. Posch, and A. Gorg. 1994. Positional reproducibility of protein spots in two-dimensional polyacrylamide gel electrophoresis using immobilised pH gradient isoelectric focusing in the first dimension: an interlaboratory comparison. Electrophoresis 15:1205-1211. [DOI] [PubMed] [Google Scholar]
- 6.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 7.Davis, M. J., P. J. Coote, and C. P. O'Byrne. 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 142:2975-2982. [DOI] [PubMed] [Google Scholar]
- 8.Duche, O., F. Tremoulet, P. Glaser, and J. Labadie. 2002. Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 68:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffes, F., P. Jenoe, and P. Boyaval. 2000. Use of two-dimensional electrophoresis to study differential protein expression in divercin V41-resistant and wild-type strains of Listeria monocytogenes. Appl. Environ. Microbiol. 66:4318-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dykes, G. A., and J. W. Hastings. 1998. Fitness costs associated with class IIa bacteriocin resistance in Listeria monocytogenes B73. Lett. Appl. Microbiol. 26:5-8. [DOI] [PubMed] [Google Scholar]
- 11.Esvan, H., J. Minet, C. Laclie, and M. Cormier. 2000. Protein variations in Listeria monocytogenes exposed to high salinities. Int. J. Food Microbiol. 55:151-155. [DOI] [PubMed] [Google Scholar]
- 12.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fillinger, S., S. Boschi-Muller, S. Azza, E. Dervyn, G. Branlant, and S. Aymerich. 2000. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 275:14031-14037. [DOI] [PubMed] [Google Scholar]
- 14.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 15.Gorg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037-1053. [DOI] [PubMed] [Google Scholar]
- 16.Gormon, T., and L. Phan-Thanh. 1995. Identification and classification of Listeria by two-dimensional protein mapping. Res. Microbiol. 146:143-154. [DOI] [PubMed] [Google Scholar]
- 17.Gravesen, A., M. Ramnath, K. B. Rechinger, N. Andersen, L. Jänsch, Y. Hechard, J. W. Hastings, and S. Knøchel. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148:2361-2369. [DOI] [PubMed] [Google Scholar]
- 18.Gravesen, A., P. Warthoe, S. Knøchel, and K. Thirstrup. 2000. Restriction fragment differential display of pediocin-resistant Listeria monocytogenes 412 mutants shows consistent overexpression of a putative beta-glucoside-specific PTS system. Microbiology 146:1381-1389. [DOI] [PubMed] [Google Scholar]
- 19.Hebraud, M., and J. Guzzo. 2000. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol. Lett. 190:29-34. [DOI] [PubMed] [Google Scholar]
- 20.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a distinct functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jungblut, P. R. 2001. Proteome analysis of bacterial pathogens. Microbes Infect. 3:831-840. [DOI] [PubMed] [Google Scholar]
- 22.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 23.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 24.Lou, Y., and A. E. Yousef. 1997. Adaptation to sublethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl. Environ. Microbiol. 63:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molloy, M. P. 2000. Two-dimensional electrophoresis of membrane proteins using immobilized pH gradients. Anal. Biochem. 280:1-10. [DOI] [PubMed] [Google Scholar]
- 26.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 27.Nouwens, A. S., S. J. Cordwell, M. R. Larsen, M. P. Molloy, M. Gillings, M. D. Willcox, and B. J. Walsh. 2000. Complementing genomics with proteomics: the membrane subproteome of Pseudomonas aeruginosa PAO1. Electrophoresis 21:3797-3809. [DOI] [PubMed] [Google Scholar]
- 28.O'Driscoll, B., C. G. Gahan, and C. Hill. 1997. Two-dimensional polyacrylamide gel electrophoresis analysis of the acid tolerance response in Listeria monocytogenes LO28. Appl. Environ. Microbiol. 63:2679-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsen, I. T., M. K. Sliwinski, and M. H. Saier, Jr. 1998. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 277:573-592. [DOI] [PubMed] [Google Scholar]
- 31.Phan-Thanh, L., and F. Mahouin. 1999. A proteomic approach to study the acid response in Listeria monocytogenes. Electrophoresis 20:2214-2224. [DOI] [PubMed] [Google Scholar]
- 32.Phan-Thanh, L., F. Mahouin, and S. Alige. 2000. Acid responses of Listeria monocytogenes. Int. J. Food Microbiol. 55:121-126. [DOI] [PubMed] [Google Scholar]
- 33.Rabilloud, T., C. Valette, and J. J. Lawrence. 1994. Sample application by in-gel rehydration improves the resolution of two-dimensional electrophoresis with immobilized pH gradients in the first dimension. Electrophoresis 15:1552-1558. [DOI] [PubMed] [Google Scholar]
- 34.Ramnath, M., M. Beukes, K. Tamura, and J. W. Hastings. 2000. Absence of a putative mannose-specific phosphotransferase system enzyme IIAB component in a leucocin A-resistant strain of Listeria monocytogenes, as shown by two-dimensional sodium dodecyl sulfate- polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 66:3098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravishankar, S., M. A. Harrison, and L. Wicker. 2000. Protein profile changes in acid adapted Listeria monocytogenes exhibiting cross-protection against an activated lactoperoxidase system in tryptic soy broth. J. Food Safety 20:27-42. [Google Scholar]
- 36.Rechinger, K. B., H. Siegumfeldt, I. Svendsen, and M. Jakobsen. 2000. “Early” protein synthesis of Lactobacillus delbrueckii ssp. bulgaricus in milk revealed by [35S]methionine labeling and two-dimensional gel electrophoresis. Electrophoresis 21:2660-2669. [DOI] [PubMed] [Google Scholar]
- 37.Santoni, V., M. Molloy, and T. Rabilloud. 2000. Membrane proteins and proteomics: un amour impossible? Electrophoresis 21:1054-1070. [DOI] [PubMed] [Google Scholar]
- 38.Solem, C., B. J. Koebmann, and P. R. Jensen. 2003. Glyceraldehyde-3-phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. J. Bacteriol. 185:1564-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tremoulet, F., O. Duche, A. Namane, B. Martinie, The European Listeria Genome Consortium, and J. C. Labadie. 2002. Comparison of protein patterns of Listeria monocytogenes grown in biofilm or in planktonic mode by proteomic analysis. FEMS Microbiol. Lett. 210:25-31. [DOI] [PubMed] [Google Scholar]
- 40.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wemekamp-Kamphuis, H. H., A. K. Karatzas, J. A. Wouters, and T. Abee. 2002. Enhanced levels of cold shock proteins in Listeria monocytogenes LO28 upon exposure to low temperature and high hydrostatic pressure. Appl. Environ. Microbiol. 68:456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willemoes, M., M. Kilstrup, P. Roepstorff, and K. Hammer. 2002. Proteome analysis of a Lactococcus lactis strain overexpressing gapA suggests that the gene product is an auxiliary glyceraldehyde 3-phosphate dehydrogenase. Proteomics 2:1041-1046. [DOI] [PubMed] [Google Scholar]