Abstract
The microbial diversity occurring in Stilton cheese was evaluated by 16S ribosomal DNA analysis with PCR-denaturing gradient gel electrophoresis. DNA templates for PCR experiments were directly extracted from the cheese as well as bulk cells harvested from a variety of viable-count media. The variable V3 and V4-V5 regions of the 16S genes were analyzed. Closest relatives of Lactococcus lactis, Enterococcus faecalis, Lactobacillus plantarum, Lactobacillus curvatus, Leuconostoc mesenteroides, Staphylococcus equorum, and Staphylococcus sp. were identified by sequencing of the DGGE fragments. Fluorescently labeled oligonucleotide probes were developed to detect Lactococcus lactis, Lactobacillus plantarum, and Leuconostoc mesenteroides in fluorescence in situ hybridization (FISH) experiments, and their specificity for the species occurring in the community of Stilton cheese was checked in FISH experiments carried out with reference cultures. The combined use of these probes and the bacterial probe Eub338 in FISH experiments on Stilton cheese sections allowed the assessment of the spatial distribution of the different microbial species in the dairy matrix. Microbial colonies of bacteria showed a differential location in the different parts of the cheese examined: the core, the veins, and the crust. Lactococci were found in the internal part of the veins as mixed colonies and as single colonies within the core. Lactobacillus plantarum was detected only underneath the surface, while Leuconostoc microcolonies were homogeneously distributed in all parts observed. The combined molecular approach is shown to be useful to simultaneously describe the structure and location of the bacterial flora in cheese. The differential distribution of species found suggests specific ecological reasons for the establishment of sites of actual microbial growth in the cheese, with implications of significance in understanding the ecology of food systems and with the aim of achieving optimization of the fermentation technologies as well as preservation of traditional products.
Stilton is an internally mold-ripened semisoft blue cheese obtained from pasteurized cows' milk; the acidification is carried out by the addition of Lactococcus lactis as a starter culture, while the ripening is promoted by the development of molds (Penicillium roqueforti) as well as yeasts and other bacteria, so that the ripened product has a complex as well as typical microbial composition (24). The molds are usually added as spores at the beginning of the manufacture. After about 6 weeks of ripening, the unpressed curd molds are pierced with stainless steel needles in order to allow air entry. Mold development along the veins is achieved in a 3-week ripening time, giving the typical blue appearance to the cheese. Stilton cheese is granted the status of a protected designation origin product, and the producing dairies are subject to regular audit by an independent inspection agency accredited to European Standard EN 45011. Only cheeses produced in the counties of Nottinghamshire, Leicestershire, and Derbyshire in the United Kingdom can be named Stilton.
The specific characteristics of typical products arise mainly from the specific raw materials employed, the area of production, the environmental conditions, and the traditional tools and manufacture. Further knowledge is needed to understand the relationships between microbiological and biochemical properties which lead to the development of typical textures and flavors. The complex microflora of Stilton cheese is responsible for cheese ripening as well as the typical aroma development. However, no published studies have reported on the structure of the microbial community in Stilton cheese, although this may represent important knowledge for improving process and ripening conditions in order to enhance the quality of the final product while preserving its typical nature.
A few studies have been carried out on the microbial compositions of food matrices by using molecular techniques such as 16S ribosomal DNA (rDNA) PCR-denaturing gradient gel electrophoresis (PCR-DGGE) analysis (7, 8, 13, 38), on the basis that a more reliable image of the community could be thus obtained. However, we previously demonstrated that while these techniques are valuable, there is a need to use a combined system, including cultivation, in order to overcome the bias of the “culture-independent-only” approach (18). Fluorescence in situ hybridization (FISH) with 16S rRNA probes is widely applied in microbial ecology, presently providing microbial identification, physical detection of uncultivable microorganisms (32, 35), and distribution of microbial populations in several environments (4, 5, 31). Despite the considerable background of knowledge, until recently FISH has not been used in food microbiology for the physical location of bacteria in situ in food, although important ecological information on the microflora development in fermented food matrices could be obtained. In a recent study (17), we presented a method for FISH which can be used for fragile matrices like cheese. In the present study a combined molecular approach was used to evaluate the microbial diversity occurring in Stilton cheese via analysis of three 16S rDNA variable regions by PCR-DGGE. FISH was then applied in order to localize the microbial species found in the cheese matrix by using rRNA-labeled oligonucleotide probes developed in this study. We discuss the results, focusing on the suitability of the whole approach for in situ studies on microbial species development in food matrices.
MATERIALS AND METHODS
Microbial strains and culture conditions.
The following strains were used in this study: Lactobacillus plantarum NCDO 1193 (National Collection of Dairy Organisms, Aberdeen, United Kingdom), Lactococcus lactis subsp. lactis NCIMB 8586 (National Collection of Industrial and Marine Bacteria, Aberdeen, United Kingdom), Leuconostoc mesenteroides subsp. mesenteroides NCIMB 10817, Enterococcus faecalis NCTC 775 (National Collection of Type Cultures, Central Public Health Laboratory, London, United Kingdom), Lactobacillus curvatus LTH 1432 (Institut für Lebensmitteltechnologie, Universitat Hohenheim, Stuttgart, Germany), and Staphylococcus equorum DSM 20674 (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). All of the cultures were grown at 30°C. Lactococcus and Enterococcus were grown in M17 broth (Oxoid), Lactobacillus and Leuconostoc were grown in MRS broth (Oxoid), and Staphylococcus was in brain heart infusion broth (Oxoid).
Microbial enumeration and collection of cells in bulk.
Sixteen samples of commercially available Stilton cheese, from different areas of production, were analyzed. The samples were processed immediately after purchase. Serial dilutions of Stilton cheese in quarter-strength Ringer's solution (Oxoid) were used for microbial enumeration with the following media: nutrient agar (Oxoid), MRS agar (Oxoid) for lactic acid bacteria, Rogosa agar (Oxoid) for lactobacilli, M17 agar (Oxoid) for streptococci, and mannitol salt agar (MSA; Oxoid) for staphylococci. Portions (0.1 ml) of appropriate dilutions were spread plated in triplicate. Counts on MRS agar, M17 agar, MSA, and Rogosa agar were obtained after incubation for 48 h at 30°C. Nutrient agar plates were incubated at 10°C for 5 days for the detection of the psychrophilic microflora. Two series of MRS agar plates were inoculated and incubated under aerobic and anaerobic conditions, respectively. Results were calculated as the means of three determinations. After the microbial counts, the plates were used for bulk formation as previously described (19). Bulk cell suspensions (1 ml) from the countable plates for each medium were used for the DNA extraction as described below.
DNA extraction from bulk cells and cheese.
Cheese suspension in 1× phosphate-buffered saline (PBS) (Oxoid) (10−1, 2 ml) or 1 ml of bulk cell suspension was centrifuged at 18,000 × g for 10 min; 500 μl of lysozyme (20 mg ml−1) in TES buffer (50 mM Tris, 1 mM EDTA, 8.7% sucrose)-5 μl of mutanolysin (5 U μl−1)-50 μl of RNase (10 mg ml−1) was added to the pellet, and the mixture was shaken by vortexing for 1 min and then incubated at 37°C. After 1 h of incubation, 50 μl of proteinase K (10 mg ml−1) was added, and the samples were further incubated at 50°C for 50 min and then at 65°C for 10 min. Prewarmed NTS buffer (0.2 M NaCl, 0.1 M Tris, 2% sodium dodecyl sulfate [SDS]) (300 μl) was added, and the samples were incubated at 65°C for 10 min. Successively, 5 M NaCl (300 μl) was added and the samples were maintained at 4°C for 15 min and then centrifuged at 29,000 × g for 15 min at 4°C. The supernatants were precipitated with 0.7 volume of cold isopropanol at −20°C for 30 min and further centrifuged at 29,000 × g for 15 min at 4°C. The pellets were washed with 70% ethanol, resuspended in 500 μl of water, and purified once with phenol (pH 8.0) and twice with chloroform-isoamyl alcohol (24:1), each time with centrifugation at 29,000 × g for 15 min at room temperature. The aqueous phase was finally precipitated with 0.7 volume isopropanol by centrifugation at 29,000 × g for 15 min at 4°C, and the pellet washed with 70% ethanol and resuspended in 50 μl of TE buffer (10 mM Tris, 1 mM EDTA).
PCR amplification.
Two sets of primers were used, amplifying variable regions of 16S rDNA. The primers V3F and V3R amplified the variable V3 region (34), while the primers V4F and V5R amplified the regions V4 and V5 (41), giving PCR products of about 200 and 400 bp, respectively. To the forward primers, a GC clamp was added as described by Muyzer et al. (34). Amplifications were performed in a programmable heating incubator (Techne; Progene). Each mixture (final volume, 25 μl) contained 20 ng of template DNA, each primer at a concentration of 0.2 μM, each deoxynucleoside triphosphate at a concentration of 0.25 mM, 2.5 mM MgCl2, 2.5 μl of 10× PCR buffer (Invitrogen), and 2.5 U of Taq polymerase (Invitrogen). The same conditions were used for both pairs of primers. Template DNA was denatured for 5 min at 94°C. A touchdown PCR was performed as previously described (18). The initial annealing temperature was 66°C, and this was decreased 1°C every cycle for 10 cycles; finally, 20 cycles were performed at 56°C. The extension for each cycle was carried out at 72°C for 3 min, while the final extension was at 72°C for 10 min. Aliquots (2 μl) of PCR products were routinely checked on 2% agarose gels.
DGGE analysis.
PCR products were analyzed by DGGE by using a Bio-Rad Dcode apparatus. Samples of 200 bp were applied to 8% (wt vol−1) polyacrylamide gels, while samples of 400 bp were run in 6.5% (wt vol−1) polyacrylamide gels in 1× TAE buffer. Parallel electrophoresis experiments were performed at 60°C by using gels containing a 30 to 50% urea-formamide denaturing gradient (100% corresponded to 7 M urea and 40% [wt vol−1] formamide). The gels were analyzed by gel electrophoresis for 10 min at 50 V followed by 6 h at 170 V, stained with ethidium bromide for 3 min, and rinsed in distilled water for 15 min. Images were acquired by ImageMaster VDS (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Bands were detected automatically and analyzed with the software ImageMaster 1D (Amersham Pharmacia Biotech).
Sequencing of DGGE fragments.
DGGE bands to be sequenced were purified in water as described by Ampe et al. (7). Eluted DNA (1 μl) of each DGGE band was reamplified by using the appropriate primers and the conditions described above. PCR products that gave a single band comigrating with the original band were then purified with a Qiaex PCR purification kit (Qiagen) and sequenced.
Sequencing was performed by the sequencing unit of the University of Nottingham; a 373 DNA sequencer (Perkin-Elmer Applied Biosystems) was used with the Taq Dye Deoxy terminator cycle sequencing kit (Perkin-Elmer Applied Biosystems). Primers V3R and V5R were used for sequencing of the V3 and V4-V5 fragments, respectively.
To determine the closest known relatives of the partial 16S rDNA sequences obtained, searches were performed in public data libraries (GenBank) with the Blast search program.
Probe design and FISH.
Oligonucleotide rRNA probes were designed from the retrieved sequences, and specificities were checked in the data libraries (GenBank) with the National Center for Biotechnology Information Blast search program. The names of the probes, the labels, the region targeted, and the specificity (this study) are reported in Table 1. The probes were synthesized by Sigma Genosys (Genosys Biotechnologies Ltd., Cambridge, United Kingdom) and were all labeled at the 5′ end.
TABLE 1.
Oligonucleotide probes used in this study
| Standardized probea | Abbreviation | Target microorganism(s) in this study | Sequence | Label | Reference or source |
|---|---|---|---|---|---|
| S-D-Bact-0338-a-A-18 | Eub338 | Eubacteria | 5′-GCT GCC TCC CGT AGG AGT-3′ | Fluorescein | 3 |
| S-D-Bact-0338-a-A-18 | Eub338 | Eubacteria | 5′-GCT GCC TCC CGT AGG AGT-3′ | Cy3 | 3 |
| S-S-L.lact-0821-a-A-18 | LactV5 | Lactococcus lactis | 5′-GCT CCC TAC ATC TAG CAC-3′ | Fluorescein | This study |
| S-S-Lb.pl-0468-a-A-18 | LbpV3 | Lactobacillus plantarum | 5′-CCG TCA ATA CCT GAA CAG-3′ | Cy3 | This study |
| S-S-Lc.ms-0748-a-A-19 | LeucV5 | Leuconostoc mesenteroides | 5′-CCT CCT AAC ACC TAG TGT T-3′ | Cy3 | This study |
As described by Alm et al. (2).
Microbial culture fixation was performed from 300 μl of overnight culture by using 4% paraformaldehyde (3). About 20 μl of the fixed cell suspension in PBS was spotted on a poly-l-lysine-coated slide, dried in an oven at 46°C for 10 min, and dehydrated successively in 50, 80, and 100% ethanol solutions for 3 min each. Once the ethanol had evaporated, the specimens were used for the hybridization.
Stilton cheese sections were obtained from different zones of the matrix; particularly, the surface, the veins, and the core were taken into account. Embedding of the specimen in cold polymerizing glycol methylacrylate resin was carried out as previously described (17).
Pretreatment of all specimens with proteinase K (10 mg ml−1; Sigma) was performed at 37°C for 10 min, followed by a washing step with ice-cold 1× PBS (Oxoid). The hybridization buffer (25% formamide, 0.9 M NaCl, 0.01% SDS, 20 mM Tris-HCl [pH 7.2]) (30 μl) containing 10 ng of 16S rRNA probe (Genosys Biotechnologies) was spotted onto the dry specimen, and the slides were incubated in a dark humid chamber at 42°C for 14 h (3 h for the cultures); a washing step in a prewarmed washing buffer (20 mM Tris-HCl [pH 7.2], 0.01% SDS, 40 mM NaCl, 5 mM EDTA) was then performed at 42°C for 15 min. When more than one probe was used for the same specimen, they were mixed in the hybridization buffer at equal amounts up to 10 ng.
Microscopy.
Microscopic observations were carried out with a Zeiss Axiovert 135 fluorescence microscope, with a TILL photonics monochromator for excitation, a 4′,6′-diamidino-2-phenylindole-fluorescein isothiocyanate-tetramethyl rhodamine isocyanate multiband filter set for viewing, and single-pass filters mounted in a biopoint filter wheel for image capture with a cooled charge-coupled device camera (ORCA 2; Hamamatsu, Welwyn Garden City, United Kingdom). Openlab software (Improvision) was used for image analysis. About 250 sections arising from the different zones of the cheese were analyzed. At least 40 fields were observed for each specimen at a magnification of ×100.
Nucleotide sequence accession numbers.
The GenBank accession numbers for 16S rDNA partial sequences retrieved from DGGE bands are given in Tables 2 and 3.
TABLE 2.
Identities of bands obtained from the bacterial community of Stilton cheese as a result of DGGE analysis of the V3 region
| Banda | Origin | Closest relative | % Identity | GenBank accession no. |
|---|---|---|---|---|
| 1 | Stilton fingerprintb | Lactococcus lactis | 99 | AF498048 |
| 2 | Stilton fingerprint | E. faecalis | 96 | AF498046 |
| 3 | Stilton fingerprint | Lactobacillus curvatus | 100 | AF498052 |
| 4 | Stilton fingerprint | S. equorum | 99 | AF498053 |
| 6 | Stilton fingerprint | Lactobacillus plantarum | 100 | AF498059 |
| 2 | Bulk cells on M17 agar (dilution, 10−7) | E. faecalis | 96 | AF498047 |
| 3 | Bulk cells on M17 agar (dilution, 10−7) | Staphylococcus sp. | 99 | AF498064 |
| 4 | Bulk cells on M17 agar (dilution, 10−7) | S. equorum | 99 | AF498054 |
| 2 | Bulk cells on MRS agar (dilution, 10−7) | E. faecalis | 98 | AF498049 |
| 3 | Bulk cells on MRS agar (dilution, 10−7) | Staphylococcus sp. | 99 | AF498063 |
| 4 | Bulk cells on MRS agar (dilution, 10−7) | S. equorum | 100 | AF498055 |
| 6 | Bulk cells on MRS agar (dilution, 10−7) | Lactobacillus plantarum | 100 | AF498060 |
| 4 | Bulk cells on anaerobic MRS agar (dilution, 10−7) | S. equorum | 100 | AF498056 |
| 6 | Bulk cells on anaerobic MRS agar (dilution, 10−7) | Lactobacillus plantarum | 100 | AF498061 |
| 3 | Bulk cells on Rogosa agar (dilution, 10−6) | Lactobacillus curvatus | 100 | AF502284 |
| 6 | Bulk cells on Rogosa agar (dilution, 10−6) | Lactobacillus plantarum | 100 | AF498062 |
| 2 | Bulk cells on MSA (dilution, 10−6) | E. faecalis | 98 | AF498050 |
| 4 | Bulk cells on MSA (dilution, 10−6) | S. equorum | 100 | AF498057 |
| 2 | Bulk cells on nutrient agar (dilution, 10−6) | E. faecalis | 98 | AF498051 |
| 4 | Bulk cells on nutrient agar (dilution, 10−6) | S. equorum | 100 | AF498058 |
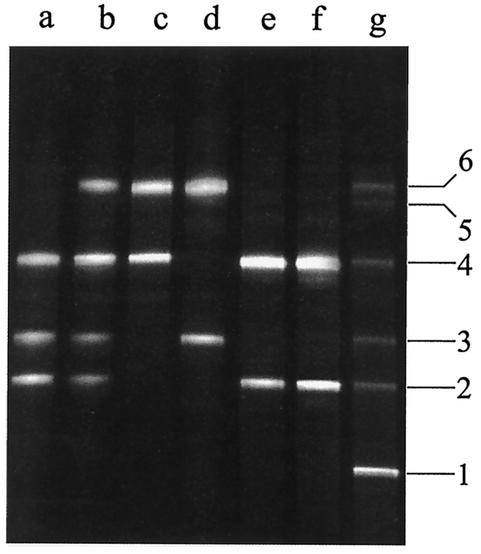
The numbers of the bands refer to those in Fig. 1.
DGGE pattern obtained after PCR amplification of DNA directly extracted from Stilton cheese.
TABLE 3.
Identities of bands obtained from the bacterial community of Stilton cheese as a result of DGGE analysis of the V4-V5 region
| Banda | Origin | Closest relative | % Identity | GenBank accession no. |
|---|---|---|---|---|
| 1 | Stilton fingerprintb | Lactococcus lactis | 99 | AF498065 |
| 3 | Stilton fingerprint | Staphylococcus sp. | 99 | AF498066 |
| 4 | Stilton fingerprint | S. equorum | 100 | AF498069 |
| 6 | Stilton fingerprint | Leuconostoc mesenteroides | 99 | AF498080 |
| 2 | Bulk cells on M17 agar (dilution, 10−7) | E. faecalis | 100 | AF498081 |
| 3 | Bulk cells on M17 agar (dilution, 10−7) | Staphylococcus sp. | 99 | AF498067 |
| 4 | Bulk cells on M17 agar (dilution, 10−7) | S. equorum | 100 | AF498073 |
| 6 | Bulk cells on M17 agar (dilution, 10−7) | Leuconostoc mesenteroides | 99 | AF498079 |
| 3 | Bulk cells on MRS agar (dilution, 10−7) | Staphylococcus sp. | 99 | AF498068 |
| 4 | Bulk cells on MRS agar (dilution, 10−7) | S. equorum | 100 | AF498072 |
| 6 | Bulk cells on MRS agar (dilution, 10−7) | Leuconostoc mesenteroides | 100 | AF498076 |
| 6 | Bulk cells on anaerobic MRS agar (dilution, 10−7) | Leuconostoc mesenteroides | 100 | AF498077 |
| 5 | Bulk cells on Rogosa agar (dilution, 10−6) | Lactobacillus plantarum | 100 | AF498075 |
| 4 | Bulk cells on MSA (dilution, 10−6) | S. equorum | 100 | AF498070 |
| 4 | Bulk cells on nutrient agar (dilution, 10−6) | S. equorum | 100 | AF498070 |
| 6 | Bulk cells on nutrient agar (dilution, 10−6) | Leuconostoc mesenteroides | 100 | AF498078 |
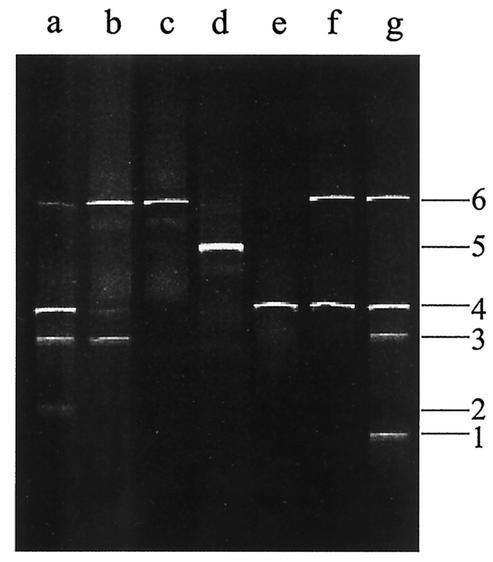
The numbers of the bands refer to those in Fig. 2.
DGGE pattern obtained after PCR amplification of DNA directly extracted from Stilton cheese.
RESULTS
Enumeration of microorganisms.
The analysis was performed on one sample of Stilton cheese chosen among the 16 previously screened (see below). The viable counts from the selective and nonselective agars are shown in Table 4. The numbers of each microbial group targeted appeared to be quite similar, with values of about 108 CFU g−1. Counts on Rogosa agar were 1 log unit lower than the other lactic flora counts. Interestingly, the psychrophilic bacterial counts were about 108 CFU g−1, indicating that a predominant section of the flora could grow at low temperatures.
TABLE 4.
Viable counts of bacterial groups for Stilton cheese
| Medium | Incubation temp (°C) | Group targeted | Log CFU g−1a (SD) |
|---|---|---|---|
| M17 agar | 30 | Mesophilic streptococci | 8.97 (0.22) |
| MRS agar | 30 | Mesophilic lactic acid bacteria | 8.87 (0.25) |
| MRS agar in anaerobiosis | 30 | Mesophilic anaerobic lactic acid bacteria | 8.85 (0.33) |
| Rogosa agar | 30 | Lactobacilli | 7.76 (0.21) |
| MSA | 30 | Staphylococci | 8.04 (0.04) |
| Nutrient agar | 10 | Psychrophilic microflora | 8.14 (0.32) |
The data are the means based on three replicates.
PCR-DGGE analysis.
The 16 samples of Stilton cheese were initially screened by analyzing the DGGE profiles shown after direct DNA extraction and PCR amplification of the V3 region of the 16S rDNA. The analysis of the V3 region by DGGE was previously shown to be useful to discriminate commercial as well as artisanal dairy products (14, 16). The variability in microbial composition, assessed by number of bands and migration position in DGGE gels, was low among the 16 Stilton cheese samples analyzed (data not shown). One sample of Stilton cheese was chosen as representative and used for the further analysis.
16S rDNA fingerprints were obtained by PCR-DGGE analysis of the variable regions V3 and V4-V5 performed on DNA extracted directly from Stilton cheese as well as from bulk cells collected on different media after the microbial counts.
The V3 fingerprint of the Stilton cheese flora obtained from the DNA directly extracted from the cheese was composed of six bands (Fig. 1, lane g). Unfortunately, purification and sequencing of band 5 did not prove successful. All of the sequences retrieved corresponded to portions of 16S rDNA of Bacteria. The closest relatives, the percent identities, and the accession numbers of the sequences analyzed are reported in Table 2. As a result of the sequencing of the V3 region fragments, the Stilton cheese flora was found to consist of closest relatives of Lactococcus lactis (Fig. 1, lane g, band 1), Enterococcus faecalis (band 2), Lactobacillus curvatus (band 3), S. equorum (band 4), and Lactobacillus plantarum (band 6). The same procedure was followed with the primers amplifying the region V4-V5. The profiles obtained are shown in Fig. 2, while the results for the sequences are reported in Table 3. The profile obtained by the direct analysis of the Stilton cheese consisted of four different bands (Fig. 2, lane g) giving closest relatives of Lactococcus lactis (band 1), Staphylococcus sp. (band 3), S. equorum (band 4), and Leuconostoc mesenteroides (band 6). The sequence analysis was extended to the profiles obtained from PCR-DGGE analysis of the DNA extracted from bulk cells from the countable plates. In Fig. 1 the V3 profiles of the bulk cells from the countable plates of different media are also reported. By analysis of the V3 region, closest relatives of E. faecalis, Staphylococcus sp., and S. equorum (listed as appearing in the gel from the bottom to the top) were found on M17 agar (Fig. 1, lane a); E. faecalis, Staphylococcus sp., S. equorum, and Lactobacillus plantarum were found on MRS agar (Fig. 1, lane b); S. equorum and Lactobacillus plantarum were found on MRS agar when incubated anaerobically (Fig. 1, lane c); and Lactobacillus curvatus and Lactobacillus plantarum were found on Rogosa agar (Fig. 1, lane d). The bulk cells collected from MSA and nutrient agar showed the same profile, consisting of E. faecalis and S. equorum (Fig. 1, lanes e and f). As shown in Fig. 1, comigration of two different species (Staphylococcus sp. and Lactobacillus curvatus) was observed by the correspondence of band 3. The analysis of bulk cells was also repeated for the V4-V5 region to identify the species occurring on the countable plates after cultivation (Fig. 2, lanes a to f). 16S rDNA sequences giving closest relatives of E. faecalis, Staphylococcus sp., S. equorum, and Leuconostoc mesenteroides were retrieved from M17 agar countable plates (Fig. 2, lane a); Staphylococcus sp., S. equorum, and Leuconostoc mesenteroides were found on MRS agar plates (Fig. 2, lane b); only Leuconostoc mesenteroides was found on MRS agar when incubated anaerobically (Fig. 2, lane c); Lactobacillus plantarum was found on Rogosa agar (Fig. 2, lane d); S. equorum was found on MSA (Fig. 2, lane e); and S. equorum and Leuconostoc mesenteroides were found on nutrient agar plates (Fig. 2, lane f).
FIG. 1.
Variable V3 region DGGE profiles of Stilton cheese and bulk cells. Lanes: a, M17 agar (10−7); b, MRS agar (10−7); c, MRS agar (anaerobiosis, 10−7); d, Rogosa agar (10−6); e, MSA (10−6); f, nutrient agar (10−6); g, Stilton cheese after direct DNA extraction. Bands: 1, Lactococcus lactis; 2, E. faecalis; 3, Lactobacillus curvatus-Staphylococcus sp.; 4, S. equorum; 5, unidentified; 6, Lactobacillus plantarum.
FIG. 2.
Variable V4-V5 region DGGE profiles of Stilton cheese and bulk cells. Lanes: a, M17 agar (10−7); b, MRS agar (10−7); c, MRS agar (anaerobiosis, 10−7); d, Rogosa agar (10−6); e, MSA (10−6); f, nutrient agar (10−6); g, Stilton cheese after direct DNA extraction. Bands: 1, Lactococcus lactis; 2, E. faecalis; 3, Staphylococcus sp.; 4, S. equorum; 5, Lactobacillus plantarum; 6, Leuconostoc mesenteroides.
FISH.
The suitability of the probes retrieved from the sequencing of DGGE fragments was checked by FISH experiments performed with culture collection strains. The conditions described in Materials and Methods were found to be the optimum, providing a good yield in fluorescence as well as specificity for the species of interest. The Eub338 probe gave hybridization with all of the cultures assayed. The LactV5 probe hybridized with Lactococcus lactis only, the LeucV5 probe hybridized with Leuconostoc only, and the LbpV3 probe hybridized with Lactobacillus plantarum only. None of the newly designed probes hybridized with E. faecalis, Lactobacillus curvatus, and S. equorum or showed cross-reactions in FISH.
Bacterial community location in Stilton cheese.
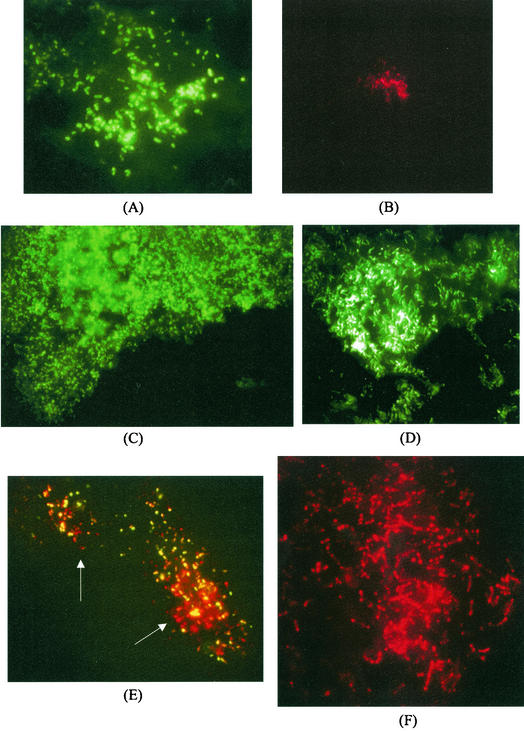
The developed probes and the bacterial probe were used in FISH experiments with embedded cheese sections in order to locate the different microbial species within the food matrix. Sections of Stilton cheese arising from different parts of the cheese mold, i.e., the core, the veins, and the crust, were examined. The bacterial probe Eub338 was used for the detection of the microbial colonies; cells were identified simultaneously by using the specific probes. The sections arising from the core had a simple composition in microbial species: hybridization reactions with the Eub338 probe and LactV5 probe showed Lactococcus lactis colonies to be still present and representing 70% of the colonies detected. Fifteen percent of the colonies hybridized with Leuconostoc probe LeucV5, while the rest of the colonies remained unidentified, although all of them were composed of coccus-like microorganisms. The colonies of Lactococcus and Leuconostoc found are depicted in Fig. 3A and B, respectively. An appreciable difference in microcolony sizes was noticed: the Leuconostoc formed small confined microcolonies, while Lactococcus colonies were large and spreading, often occupying more than one field of observation. The composition of the microflora across the veins and the surface was more complex. Extensive colonies of cocci, often extending over 5 to 10 fields of observation, were detected with the bacterial probe; however, none of the specific probes developed gave a positive hybridization result. As shown in Fig. 3C, these large colonies were found lying along the margin of the vein, almost producing a border. In addition, large colonies of rods (Fig. 3D) could be detected in the neighborhood of the veins; however, they did not hybridize with the LbpV3 probe for Lactobacillus plantarum, suggesting that these may be microcolonies of Lactobacillus curvatus, the other Lactobacillus species found. Leuconostoc microcolonies were found in the most internal part of the vein. A few Lactococcus lactis microcolonies were also found; however, unlike in the core, they were in most cases mixed with other unidentified coccus-shaped microorganisms (Fig. 3E). The surface presented an external layer of large yeast-like cells not hybridizing with the bacterial probe but detectable when excited at 488 nm because of a weak signal from autofluorescence (results not shown). The external part was also characterized by the presence of large colonies of unidentified cocci similar to the ones found along the veins, although less extended in size. Just under the crust, different microbial species could be detected. Lactobacillus plantarum microcolonies were frequently and randomly found; they were usually large and concentrated rather than spreading (Fig. 3F). Leuconostoc microcolonies were also found, as were microcolonies of unidentified rods not hybridizing with the LbpV3 probe. Moreover, none of the coccus-like colonies found under the crust were identified as Lactococcus lactis by FISH.
FIG.3.
FISH of Stilton cheese section. (A) Lactococcus lactis microcolony from the core detected by using probe LactV5 labeled with fluorescein. (B) Leuconostoc mesenteroides microcolony from the core detected by probe LeucV5 labeled with Cy3. (C) Microcolony of cocci along the vein detected by probe Eub338 labeled with fluorescein. (D) Colony of rods underneath the veins detected by probe Eub338 labeled with fluorescein. (E) Mixed microcolony of Lactococcus lactis and other bacteria from the veins simultaneously detected by hybridization with probes LactV5 labeled with fluorescein and Eub338 labeled with Cy3. The arrows indicate the locations of nonlactococci. (F) Microcolony of Lactobacillus plantarum underneath the crust detected by using probe LbpV3 labeled with Cy3.
DISCUSSION
A combined molecular approach was applied in this study; the goal was to obtain information about the complex bacterial flora that develops in Stilton cheese and to examine the spatial distribution of the species found within the cheese matrix.
Microbiological counts showed a significant presence of all of the microbial groups targeted; however, the 16S rDNA analysis of the colonies in bulk cells from cultured plates revealed that only a small number of species was actually recovered from the plates. The identification of the microbial species occurring in the Stilton cheese was performed by sequencing the DGGE fragments belonging to regions V3 and V4-V5 of the 16S rDNA. The structure of the microbial community was shown to be complex. Stilton cheese is made from pasteurized milk and is thus supposed to be free of undesirable bacteria; pasteurization was introduced in 1989 as a result of a food poisoning outbreak in England associated with the consumption of Stilton cheese (26). Lactococcus lactis is commonly used as a starter in the manufacture in order to provide rapid acidification and curd production. A closest relative of Lactococcus lactis was detected in the community profile of the Stilton cheese from analyzing both the V3 and the V4-V5 regions of the 16S rDNA. However, the fragment corresponding to Lactococcus lactis was never found in the profiles retrieved from the culture media. The analysis of 16S rDNA from bulk cells of dilutions of 10−5 (plates outside the countable range) revealed the presence of a band comigrating with the Lactococcus fragment (results not shown). If only a traditional approach had been used, no lactococci would have been isolated from the plates, because their colonies were present not on the plates within the countable range but on the confluent plates, which are not usually taken into account for the isolation and enumeration of bacteria.
Considerable amounts of nonstarter lactic acid bacteria were also found. Closest relatives of Lactobacillus plantarum and Lactobacillus curvatus were detected; however, Lactobacillus curvatus was detected by amplifying the V3 region only. Both species usually occur in dairy ecosystems: Lactobacillus plantarum is recognized as contributing to flavor development (1, 6) and as a protective antimicrobial agent in cheese (15, 44), while Lactobacillus curvatus has been associated with undesirable biofilm development in the dairy environment (42, 51). Leuconostoc mesenteroides was also detected, but only by the amplification of the V4-V5 region; it is often found in food matrices and in many cheeses (12, 29, 37, 49). Interestingly, Leuconostoc mesenteroides subsp. cremoris has been shown to stimulate the growth of P. roqueforti, the mold used in Stilton ripening (23). The organism was detected from M17, MRS, and nutrient agar plates at the highest dilution of 10−7, suggesting a considerable contribution to the complex ecosystem. E. faecalis is often isolated from dairy samples (9, 19, 29, 30); its role in this environment is not clearly defined, although it has been suggested as an adjunct culture in cheese production (36).
S. equorum was recovered from multiple culture plates and thus shown to be one of the predominant species in Stilton cheese. It has been previously found in milk and cheese (28), and its physiological properties would suggest a further contribution to cheese ripening (11, 43). Finally, closest relatives of Staphylococcus sp. were also found; however, identification at the species level could not be achieved, probably due to the low level of variability of the regions of 16S rDNA considered among species of staphylococci.
The results obtained by analyzing the Stilton cheese on the basis of the sequence variability of regions V3 and V4-V5 were remarkably different. Analysis of the V4-V5 region from the DNA directly extracted from the cheese did not allow Lactobacillus plantarum and Lactobacillus curvatus to be detected, while both amplicons were picked up in the V3 region profile arising from the same DNA template. However, Lactobacillus plantarum was detected as a unique band in the V4-V5 bulk cell profile from Rogosa agar. On the other hand, Leuconostoc mesenteroides was detectable only by analyzing the V4-V5 region. None of the same templates gave detectable Leuconostoc fragments after the amplification of the V3 region. In addition, E. faecalis was undetected in the V4-V5 profile of Stilton cheese, and Staphylococcus sp. was undetected in the V3 profile of the cheese, although both were recovered from the bulk cell profiles of the respective regions. The templates and the amounts of DNA used in the PCRs were exactly the same for the analysis of both the V3 and V4-V5 regions; however, the community structures obtained were different. The inconsistency of the results is probably due to preferential amplification from the two pairs of primers used. Selective amplification has been assessed previously (19, 33, 39, 40, 47, 50), and the results obtained in this study suggest that a more careful interpretation of data obtained by analyzing 16S regions of mixed bacterial populations is needed.
The identification of the microbial species from DGGE profiles of bulk cells from the countable plates (19) has been shown to be useful and rapid. This approach allowed the absence of Lactococcus lactis on the countable plates to be appreciated immediately. The analysis of the bulk cells also highlighted the occurrence of Staphylococcus sp. in the analysis of the V3 region and of Lactobacillus plantarum and E. faecalis in the analysis of the V4-V5 region, while these species were undetected in the corresponding Stilton cheese profiles.
It is noteworthy, however, that some of the media were not very selective. Staphylococci were found on M17 and MRS agars, enterococci occurred on MSA, and leuconostocs were found on M17 agar plates. The lack of selectivity of culture media to study complex environmental communities from food has been shown previously (7, 19, 38).
The other important goal of this study was to examine the spatial distribution of the bacteria within the cheese matrix. This was achieved by means of FISH experiments performed on embedded cheese sections by using the universal probe Eub338 (3) in combination with other 16S rRNA probes developed in this study.
The specific oligonucleotide probes were designed from the sequences retrieved from the DGGE analysis and targeted regions of the 16S rRNAs of Lactococcus lactis, Leuconostoc mesenteroides, and Lactobacillus plantarum. These probes were not intended to be specific beyond the environment of study, and the specificities were checked only among the species occurring in the cheese. The aim was not to develop absolutely specific probes but only to develop probes capable of distinguishing, and then locating, some of the species detected in this study in Stilton cheese. Indeed, the probes have been developed from DGGE-retrieved sequences, which guaranteed a significant difference in base pair composition of the fragments, since they could be separated according to their sequence in DGGE. Other probes from 16S rRNA have been developed for the identification of lactic acid bacteria (25); however, their suitability for in situ hybridization experiments with 16S rRNA has never been demonstrated. The suitability of 16S rRNA probes for FISH is related to the probe accessibility to 16S rRNA in its natural conformation (21, 22).
The combined use of the universal Eub338 probe and the specific probes in FISH experiments on Stilton cheese sections showed a differential spatial distribution of the bacterial flora within the dairy matrix. A significant difference between the core and the rest of the cheese was detected. The colony density in the core was about fivefold lower than that in the surface and the veins, although in the microenvironment in the latter two, bacteria were outcompeted by the mold development. In the core, most of the microcolonies were Lactococcus lactis, even though a considerable amount of Leuconostoc mesenteroides was also detected. Lactococci were detected by direct DNA isolation from cheese and PCR-DGGE analysis. Interestingly, despite the fact that lactococci were undetected from the DGGE bulk cell profiles from the M17 agar plates, their colonies in the matrix were still detectable by FISH. The Lactococcus cells did not look damaged and yielded positive FISH reactions. Failure to recover the species in high numbers on selective agars does, however, suggest some measure of metabolic impairment of the cells.
Rod-shaped bacteria were not found in the core, but conspicuous amounts of Lactobacillus plantarum and, presumptively, Lactobacillus curvatus were found underneath the crust. Across the veins, moreover, a few microcolonies of Lactobacillus plantarum could be observed, while microcolonies of shorter rods resembling Lactobacillus curvatus were much more abundant. On the surface, and much more along the internal margin of the veins, very large colonies of non-Lactococcus cocci were detected; we suppose these to be staphylococci even though identification has not been achieved. The staphylococci would grow better in this part of the cheese, where more oxygen is available, since they are facultative anaerobes. Moreover, the pH in these zones is thought to be high due to mold development, and the staphylococci are not as acid tolerant as lactic acid bacteria. The colonies of cocci along the veins might also be brought in while the cheese is pierced with the needle, but the presence of the lactobacilli in the internal part of the veins and underneath the surface of the cheese suggests that they arise from the milk. If so, it is interesting to speculate why they did not develop in the core of the cheese and only towards the edges. This is probably due to ecological reasons: they may find optimal growth conditions in the presence of other bacterial entities and molds releasing metabolites useful for their own nutrition. The lactococci are used as a starter and are thought to be homogeneously distributed in the cheese; their occurrence in the parts of the cheese that are least rich in bacteria only suggests that they have outcompeted the other bacteria at this site. This probably represents the community dynamic prior to piercing. After air is introduced, the growth of the molds produces a changed environment which allows a new equilibrium to develop. Interestingly, a few microcolonies of lactococci were found in the vein sections, and they were mixed with other cocci, indicating a possible commensalism in that part of the dairy matrix. The differential spatial distribution of the microbial species in the cheese has a meaningful impact on knowledge in dairy science. The location of the different species in different zones of the matrix certainly implies differential utilization of the dairy nutrients, consequently producing an unequal distribution of flavor and antimicrobial compounds in the cheese. This in turn would affect local flora development and may in part explain the differential distribution of species seen. When poor-quality products arise, it may be due to the lack of development of one or more of these microenvironments.
Other studies have focused on monitoring the spatial distribution of the bacterial flora in food, but they were either PCR dependent (7, 20) or based on antibody-mediated detection of the bacteria in situ (45, 46). Scanning electron microscopy, as well as fluorescence and light microscopy, have been used to examine microbial diversity in cheese (27, 52), although these techniques provide only nonspecific information. Recently, confocal microscopy has been also suggested as a tool for studying the viability of bacteria in situ in food (10, 48). However, 16S rRNA-FISH analysis has the advantage of potentially providing both specific and nonspecific detection in a single step. Furthermore, the probe target in FISH is rRNA, which is thought to be far more available in cells than specific genomic DNA fragments. The results of this study are interesting not only for the contribution to the knowledge on the microflora of the Stilton cheese but also because the working application of the whole approach may represent a tool of utmost importance in ecological studies looking at specific microflora development in cheese. Moreover, this kind of approach can play an important role in the quality control and preservation of artisanal as well as industrial fermented foods.
Acknowledgments
This research was supported by the European Commission under the Fifth Framework Program (FP5) involving the University of Nottingham as a Marie Curie Training Site (contract no. QLK1-CT-2000-60022).
Danilo Ercolini thanks Salvatore Coppola for support and encouragement. Strains NCDO 1193, LTH 1432, and DSM 20674 were provided by Dipartimento di Scienza degli Alimenti, Universita' “Federico II,” Naples, Italy.
REFERENCES
- 1.Albenzio, M., M. R. Corbo, S. U. Rehman, P. F. Fox, M. De Angelis, A. Corsetti, A. Sevi, and M. Gobetti. 2001. Microbiological and biochemical characteristics of Canestrato Pugliese cheese made from raw milk, pasteurized milk or by heating the curd in hot whey. Int. J. Food Microbiol. 67:35-48. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231-236. [DOI] [PubMed] [Google Scholar]
- 6.Amarita, F., T. Requena, G. Taborda, L. Amigo, and C. Pelaez. 2001. Lactobacillus casei and Lactobacillus plantarum initiate catabolism of methionine transamination. J. Appl. Microbiol. 90:971-978. [DOI] [PubMed] [Google Scholar]
- 7.Ampe, F., N. ben Omar, C. Moizan, C. Wacher, and J.-P. Guyot. 1999. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 65:5464-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ampe, F., A. Sirvent, and N. Zakhia. 2001. Dynamics of the microbial community responsible for traditional sour cassava starch fermentation studied by denaturing gradient gel electrophoresis and quantitative rRNA hybridization. Int. J. Food Microbiol. 65:45-54. [DOI] [PubMed] [Google Scholar]
- 9.Arizcun, C., Y. Barcina, and P. Torre. 1997. Identification and characterization of proteolytic activity of Enterococcus spp. isolated from milk and Roncal and Idiazabal cheese. Int. J. Food Microbiol. 38:17-24. [DOI] [PubMed] [Google Scholar]
- 10.Auty, M. A. E., G. E. Gardiner, S. J. McBrearty, E. O. O'Sullivan, D. M. Mulvihill, J. K. Collins, G. F. Fitzgerald, C. Stanton, and R. P. Ross. 2001. Direct in situ viability assessment of bacteria in probiotic dairy products using viability staining in conjunction with confocal scanning laser microscopy. Appl. Environ. Microbiol. 67:420-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnarme, P., C. Lapadatescu, M. Yvon, and H. E. Spinnler. 2001. l-Methionine degradation potentialities of cheese-ripening microorganisms. J. Dairy Res. 68:663-674. [DOI] [PubMed] [Google Scholar]
- 12.Cibik, R., E. Lepage, and P. Talliez. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional French cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267-278. [DOI] [PubMed] [Google Scholar]
- 13.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppola, S., G. Blaiotta, D. Ercolini, and G. Moschetti. 2001. Molecular evaluation of microbial diversity occurring in different types of Mozzarella cheese. J. Appl. Microbiol. 90:414-420. [DOI] [PubMed] [Google Scholar]
- 15.Ennahar, S., O. Assobhel, and C. Hasselmann. 1998. Inhibition of Listeria monocytogenes in a smear-surface soft cheese by Lactobacillus plantarum WHE 92, a pediocin AcH producer. J. Food Prot. 61:186-191. [DOI] [PubMed] [Google Scholar]
- 16.Ercolini, D., G. Blaiotta, G. Moschetti, and S. Coppola. 2002. Molecular typing of cheeses on the basis of their microflora as detected by PCR-DGGE analysis. Ann. Microbiol. 52:81-87. [Google Scholar]
- 17.Ercolini, D., P. J. Hill, and C. E. R. Dodd. 2002. Development of a fluorescence in situ hybridisation method for cheese using a 16S rRNA probe. J. Microbiol. Methods 52:267-271. [DOI] [PubMed] [Google Scholar]
- 18.Ercolini, D., G. Moschetti, G. Blaiotta, and S. Coppola. 2001. Behavior of variable V3 region from 16S rDNA of important lactic acid bacteria in denaturing gradient gel electrophoresis. Curr. Microbiol. 42:199-202. [DOI] [PubMed] [Google Scholar]
- 19.Ercolini, D., G. Moschetti, G. Blaiotta, and S. Coppola. 2001. The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo Mozzarella cheese production: bias of “culture dependent” and “culture independent” approaches. Syst. Appl. Microbiol. 24:610-617. [DOI] [PubMed] [Google Scholar]
- 20.Fitzsimons, N. A., T. M. Cogan, S. Condon, and T. Beresford. 2001. Spatial and temporal distribution of non-starter lactic acid bacteria in Cheddar cheese. J. Appl. Microbiol. 90:600-608. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs, B. M., F. O. Glockner, J. Wulf, and R. Amann. 2000. Unlabeled oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, T. K., and M. Jacobsen. 1997. Possible role of microbial interactions for growth and sporulation of P. roqueforti in Danablu. Lait 77:479-488. [Google Scholar]
- 24.Johnson, M. E. 2001. Cheese products, p. 345-384. In E. H. Marth and J. L. Steele (ed.), Applied dairy microbiology, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 25.Klijn, N., A. H. Weerkamp, and W. M. de Vos. 1991. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl. Environ. Microbiol. 57:390-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maguire, H. C., M. Boyle, M. J. Lewis, J. Pankhurst, A. A. Wieneke, M. Jacob, J. Bruce, and M. O'Mahony. 1991. A large outbreak of food poisoning of unknown aetiology associated with Stilton cheese. Epidemiol. Infect. 106:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcellino, N., and D. R. Benson. 1992. Scanning electron and light microscopy study of microbial succession on Bethlehem St. Nectaire cheese. Appl. Environ. Microbiol. 58:3448-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meugnier, H., M. Bes, C. Vernozy-Rozand, C. Mazuy, Y. Brun, J. Freney, and J. Fleurette. 1996. Identification and ribotyping of Staphylococcus xylosus and Staphylococcus equorum strains isolated from goat milk and cheese. Int. J. Food Microbiol. 31:325-331. [DOI] [PubMed] [Google Scholar]
- 29.Morea, M., F. Baruzzi, and P. S. Cocconcelli. 1999. Molecular and physiological characterization of dominant bacterial populations in traditional mozzarella cheese processing. J. Appl. Microbiol. 87:574-582. [DOI] [PubMed] [Google Scholar]
- 30.Moschetti, G., G. Blaiotta, M. Aponte, P. Catzeddu, F. Villani, P. Deiana, and S. Coppola. 1998. Random amplified polymorphic DNA and amplified ribosomal DNA spacer polymorphism: powerful methods to differentiate Streptococcus thermophilus strains. J. Appl. Microbiol. 85:25-36. [DOI] [PubMed] [Google Scholar]
- 31.Moter, A., and U. B. Gobel. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 41:85-112. [DOI] [PubMed] [Google Scholar]
- 32.Moter, A., G. Leist, R. Rudolph, K. Schrank, B.-K. Choi, M. Wagner, and U. B. Gobel. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144:2459-2467. [DOI] [PubMed] [Google Scholar]
- 33.Muyzer, G. 1999. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen, A. T., W-T. Liu, C. Filipe, L. Grady, S. Molin, and D. Stahl. 1999. Identification of a novel group of bacteria in sludge from a deteriorated biological phosphorus removal reactor. Appl. Environ. Microbiol. 65:1251-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oumer, B. A., P. Gaya, E. Fernandez-Garcia, R. Marciaca, S. Garde, M. Medina, and M. Nunez. 2001. Proteolysis and formation of volatile compounds in cheese manufactured with a bacteriocin-producing adjunct culture. J. Dairy Res. 68:117-129. [DOI] [PubMed] [Google Scholar]
- 37.Perez, G., E. Cardell, and V. Zarate. 2002. Random amplified polymorphic DNA analysis for differentiation of Leuconostoc mesenteroides subspecies isolated from Tenerife cheese. Lett. Appl. Microbiol. 34:82-85. [DOI] [PubMed] [Google Scholar]
- 38.Randazzo, C. L., S. Torriani, A. D. L. Akkermans, W. M. de Vos, and E. E. Vaughan. 2002. Diversity, dynamics and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somers, E. B., M. E. Johnson, and A. C. Wong. 2001. Biofilm formation and contamination of cheese by nonstarter lactic acid bacteria in dairy environment. J. Dairy Sci. 84:1926-1936. [DOI] [PubMed] [Google Scholar]
- 43.Søndergaard, A. K., and L. H. Stahnke. 2002. Growth and aroma production by Staphylococcus xylosus, S. carnosus and S. equorum—a comparative study in model systems. Int. J. Food Microbiol. 75:99-109. [DOI] [PubMed] [Google Scholar]
- 44.Stecchini, M. L., I. Sarais, and M. de Bertoldi. 1991. The influence of Lactobacillus plantarum culture inoculation on the fate of Staphylococcus aureus and Salmonella typhimurium in Montasio cheese. Int. J. Food Microbiol. 14:99-109. [DOI] [PubMed] [Google Scholar]
- 45.Stringer, S. C., B. J. Chaffey, C. E. R. Dodd, M. R. A. Morgan, and W. M. Waites. 1995. Specific antibody-mediated detection of Brochothrix thermosphacta in situ in British fresh sausages. J. Appl. Bacteriol. 78:335-340. [DOI] [PubMed] [Google Scholar]
- 46.Stringer, S. C., C. E. R. Dodd, M. R. A. Morgan, and W. M. Waites. 1995. Locating nisin-producing Lactococcus lactis in a fermented meat system. J. Appl. Bacteriol. 78:341-348. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi, K., and J. F. Frank. 2001. Confocal microscopy and microbial viability detection for food research. J. Food Prot. 64:2088-2102. [DOI] [PubMed] [Google Scholar]
- 49.Villani, F., G. Moschetti, G. Blaiotta, and S. Coppola. 1997. Characterization of strains of Leuconostoc mesenteroides by analysis of soluble whole-cell protein pattern, DNA fingerprinting and restriction of ribosomal DNA. J. Appl. Microbiol. 82:578-588. [DOI] [PubMed] [Google Scholar]
- 50.Von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 51.Wong, A. C. 1998. Biofilms in food processing environments. J. Dairy Sci. 81:2765-2770. [DOI] [PubMed] [Google Scholar]
- 52.Yiu, S. H. 1985. A fluorescence microscopic study of cheese. Food Microstruct. 4:99-106. [Google Scholar]