Abstract
It has been demonstrated that for a nonpathogenic, leaf-associated bacterium, effectiveness in the control of bacterial speck of tomato is correlated with the similarity in the nutritional needs of the nonpathogenic bacterium and the pathogen Pseudomonas syringae pv. tomato. This relationship was investigated further in this study by using the pathogen Xanthomonas campestris pv. vesicatoria, the causal agent of bacterial spot of tomato, and a collection of nonpathogenic bacteria isolated from tomato foliage. The effects of inoculation of tomato plants with one of 34 nonpathogenic bacteria prior to inoculation with the pathogen X. campestris pv. vesicatoria were quantified by determining (i) the reduction in disease severity (number of lesions per square centimeter) in greenhouse assays and (ii) the reduction in leaf surface pathogen population size (log10 of the number of CFU per leaflet) in growth chamber assays. Nutritional similarity between the nonpathogenic bacteria and X. campestris pv. vesicatoria was quantified by using either niche overlap indices (NOI) or relatedness in cluster analyses based upon in vitro utilization of carbon or nitrogen sources reported to be present in tomato tissues or in Biolog GN plates. In contrast to studies with P. syringae pv. tomato, nutritional similarity between the nonpathogenic bacteria and the pathogen X. campestris pv. vesicatoria was not correlated with reductions in disease severity. Nutritional similarity was also not correlated with reductions in pathogen population size. Further, the percentage of reduction in leaf surface pathogen population size was not correlated with the percentage of reduction in disease severity, suggesting that the epiphytic population size of X. campestris pv. vesicatoria is not related to disease severity and that X. campestris pv. vesicatoria exhibits behavior in the phyllosphere prior to lesion formation that is different from that of P. syringae pv. tomato.
Biological control of plant diseases involves interactions between a biological control agent and a plant pathogen; hence, an understanding of the ecology of the pathogen and the biological control agent is essential to the development of a biological control strategy. Many foliar bacterial pathogens have been reported to colonize plant surfaces prior to infection (6), and the sizes of these populations have subsequently been related to disease severity or incidence (11, 24, 29). These findings suggest that strategies which reduce the leaf surface population size of a pathogen might reduce the subsequent severity or incidence of the disease. While data demonstrating such a phenomenon in the biological control of a plant pathogen are scarce, this relationship has been demonstrated with ice nucleation-active (Ice+) bacteria. The incidence of frost injury to susceptible plant tissues is proportional to the logarithm of the population size of Ice+ Pseudomonas syringae on each leaf (17); hence, the incidence of frost injury can be reduced by prior application of Ice− P. syringae strains that reduce the population size of Ice+ P. syringae strains (12-16).
The success of this strategy in the biological control of frost injury suggested that this approach might be effective not only against Ice+ P. syringae strains but also against other phytopathogenic bacteria which colonize leaf surfaces. Preemptive competitive exclusion due to the prior utilization of limiting nutritional resources by the Ice− bacteria in the phyllosphere is an important aspect of the mechanism involved in the reduction of population sizes of Ice+ P. syringae and frost injury (14, 15, 34). Wilson and Lindow (34) demonstrated that on nitrogen-sufficient plants, the leaf-associated bacterial populations were limited primarily by carbon and secondarily by nitrogen and that preemptive exclusion of Ice+ P. syringae by the Ice− strains was probably due to prior utilization of carbon sources in the phyllosphere. Wilson and Lindow also demonstrated that the level of coexistence between Ice+ P. syringae and various nonpathogenic leaf-associated bacteria in the phyllosphere was inversely correlated with the similarity of the nutritional needs of Ice+ and Ice− P. syringae strains, which was quantified by using a niche overlap index (NOI) based upon in vitro carbon source utilization profiles (34, 35). These studies suggested that nutritional similarity between a biological control agent and a pathogen may be an important determinant of the ability of nonpathogenic leaf-associated bacteria to reduce pathogen population size and, hence, disease severity. Further, the studies suggested that the effectiveness of a given nonpathogenic bacterium in biological control through preemptive competitive exclusion should be proportional to the nutritional similarity between that bacterium and the target pathogen (35).
The importance of nutritional similarity in the biological control of foliar bacterial diseases has been investigated previously with P. syringae pv. tomato, the causal agent of bacterial speck of tomato (8). Using a collection of nonpathogenic leaf-associated bacteria and Tn5-generated catabolic mutants of P. syringae strain TLP2, a biological control agent of P. syringae pv. tomato (33), Ji and Wilson (8) observed nutritional similarity between the nonpathogenic bacteria or the catabolic mutants and P. syringae pv. tomato to be positively correlated with reductions in bacterial speck severity.
In order to determine whether the significance of nutritional similarity is also applicable to the biological control of other leaf-associated phytopathogenic bacteria or whether it applies only to the biological control of Ice+ P. syringae and P. syringae pathovars, we decided to examine the tomato pathogen Xanthomonas campestris pv. vesicatoria, the causal agent of bacterial spot of tomato. Leben (10) was the first to observe that X. campestris pv. vesicatoria multiplied on tomato seedlings for several days before symptom occurrence. Since then it has been reported several times that X. campestris pv. vesicatoria is capable of epiphytic growth (multiplication on the leaf surface) and survival on the surfaces of host species prior to the appearance of symptoms (1, 9, 18, 19, 22, 28, 31). These observations suggest that X. campestris pv. vesicatoria colonizes the leaf surface prior to invading the leaf and that a reduction in the population size of X. campestris pv. vesicatoria on the leaf surface through prior application of preemptive biological-control agents might result in a reduction in disease severity.
This study tested the hypothesis that the effectiveness of a given nonpathogenic bacterium in the reduction of the population size of the pathogen and the suppression of the severity of bacterial spot is positively correlated with the nutritional similarity between the nonpathogenic bacterium and X. campestris pv. vesicatoria. Since bacterial populations in the phyllosphere are limited primarily by carbon and secondarily by nitrogen (34), nutritional similarity between the nonpathogenic bacteria and X. campestris pv. vesicatoria was estimated by using both carbon and nitrogen sources. Determination of such a relationship suggests that the role of nutritional similarity in determining biological control efficacy is applicable to different phytopathogenic bacterial species and that preemptive competitive exclusion may be an appropriate strategy for the biological control of several foliar bacterial diseases.
MATERIALS AND METHODS
Bacterial strains.
The nonpathogenic bacteria used in this study were isolated from nonsymptomatic tomato foliage. Strains whose designations begin with “B” were isolated in Florida (J. B. Jones, University of Florida, Gainesville). The “Xc” strains were isolated in Georgia (R. Gitaitis, University of Georgia, Tifton), the “BT” strains were isolated in Alabama (P. A. Backman, Auburn University, Auburn, Ala.), and the “Cu” strains were isolated in Alabama (this study). All strains were identified using the Sherlock GC-FAME system (MIDI, Newark, Del.). This collection of nonpathogenic bacteria from tomato foliage was supplemented with P. syringae strains TLP2 and CIT7 (13) and Pseudomonas fluorescens strain A506 (36) from S. E. Lindow (University of California, Berkeley). This study employed a total of 34 bacteria from diverse genera, including Xanthomonas, Serratia, Bacillus, Cellulomonas, Comamonas, Pseudomonas, Erwinia, Clavibacter, and Pantoea. Strains were stored in 20% (vol/vol) glycerol in tryptic soy broth at −80°C. X. campestris pv. vesicatoria strain AD17 was isolated from an infected tomato plant in Alabama and was determined to be from tomato race T1/pepper race P2 (T1P2) subgroup A (2). A spontaneous mutant of AD17, designated AD17rif3, which is resistant to 100 μg of rifampin per ml and was not significantly less virulent than the wild type (data not shown), was selected for use throughout this study.
Preparation of bacterial inocula.
All bacterial cultures were grown on tryptic soy agar (TSA) (Difco Laboratories, Detroit, Mich.) or TSA amended with 100 μg of cycloheximide per ml and 100 μg of rifampin per ml for 48 h at 28°C. Bacterial cells were scraped from the plate and suspended in potassium phosphate buffer (PPB; 0.01 M, pH 7.0). The cell suspensions were adjusted to the appropriate concentrations by using a spectrophotometer and a standard curve of optical density (λ = 600 nm) and concentration (number of CFU per milliliter) of X. campestris pv. vesicatoria.
Assessment of the reduction in disease severity.
One group of four 5-week-old tomato plants (Lycopersicon esculentum cv. Agriset 761) was mist inoculated with a suspension of each of the nonpathogenic bacterial strains (∼107 CFU/ml) or with sterile PPB (pathogen-only control). The plants were arranged in a randomized complete block design in a plastic tent and incubated under conditions of continuous high relative humidity. After 48 h, all the plants were inoculated with a suspension of the pathogen X. campestris pv. vesicatoria strain AD17rif3 (∼108 CFU/ml). The plants were incubated under conditions of high relative humidity, at a temperature of 25 to 30°C, and with a light period of approximately 12 h until symptom development. Disease severity was quantified by determining the number of lesions per unit leaf area (i.e., number of lesions per square centimeter) on 10 leaflets from each of four plants collected 10 days after pathogen inoculation. The disease severity data were subjected to log transformation, and analysis of variance was performed using the analysis of variance or general linear models (PROC/GLM) of the Statistical Analysis System (SAS Institute, Cary, N.C.). The means were compared by using Duncan's multiple-range test at a P of 0.05. Biological control efficacy was quantified as the percentage of reduction in disease severity compared to that with the pathogen-only control. The experiment was repeated three times to allow for a determination of the mean reduction in disease severity.
Assessment of the reduction in pathogen population size.
One group of three 5-week-old tomato plants (cv. Agriset) was inoculated with a suspension of each of the nonpathogenic bacteria (∼107 CFU/ml) or with sterile PPB (the pathogen-only control). The plants were incubated under the conditions described above for disease assessment. After 48 h, all plants were inoculated with a suspension of X. campestris pv. vesicatoria strain AD17rif3 (∼106 CFU/ml). The initial pathogen population size was determined from 10 leaflets collected from the pathogen-only control (immediately after pathogen inoculation), and the final pathogen population size was determined from 10 leaflets collected from each treatment group 8 days after inoculation with the pathogen. Enumeration of leaf surface pathogen population sizes was performed as described previously (8). The percentage of reduction in pathogen population size on the plants treated with each nonpathogenic bacterium compared to the population size of the pathogen-only control (y) was calculated by using the formula y = 100[(a − b)(a − x)], where a is the final mean pathogen population size on the pathogen-only control plants (number of CFU per leaf), b is the final mean pathogen population size on the non-pathogen-treated plants (number of CFU per leaf), and x is the mean population size on the pathogen-only control plants immediately following inoculation (number of CFU per leaf) (i.e., the increase in pathogen population size on the non-pathogen-treated plants as a percentage of the increase in pathogen population size on the pathogen-only plants). The experiment was conducted three times, and the mean percentage of reduction in population size was used in the subsequent correlation analyses.
Determination of nutritional similarity: NOIs.
In vitro carbon and nitrogen source utilization profiles were determined for X. campestris pv. vesicatoria strain AD17rif3 by using compounds reported to be present in tomato tissues (3, 4, 23, 27) (Table 1) and presumed to also be present in the tomato phyllosphere. X. campestris pv. vesicatoria strain AD17rif3 was grown on 1/10-strength TSA for 72 h at 28°C. A 10-ml volume of bacterial suspension (∼108 CFU/ml) in PPB was inoculated onto minimal medium A (21) containing individual compounds as a sole source of carbon (with ammonium sulfate added as a nitrogen source) or nitrogen (with glucose added as a carbon source) at a concentration of approximately 10 mM. Plates were incubated for 10 days at 28°C. Growth on the sole carbon or nitrogen source plates was compared with that on a negative-control (minimal medium A with no carbon or nitrogen source) plate to determine which compounds were utilized and supported an increase in biomass. Carbon and nitrogen source utilization profiles of the nonpathogenic bacteria were subsequently determined by using the carbon or nitrogen sources which supported the growth of X. campestris pv. vesicatoria. Nutritional profiles were also obtained for X. campestris pv. vesicatoria strain AD17rif3 and the nonpathogenic bacteria by using Biolog, Inc. (Hayward, Calif.), GN microplates and recommended procedures.
TABLE 1.
In vitro sole carbon and nitrogen source utilization profiles for X. campestris pv. vesicatoria strain AD17rif3 based on carbon and nitrogen sources reported to be present in tomato tissues and presumed to be present in tomato leaf exudates
| Source compound | Compounds utilized or not utilized by X. campestris pv. vesicatoria strain AD17rif3
|
|
|---|---|---|
| Utilized | Not utilized | |
| Carbon | trans-aconitic acid, alanine, aspartic acid, citric acid, fructose, fumaric acid, glucose, glutamic acid, histidine, malic acid, oxaloacetic acid, serine, succinic acid, sucrose, threonine | Acetic acid, β-alanine, arginine, ascorbic acid, asparagine, citrulline, cysteine, dihydroxytartaric acid, ethanolamine, formic acid, GABA, galacturonic acid, glutaric acid, glycine, glycolic acid, isoleucine, lactic acid, lysine, leucine, maleic acid, malonic acid, methionine, mevalonic acid, myoinositol, oxalic acid, phenylalanine, pipecolic acid, proline, quinic acid, starch, tartaric acid, tryptophan, tyrosine, valine |
| Nitrogen | Alanine, arginine, asparagine, aspartic acid, citrulline, ethanolamine, GABA, glutamic acid, glutamine, histidine, isoleucine, leucine, methionine, phenylalanine, pipecolic acid, proline, tyrosine | β-Alanine, cysteine, glycine, lysine, quinic acid, serine, threonine, valine |
Wilson and Lindow (34, 35) estimated nutritional similarity using NOIs calculated with the formula NOI = the number of carbon sources used by both the nonpathogenic bacterium and the pathogen/the total number of carbon sources used by the pathogen.In this study, nutritional similarity between the nonpathogenic bacteria and X. campestris pv. vesicatoria was estimated by using carbon and nitrogen utilization data and the Biolog GN profiles (Table 2) with the formulae NOIC = the number of carbon sources used by both the nonpathogenic bacterium and X. campestris pv. vesicatoria/the number of carbon sources used by X. campestris pv. vesicatoria; NOIN = the number of nitrogen sources used by both the nonpathogenic bacterium and X. campestris pv. vesicatoria/the number of nitrogen sources used by X. campestris pv. vesicatoria; and NOIBiolog = the number of compounds used by both the nonpathogenic bacterium and X. campestris pv. vesicatoria/the number of compounds used by X. campestris pv. vesicatoria.
TABLE 2.
Nutritional similarities between nonpathogenic tomato leaf-associated bacteria and the bacterial spot pathogen X. campestris pv. vesicatoria strain AD17rif3
| Strain | Gram reactiona | Genus | Nutritional similarity (%)b
|
||
|---|---|---|---|---|---|
| NOIC | NOIBiolog | NOIN | |||
| Xc84-1 | − | Xanthomonas | 0.93 | 0.78 | 1.00 |
| Xc84-2a | − | Xanthomonas | 0.80 | 0.88 | 1.00 |
| Xc84-2b | − | Xanthomonas | 0.80 | 0.96 | 1.00 |
| Xc88-34 | − | Xanthomonas | 0.80 | 0.98 | 1.00 |
| Xc88-37 | − | Xanthomonas | 0.80 | 0.94 | 1.00 |
| Xc89-22a | − | Xanthomonas | 0.87 | ND | 1.00 |
| Xc89-22b | − | Xanthomonas | 0.80 | ND | 0.94 |
| Xc89-301 | − | Xanthomonas | 0.87 | 0.94 | 1.00 |
| BT1 | + | Cellulomonas | 0.73 | ND | 0.94 |
| BT2 | − | Xanthomonas | 0.80 | 0.92 | 0.59 |
| BT3a | − | Xanthomonas | 0.80 | 0.88 | 0.35 |
| BT3b | + | Bacillus | 0.80 | ND | 0.80 |
| BT5 | − | Serratia | 1.00 | 0.96 | 1.00 |
| BT7 | + | Bacillus | 0.67 | ND | 1.00 |
| BT9 | + | Cellulomonas | 0.87 | ND | 0.41 |
| BT10 | + | Bacillus | 0.27 | ND | 1.00 |
| BT11 | − | Stenotrophomonas | 0.27 | ND | 0.47 |
| BT13 | − | Xanthomonas | 0.87 | 0.88 | 0.06 |
| BT25 | − | Serratia | 1.00 | 0.96 | 1.00 |
| B2 | − | Erwinia | 0.93 | 0.74 | 1.00 |
| B11 | − | Comamonas | 0.80 | 0.55 | 0.82 |
| B18 | + | Clavibacter | 0.73 | ND | 1.00 |
| B26 | + | Clavibacter | 0.73 | ND | 1.00 |
| B36 | 0 | Bacillus | 0.73 | ND | 0.35 |
| B41 | − | Pseudomonas | 0.67 | 0.53 | 1.00 |
| B52 | − | Pseudomonas | 0.87 | 0.47 | 1.00 |
| B56 | − | Pseudomonas | 1.00 | 0.64 | 1.00 |
| TLP2 | − | Pseudomonas | 0.87 | 0.73 | 1.00 |
| A506 | − | Pseudomonas | 0.87 | 0.70 | 1.00 |
| Cit7 | − | Pseudomonas | 0.87 | 0.51 | 0.88 |
| Cu1 | + | Bacillus | 0.74 | 0.30 | 0.88 |
| Cu21 | − | Pseudomonas | 0.93 | 0.43 | 0.94 |
| Cu26 | − | Pantoea | 0.73 | 0.80 | 0.94 |
| Cu34 | − | Pseudomonas | 0.93 | 0.57 | 1.00 |
+, −, or 0 indicates a gram-positive, gram-negative, or neutral reaction, respectively.
Determination of nutritional similarity was based on in vitro tomato carbon source utilization profiles (NOIC), Biolog GN plate carbon source profiles (NOIBiolog), or in vitro tomato nitrogen source utilization profiles (NOIN). ND, not determined.
Determination of nutritional similarity: cluster analysis.
Cluster analysis was included in this study with X. campestris pv. vesicatoria, though it was not included in the previous study with P. syringae pv. tomato (8), because it provides an additional, slightly different perspective on nutritional similarity than does the NOI. The NOI focuses specifically on the overlap in nutrient use between the biological control agent and the pathogen but does not address the use of nutrients outside this region of overlap, while cluster analysis takes into account the complete list of nutrients utilized by both organisms. The use of both analytical approaches provides a more thorough assessment of nutritional similarity between the biological control agent and the pathogen.
The carbon and nitrogen source utilization data were subjected to cluster analysis by Ward's clustering method in the JMP statistical software package (SAS Institute). The Biolog GN nutritional profiles were subjected to cluster analysis with Biolog ML3N software (Biolog, Inc.). Nutritional similarity between the nonpathogenic bacteria and the pathogen was estimated from these dendrograms, in which the x axis represents relatedness by a relative linear measurement in centimeters.
Relationship between nutritional similarity and reduction in disease severity or pathogen population size.
Regression analyses were used to assess the relationship between a similarity in nutritional needs (i.e., sole carbon source utilization [NOIC or relatedness], carbon source utilization in the Biolog GN microplates [NOIBiolog or relatedness], or sole nitrogen source utilization [NOIN or relatedness]) and the percentage of reduction in disease severity or pathogen population size, for which the mean percentage of reduction from the three repeated experiments was used. Multiple-correlation analyses were employed to determine whether reductions in disease severity or pathogen population size were correlated with the utilization of any individual carbon or nitrogen source by the nonpathogenic bacteria.
RESULTS
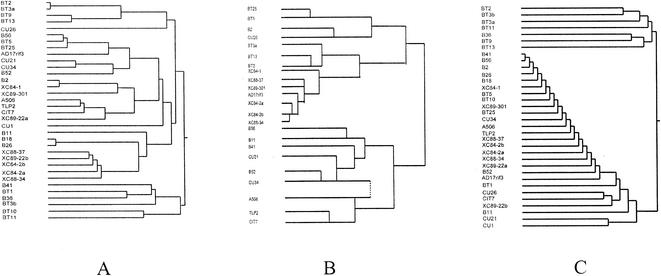
Qualitative examination of the dendrograms from cluster analyses suggested that a similarity in the nutritional needs of the nonpathogenic bacteria and X. campestris pv. vesicatoria, estimated from the sole carbon source or the Biolog GN carbon source utilization profiles, was not correlated with reductions in either disease severity or pathogen population size. In the dendrogram from cluster analysis of the sole carbon source utilization data (Fig. 1A), many of the strains that provided the largest mean reductions in disease severity (e.g., B2, Xc89-301, B41, and BT1) or population size (e.g., Cu26, Cit7, Xc84-2b, Xc84-2a, and B36) clustered in groups different from that of the pathogen. The only strains which clustered close to the pathogen were Cu34 (Pseudomonas sp.), which provided a large mean reduction in disease severity, and B56 (Pseudomonas putida), which provided a large mean reduction in population size. A similar situation occurred with the dendrogram from cluster analysis of the Biolog GN carbon source utilization data with the ML3N software (Fig. 1B). (Note that gram-positive strains were not included in this analysis.) Although some of the strains which provided large reductions in mean population size (e.g., Xc84-2a, Xc84-2b, and Xc88-34) clustered very close to the pathogen, other strains which provided equally large reductions in mean population size (e.g., Cu26, B56, and Cit7) clustered far from the pathogen.
FIG. 1.
(A and C) Cluster analysis of in vitro carbon (A) and nitrogen (C) source utilization data for all 34 nonpathogenic leaf-associated bacteria and X. campestris pv. vesicatoria strain AD17rif3 with Ward's clustering method in JMP software (SAS Institute). (B) Cluster analysis of Biolog GN carbon source utilization data for gram-negative nonpathogenic leaf-associated bacteria and X. campestris pv. vesicatoria strain AD17rif3 with Biolog ML-Clust 3 N software.
In contrast to the carbon source data, in which no qualitative correlations were apparent, some of the strains which provided large reductions in disease severity (e.g., B41, B2, Xc89-301, Cu34, and even BT1) clustered quite close to the pathogen when nitrogen source utilization data were analyzed (Fig. 1C). No such qualitative correlation was observed for population reduction, however, since some strains which provided large reductions in pathogen population size (e.g., B56, Xc84-2b, Xc84-2a, and Xc88-34) clustered close to the pathogen, while others (e.g., B36, Cu26, and Cit7) clustered much farther away.
Based on the data for in vitro utilization of the tomato carbon sources and quantifications with the formula for NOIC, the nutritional similarity between X. campestris pv. vesicatoria strain AD17rif3 and the 34 nonpathogenic bacteria ranged from a low NOI value of 0.27 to a high value of 1.0 (Table 2). When the data for Biolog GN carbon source utilization or in vitro utilization of the tomato nitrogen sources were used to estimate nutritional similarity, the NOI values for the bacterial strains ranged from 0.30 to 0.98 and from 0.06 to 1.0, respectively (Table 2).
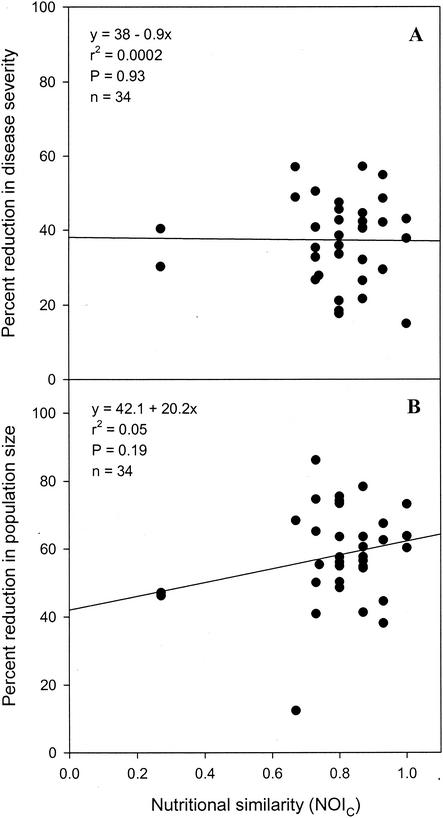
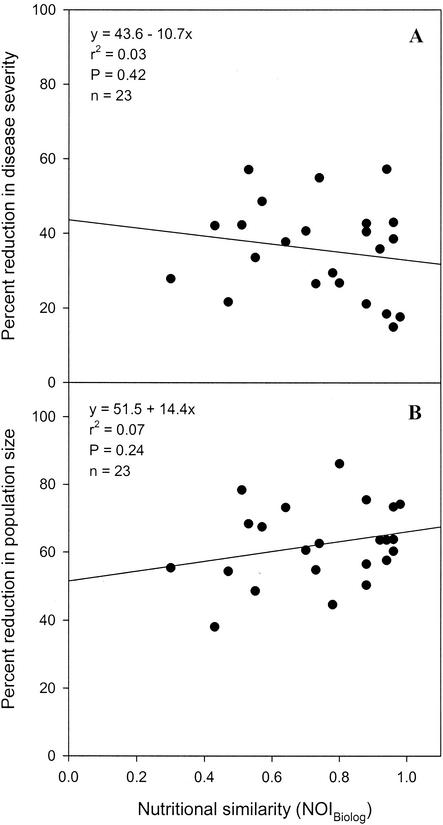
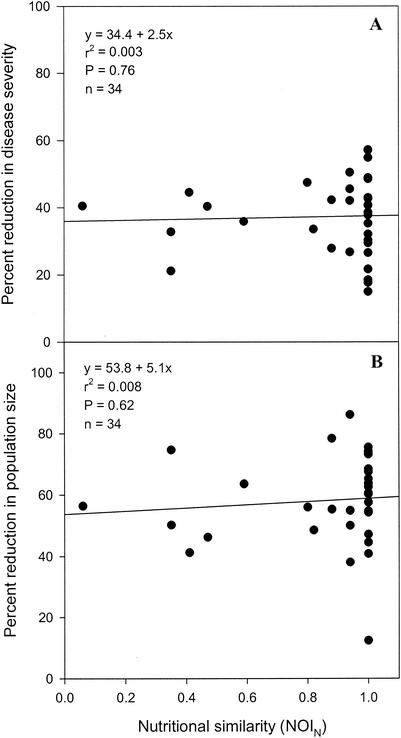
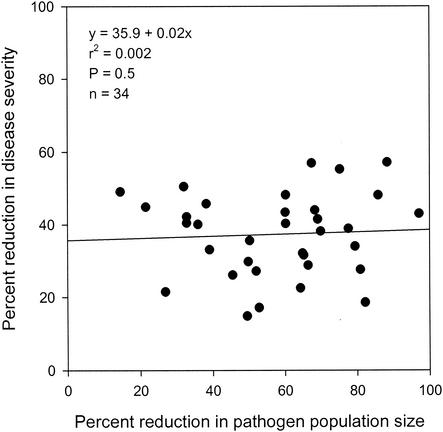
Regression analyses indicated that the percentage of reduction in the severity of bacterial spot was not significantly correlated with nutritional similarity, which was estimated from carbon source utilization data obtained with the formula for either NOIC (Fig. 2A) (P = 0.93) or NOIBiolog (Fig. 3A) (P = 0.42) or estimated from nitrogen source utilization data obtained with the formula for NOIN (Fig. 4A) (P = 0.76). A significant correlation was also not observed between the percentage of reduction in the severity of bacterial spot and nutritional similarity when nutritional similarity was quantified by using the relatedness in cluster analyses of tomato carbon source, Biolog GN carbon source, or tomato nitrogen source utilization data (Table 3). Similarly, regression analyses indicated that the percentage of reduction in X. campestris pv. vesicatoria population size was not significantly correlated with nutritional similarity, as determined by using either NOI (Fig. 2B, 3B, and 4B) or relatedness data (Table 3). Further, the percentage of reduction in the severity of bacterial spot was not significantly correlated with the percentage of reduction in the population size of X. campestris pv. vesicatoria (Fig. 5) (P = 0.5).
FIG. 2.
Regression analysis of nutritional similarity (estimated from in vitro carbon source utilization profiles) between nonpathogenic bacteria and the pathogen X. campestris pv. vesicatoria strain AD17rif3 and percentages of reduction in the spot severity (A) and the population size (B) of the pathogen.
FIG. 3.
Regression analysis of nutritional similarity (estimated from Biolog GN carbon source utilization profiles) between nonpathogenic bacteria and the pathogen X. campestris pv. vesicatoria strain AD17rif3 and percentages of reduction in the spot severity (A) and the population size (B) of the pathogen.
FIG. 4.
Regression analysis of nutritional similarity (estimated from in vitro nitrogen source utilization profiles) between nonpathogenic bacteria and the pathogen X. campestris pv. vesicatoria strain AD17rif3 and percentages of reduction in the spot severity (A) and the population size (B) of the pathogen.
TABLE 3.
Regression analyses of the relationship of the percentage of reduction in disease severity or pathogen population size to mututional similarity between nonpathogenic bacteria and the pathogen X. campestris pv. vesicatoriaa
| Relatedness method | Value for:
|
|||||
|---|---|---|---|---|---|---|
| Disease severity
|
Population size
|
|||||
| y | r2 | P | y | r2 | P | |
| RelatednessC | 30.8 + 0.8x | 0.01 | 0.54 | 68.2 − 1.2x | 0.02 | 0.40 |
| RelatednessBiolog | 31.5 + 0.78x | 0.03 | 0.45 | 64.2 − 0.29x | 0.004 | 0.77 |
| RelatednessN | 35.3 + 0.3x | 0.01 | 0.84 | 70.8 − 1.8x | 0.06 | 0.18 |
The mean percentage of reduction in the severity of bacterial spot of tomato disease or the population size of the pathogen X. campestris pv. vesicatoria was determined from the results of three replicate experiments. The regression analyses were estimated from cluster analyses of the relatedness of carbon or nitrogen source utilization data. Nutritional similarity was quantified by using relatedness data estimated from cluster analyses of tomato carbon source (RelatednessC), Biolog GN carbon source (RelatednessBiolog), or tomato nitrogen source (RelatednessN) utilization data.
FIG. 5.
Regression analysis of the percentage of reduction in bacterial spot severity and the percentage of reduction in the leaf surface population size of the pathogen X. campestris pv. vesicatoria strain AD17rif3 following inoculation of tomato leaves with various nonpathogenic bacteria.
Multiple-correlation analysis of the disease severity reduction data and nutrient utilization profiles based on the 49 tomato carbon sources and the 25 tomato nitrogen sources indicated that the utilization of histidine (canonical coefficient, 0.42) and serine (canonical coefficient, 0.48) as the sole carbon sources was weakly correlated (P = 0.11) with a reduction in disease severity. Further, the difference in the mean percentages of reduction in disease severity for the nonpathogenic bacteria that utilized both histidine and serine and those that did not, when analyzed by using Student's t test, was weakly significant (P = 0.07). Multiple-correlation analysis of the population reduction data and the 49 carbon and 25 nitrogen source utilization profiles indicated that the utilization of three compounds, aspartic acid (canonical coefficient, 0.35), γ-aminobutyric acid (GABA) (canonical coefficient, 0.32), and methionine (canonical coefficient, 0.35), as sole nitrogen sources was correlated significantly (P = 0.052) with the percentage of reduction in the pathogen population size.
DISCUSSION
In contrast to expectations, in this study no correlation was observed between the reduction in severity of bacterial spot and the nutritional similarity of the nonpathogenic bacteria with the pathogen X. campestris pv. vesicatoria, regardless of whether nutritional similarity was estimated from in vitro sole carbon source utilization profiles, Biolog GN carbon source utilization profiles, or in vitro sole nitrogen source utilization profiles or whether NOI (8, 34, 35) or relatedness data were used for cluster analysis. A correlation was also not observed between a reduction in the pathogen population size and the nutritional similarity of the nonpathogenic bacteria and the pathogen X. campestris pv. vesicatoria, regardless of how nutritional similarity was estimated. These findings differ from those with Ice+ P. syringae strains and P. syringae pv. tomato, which indicated that nutritional similarity was correlated with the exclusion of Ice+ P. syringae populations by Ice− bacterial strains (34, 35) and with reductions in bacterial speck severity (8). However, the findings are consistent with the studies of Xanthomonas translucens pv. translucens (30), which demonstrated that nutritional similarity was not predictive of the ability of epiphytic bacteria to reduce either pathogen population size or disease severity.
It was further determined that, despite reports of epiphytic populations of X. campestris pv. vesicatoria preceding lesion formation (1, 9, 18, 19, 22, 28, 31), the percentage of reduction in leaf surface pathogen population size was not correlated with the percentage of reduction in disease severity, suggesting that the epiphytic population size of X. campestris pv. vesicatoria is not proportionally related to the subsequent severity of the disease. This suggestion contrasts with the relationship observed between the population size of Ice+ P. syringae and the incidence of frost injury (17), between P. syringae pv. syringae population size and brown spot incidence and severity (11, 24), between X. translucens pv. translucens population size and bacterial leaf streak severity (29), and between the relationship inferred to occur between P. syringae pv. tomato population size and bacterial speck severity (8). In the absence of a proportional relationship between pathogen population size and subsequent disease severity, even if nutritional similarity were correlated with reduction in pathogen population size, one cannot expect a correlation between nutritional similarity and reduction in disease severity.
These observations suggest that the pathogen X. campestris pv. vesicatoria behaves differently in the phyllosphere than Ice+ P. syringae (17), P. syringae pathovars (8, 11, 24), or even X. translucens pv. translucens (29). More specifically, there appears to be a difference in significance of leaf surface population sizes. While P. syringae pathovars apparently must achieve relatively high population sizes on leaf surfaces prior to invasion (7, 11, 24), this does not appear to be the case for X. campestris pv. vesicatoria on tomato. Possibly, the pathogen is able to access the interior of the leaf at much lower leaf surface population sizes, which suggests that the severity of an infection by X. campestris pv. vesicatoria might be predicted more accurately by internal population size than by leaf surface or epiphytic population size. But what of the large epiphytic populations of X. campestris pv. vesicatoria that have been reported (1, 9, 18, 19, 22, 28, 31)? These populations are probably not the cause of infection but the result of infection; in other words, they result from the egress of bacteria onto the leaf surface from substomatal chambers (20) which are the sites of developing lesions. If the leaf surface populations in this study were partly or predominantly the result of egress from substomatal chambers, these populations would not necessarily be affected by prior nutrient use in a predictable manner and one would not expect a correlation between nutrient similarity and reduction in population size.
A further possible explanation for the lack of correlation between nutritional similarity and reduction in X. campestris pv. vesicatoria population size may be the differential levels of abundance or preferential use of nutritional substances in the phyllosphere, either of which would impact estimates of nutritional similarity. While these factors did not obscure the correlation between nutritional similarity and efficacy observed with P. syringae pv. tomato (8), the range of tomato carbon sources utilized by X. campestris pv. vesicatoria (15 out of 49 compounds [this study]) is significantly smaller than that utilized by P. syringae pv. tomato [(30 out of 52 compounds) (8), rendering X. campestris pv. vesicatoria more susceptible to large differences in the levels of abundance of carbon sources. The significant positive correlation between the in vitro utilization of aspartic acid, GABA, and methionine as nitrogen sources and the reduction in leaf surface population size of X. campestris pv. vesicatoria does suggest that some nutrients are more important than others. Similarly, Stromberg et al. (30) found some nutrients to be more important than others, with successful antagonists of bacterial leaf streak of wheat, caused by X. translucens pv. translucens, utilizing sucrose and inositol more frequently than did poor antagonists. These observations clearly indicate that when the number of phyllosphere carbon or nitrogen sources utilized is small, estimates of nutritional similarity should be based on NOI values weighted for both abundance and preference.
Although preemptive exclusion of the pathogen population by a nutritionally similar nonpathogenic bacterium does not appear to be involved in the biological control of bacterial spot, as it is with bacterial speck (8), some of the nonpathogenic bacteria did provide significant reductions in the severity of bacterial spot under both greenhouse and, subsequently, field conditions (M. Wilson et al., unpublished data); hence, other mechanisms, such as antibiosis, induced host resistance, and inhibition of the expression of pathogenicity and/or virulence in the pathogen, must be involved. For example, hrp gene induction may be reduced by prior colonization by the nonpathogenic bacteria if those bacteria catabolize compounds in leaf exudates involved in hrp gene induction in X. campestris pv. vesicatoria (25, 26). The positive correlation between the utilization of histidine and serine and the reduction in spot severity suggests that these amino acids may be involved in such a phenomenon. The nonpathogenic bacteria may also affect the expression of disease in the host without suppressing the pathogen population size or pathogenicity or virulence per se. Prior inoculation of a tomato leaf with an hrpG mutant of X. campestris pv. vesicatoria significantly reduced bacterial spot severity on that leaf when it was subsequently challenged with pathogenic X. campestris pv. vesicatoria, yet there was apparently little or no reduction in either leaf surface or internal pathogen population size (W. Moss, U. Bonas, J. B. Jones, and M. Wilson, Abstr. 7th Int. Congr. Plant Pathol., abstr. 1.7.2, 1998). The hrpG mutant provided an induced local tolerance, rather than resistance, in the host, and this phenomenon may well occur with some of the nonpathogenic bacteria in this study.
Although biological control of bacterial spot of tomato apparently cannot be achieved through preemptive competitive exclusion of leaf surface pathogen populations, it should not be assumed that X. campestris pv. vesicatoria is typical of all xanthomonads. The leaf surface population sizes of other xanthomonads, including Xanthomonas campestris pv. undulosa (5) and X. translucens pv. translucens (29) on wheat and Xanthomonas campestris pv. phaseoli on bean (32), do appear to be predictive of subsequent disease severity; hence, preemptive competitive exclusion still may be an appropriate strategy in the biological control of these pathogens.
Acknowledgments
We acknowledge E. Bauske, K. Bowen, L. L. Kinkel, and S. E. Lindow for suggestions with regard to the statistical analyses; G. Beattie, J. B. Jones, and L. L. Kinkel for editorial suggestions; and W. Moss for useful discussions about X. campestris ecology.
This work was supported by a grant from the USDA NRIGCP (grant 9501182) awarded to M. Wilson.
REFERENCES
- 1.Bernal, R. F., and R. D. Berger. 1996. The spread of epiphytic populations of Xanthomonas campestris pv. vesicatoria on pepper in the field. J. Phytopathol. 144:479-484. [Google Scholar]
- 2.Bouzar, H., J. B. Jones, R. E. Stall, N. C. Hodge, G. V. Minsavage, A. Benedict, and A. M. Alvarez. 1994. Physiological, chemical, serological and pathogenic analyses of a worldwide collection of Xanthomonas campestris pv. vesicatoria strains. Phytopathology 84:663-671. [Google Scholar]
- 3.Carpena-Ruiz, R., A. Sopena, and A. M. Ramon. 1989. Extraction of free amino acids from tomato leaves. Plant Soil 119:251-254. [Google Scholar]
- 4.Davies, J. N., and G. E. Hobson. 1981. The constituents of tomato fruit—the influence of environment, nutrition, and genotype. Crit. Rev. Food Sci. Nutr. 15:205-280. [DOI] [PubMed] [Google Scholar]
- 5.Duveiller, E., and H. Maraite. 1995. Effect of temperature and air humidity on multiplication of Xanthomonas campestris pv. undulosa and symptom expression in susceptible and field-tolerant wheat genotypes. J. Phytopathol. 143:227-232. [Google Scholar]
- 6.Hirano, S. S., and C. D. Upper. 1983. Ecology and epidemiology of foliar bacterial plant pathogens. Annu. Rev. Phytopathol. 21:243-269. [Google Scholar]
- 7.Jardine, D. J., and C. T. Stephens. 1987. A predictive system for timing chemical applications to control Pseudomonas syringae pv. tomato, causal agent of bacterial speck. Phytopathology 77:823-827. [Google Scholar]
- 8.Ji, P., and M. Wilson. 2002. Assessment of the importance of similarity in carbon source utilization profiles between the biological control agent and the pathogen in biological control of bacterial speck of tomato. Appl. Environ. Microbiol. 68:4383-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, J. B., S. S. Woltz, J. P. Jones, and K. L. Portier. 1991. Population dynamics of Xanthomonas campestris pv. vesicatoria on tomato leaflets treated with copper bactericides. Phytopathology 81:714-719. [Google Scholar]
- 10.Leben, C. 1963. Multiplication of Xanthomonas vesicatoria on tomato seedlings. Phytopathology 53:778-781. [Google Scholar]
- 11.Lindemann, J., D. C. Arny, and C. D. Upper. 1984. Use of an apparent infection threshold population of Pseudomonas syringae to predict incidence and severity of brown spot of bean. Phytopathology 74:1334-1339. [Google Scholar]
- 12.Lindemann, J., and T. V. Suslow. 1987. Competition between ice nucleation-active wild type and ice nucleation-deficient deletion mutant strains of Pseudomonas syringae and P. fluorescens biovar I and biological control of frost injury on strawberry blossoms. Phytopathology 77:882-886. [Google Scholar]
- 13.Lindow, S. E. 1985. Ecology of Pseudomonas syringae relevant to the field use of Ice− deletion mutants constructed in vitro for plant frost control, p. 23-35. In H. O. Halvorson, D. Pramer, and M. Rogul (ed.), Engineered organisms in the environment: scientific issues. American Society for Microbiology, Washington, D.C.
- 14.Lindow, S. E. 1987. Competitive exclusion of epiphytic bacteria by Ice− Pseudomonas syringae mutants. Appl. Environ. Microbiol. 53:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindow, S. E. 1988. Lack of correlation of in vitro antibiosis with antagonism of ice nucleation active bacteria on leaf surfaces by non-ice nucleation active bacteria. Phytopathology 78:444-450. [Google Scholar]
- 16.Lindow, S. E. 1993. Biological control of plant frost injury: the Ice− story, p. 113-128. In L. Kim (ed.), Advanced engineered pesticides. Marcel Dekker, Inc., New York, N.Y.
- 17.Lindow, S. E., S. S. Hirano, W. R. Barchet, D. C. Arny, and C. D. Upper. 1982. The relationship between ice nucleation frequency of bacteria and frost injury. Plant Physiol. 70:1090-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mcguire, R. G., J. B. Jones, and J. W. Scott. 1991. Epiphytic populations of Xanthomonas campestris pv. vesicatoria on tomato cultigens resistant and susceptible to bacterial spot. Plant Dis. 75:606-609. [Google Scholar]
- 19.Mcguire, R. G., J. B. Jones, C. D. Stanley, and A. A. Csizinsky. 1991. Epiphytic populations of Xanthomonas campestris pv. vesicatoria and bacterial spot of tomato as influenced by nitrogen and potassium fertilization. Phytopathology 81:656-660. [Google Scholar]
- 20.Miles, W. G., R. H. Daines, and J. W. Rue. 1977. Presymptomatic egress of Xanthomonas pruni from infected peach leaves. Phytopathology 67:895-897. [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Pernezny, K., and J. Collins. 1997. Epiphytic populations of Xanthomonas campestris pv. vesicatoria on pepper: relationships to host-plant resistance and exposure to copper sprays. Plant Dis. 81:791-794. [DOI] [PubMed] [Google Scholar]
- 23.Picha, D. H. 1987. Sugar and organic acid content of cherry tomato fruit at different ripening stages. Hortscience 22:94-96. [Google Scholar]
- 24.Rouse, D. I., E. V. Nordheim, S. S. Hirano, and C. D. Upper. 1985. A model relating the probability of foliar disease incidence to the population frequencies of bacterial plant pathogens. Phytopathology 75:505-509. [Google Scholar]
- 25.Schulte, R., and U. Bonas. 1992. Expression of the Xanthomonas campestris pv. vesicatoria hrp gene cluster, which determines pathogenicity and hypersensitivity on pepper and tomato, is plant inducible. J. Bacteriol. 174:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulte, R., and U. Bonas. 1992. A Xanthomonas pathogenicity locus is induced by sucrose and sulfur-containing amino acids. Plant Cell 4:79-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senden, M. H. M. N., A. J. G. M. van der Meer, J. Limborgh, and H. T. Wolterbeek. 1992. Analysis of major tomato xylem organic acids and PITC-derivatives of amino acids by RP-HPLC and UV detection. Plant Soil 142:81-89. [Google Scholar]
- 28.Sharon, E., Y. Bashan, Y. Okon, and Y. Henis. 1982. Presymptomatic multiplication of Xanthomonas campestris pv. vesicatoria on the surface of pepper leaves. Can. J. Bot. 60:1041-1045. [Google Scholar]
- 29.Stromberg, K. D., L. L. Kinkel, and K. J. Leonard. 1999. Relationship between phyllosphere population sizes of Xanthomonas translucens pv. translucens and bacterial leaf streak severity on wheat seedlings. Phytopathology 89:131-135. [DOI] [PubMed] [Google Scholar]
- 30.Stromberg, K. D., L. L. Kinkel, and K. J. Leonard. 2000. Interactions between Xanthomonas translucens pv. translucens, the causal agent of bacterial leaf streak of wheat, and bacterial epiphytes in the wheat phyllosphere. Biol. Control 17:61-72. [Google Scholar]
- 31.Timmer, L. W., J. J. Marois, and D. Achor. 1987. Growth and survival of xanthomonads under conditions non-conducive to disease development. Phytopathology 77:1341-1345. [Google Scholar]
- 32.Weller, D. M., and A. W. Saettler. 1980. Colonization and distribution of Xanthomonas phaseoli and Xanthomonas phaseoli var. fuscans in field-grown navy beans. Phytopathology 70:500-506. [Google Scholar]
- 33.Wilson, M., H. L. Campbell, P. Ji, J. B. Jones, and D. A. Cuppels. 2002. Biological control of bacterial speck of tomato under field conditions at several locations in North America. Phytopathology 92:1284-1292. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, M., and S. E. Lindow. 1994. Ecological similarity and coexistence of epiphytic ice-nucleating (Ice+) Pseudomonas syringae strains and a non-ice-nucleating (Ice−) biological control agent. Appl. Environ. Microbiol. 60:3128-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, M., and S. E. Lindow. 1994. Coexistence among epiphytic bacterial populations mediated through nutritional resource partitioning. Appl. Environ. Microbiol. 60:4468-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, M., and S. E. Lindow. 1993. Interaction between the biological control agent Pseudomonas fluorescens strain A506 and Erwinia amylovora in pear blossoms. Phytopathology 83:117-123. [Google Scholar]