Abstract
Hybridization and restriction fragment length polymorphism data (K. G. Stuart-Keil, A. M. Hohnstock, K. P. Drees, J. B. Herrick, and E. L. Madsen, Appl. Environ. Microbiol. 64:3633-3640, 1998) have shown that pCg1, a naphthalene catabolic plasmid carried by Pseudomonas putida Cg1, is homologous to the archetypal naphthalene catabolic plasmid, pDTG1, in P. putida NCIB 9816-4. Sequencing of the latter plasmid allowed PCR primers to be designed for amplifying and sequencing the conjugal transfer region in pCg1. The mating pair formation (mpf) gene, mpfA encoding the putative precursor of the conjugative pilin subunit from pCg1, was identified along with other trb-like mpf genes. Sequence comparison revealed that the 10 mpf genes in pCg1 and pDTG1 are closely related (61 to 84% identity) in sequence and operon structure to the putative mpf genes of catabolic plasmid pWW0 (TOL plasmid of P. putida) and pM3 (antibiotic resistance plasmid of Pseudomonas. spp). A polar mutation caused by insertional inactivation in mpfA of pCg1 and reverse transcriptase PCR analysis of mRNA showed that this mpf region was involved in conjugation and was transcribed from a promoter located upstream of an open reading frame adjacent to mpfA. lacZ transcriptional fusions revealed that mpf genes of pCg1 were expressed constitutively both in liquid and on solid media. This expression did not respond to host exposure to naphthalene. Conjugation frequency on semisolid media was consistently 10- to 100-fold higher than that in liquid media. Thus, conjugation of pCg1 in P. putida Cg1 was enhanced by expression of genes in the mpf region and by surfaces where conditions fostering stable, high-density cell-to-cell contact are manifest.
Horizontal gene transfer (HGT) is the transmission of DNA between organisms of different lineages (5, 28, 53). An understanding of HGT is important because of its implication in the spread of antibiotic resistance genes (9), in the possible exchange of genes between genetically engineered and native microorganisms (10), in interactions between plants and bacteria (61), and in the dissemination of genes involved in pollutant biodegradation (18, 26, 40) and metal resistance (34, 51). It is becoming increasingly apparent that HGT has played an integral role in the evolution of bacterial species (55). One of the mechanisms of HGT is conjugation (28). Bacterial conjugation is a subject of ongoing scientific inquiry that explores how DNA is transmitted from donor to recipient cells via physical contact (31). The conjugation process may involve a combination of DNA rolling-circle replication with a type IV secretion system (32). While the role of pili in establishing cell-cell contact is widely accepted, conveyance of DNA through pili to a recipient cell is still under debate (32).
One of the most intensively studied conjugal transfer systems is the broad-host-range enteric plasmid RP4 (IncPα). RP4 conjugal transfer is encoded by two regions, Tra1 and Tra2, which account for almost half of the 60-kb plasmid DNA (41). The Tra1 region encodes the primase, relaxase, the leader operon, and one protein involved in mating pair formation (41). Tra2 genes (termed trb) code for mating pair formation proteins (29, 30). Many of the Tra2 gene products are homologous to those of the virB region of the Ti plasmid of Agrobacterium (31). The mpf (for mating pair formation; trb) gene homologues are believed to encode the mating pair formation apparatus essential for the physical contact and DNA translocation between donor and recipient cells: for instance, the trbC gene encodes the precursor of the pilin subunit of the conjugal pilus (17). Conjugal DNA transfer is abolished by inactivation of the pilin precursor gene or of any gene involved in assembling the pilus (14, 17).
Despite their early discovery (e.g., references 11, 24, 47, 54, and 56), very little is known about regulation of conjugal transfer of catabolic plasmids in pseudomonads or about their surface requirement for conjugation. Electron microscopy and conjugation frequency observations suggested that the pili encoded by pWW0 (IncP-9, TOL plasmid of P. putida [15]) are thick, flexible, involved in surface-preferred conjugation, and distinct from the rigid pili encoded by IncP-1, IncI, IncW, and IncN plasmids that show surface-obligatory conjugation (4, 54). The plasmids carried by bacteria native to our coal tar-contaminated study site are closely related to the archetypal naphthalene catabolic plasmid pDTG1 (52). This plasmid has recently been sequenced by G. Zylstra (GenBank accession no. AF491307). Several genes in the naphthalene catabolic operon (nahAc [iron sulfur protein, large subunit of naphthalene dioxygenase], nahR [transcriptional regulator for naphthalene degradation], and intergenic region between nahR and nahG [salicylate hydroxylase]) from the naphthalene catabolic plasmid pCg1 in P. putida Cg1 were previously detected with degenerate primer pairs and characterized (20, 43). Sequence analysis showed that these regions were identical to those genes of pDTG1. In this report, putative mpf genes (trb-type genes) were identified on the naphthalene catabolic plasmids pCg1 and pDTG1. Sequence comparisons showed that the mpf genes in pCg1 and pDTG1 are closely related in sequence and operon structure to those in pWW0. Furthermore, the influence of solid surfaces and induction of the mpf genes on conjugal transfer of pDTG1-like plasmids were investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. P. putida NCIB 9816-4 and P. putida Cg1 naphthalene-degrading strains have been described elsewhere (20, 42, 43, 52). P. putida KT2440-Tc was kindly provided by Niels Kroer (Department of Marine Ecology and Microbiology, National Environmental Research Institute, Roskilde, Denmark). All media and growth condition for Escherichia coli strains were prepared as described by Ausubel et al. (1). All Pseudomonas strains were grown at 22°C in mineral salts broth (MSB) and other routine media described elsewhere (21, 52). Rifampin (100 μg/ml), tetracycline (15 μg/ml), kanamycin (100 μg/ml), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg/ml) were added to broth or agar plates when necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| P. putida NCIB 9816-4 | Naphthalene degrader containing the pDTG1 plasmid | 52 |
| P. putida Cg1 | Naphthalene degrader containing the pCg1 plasmid | 52 |
| P. putida Cg1CR | pCg1 plasmid-cured derivative of P. putida Cg1, Rifr | 52 |
| P. putida KT2440-Tc | TOL plasmid-cured derivative of P. putida mt-2 with mini-Tn5 insertion of Tetr | 39 |
| P. putida Cg1pMPF | Derivative of P. putida Cg1, insertion of pMPF in pCg1, Kanr, LacZ+ | This study |
| P. putida Cg1pEM1 | Derivative of P. putida Cg1, insertion of pEM1 in pCg1, Kanr, LacZ+ | This study |
| P. putida Cg1pAH7 | Derivative of P. putida Cg1, insertion of pAH7 in pCg1, Kanr, LacZ+ | This study |
| E. coli INV-αF′ | Host for cloning and maintaining plasmids | Invitrogen |
| E. coli S17-1 λpir | trp SmrrecA thi pro hsdM+ RP4-2-Tc::Mu::Km Tn7λpir; hsdR mutant | 48 |
| Plasmids | ||
| pCR2.1 | Cloning vector, Kanr, Ampr | Invitrogen |
| pVIK110 | R6KoriV, suicide vector, LacZ translation fusion vector, Kanr | 27 |
| pVIK112 | R6KoriV, suicide vector, LacZ transcriptional fusion vector, Kanr | 27 |
| pMPF | Internal mpfA fragment in pVIK110 | This study |
| pAH7 | Putative promoter region of mpf genes cloned into pVIK110 | This study |
| pEM1 | Putative promoter region of mpf genes cloned into pVIK112 | This study |
DNA and plasmid manipulation.
Primers used in this study are listed in Table 2. Restriction fragment length polymorphism (RFLP) characterization of the mpf regions in pDTG1 and pCg1 was performed by using the primer pairs listed in Table 2. In RFLP analysis, each fragment was digested with HaeIII and HhaI (Promega) and was electrophoresed on a 3% agarose gel (Metaphor agarose; BMA). Colony PCR and cloning for sequencing were carried out by the following procedures: a 10-μl volume of sterile water was added to each PCR tube, and bacterial cells were transferred to the PCR tube with a sterile platinum needle from a freshly grown colony. The cells were lysed (95°C for 5 min) in a PTC-200 thermocycler (MJ Research Inc., Watertown, Mass.). The 50-μl volume reaction mixture in each tube contained each primer (0.5 μM), 1× PCR buffer (Gibco BRL, Gaithersburg, Md.), deoxynucleoside triphosphate (0.2 mM), MgCl2 (1.5 mM), and 1.25 U of Taq DNA polymerase (Gibco BRL). A cycling regimen of 94°C for 5 min (one cycle), 94°C for 1 min, 55°C for 30 s, 72°C for 1 min (35 cycles), and 72°C for 5 min (one cycle) was employed. Amplicons were separated by electrophoresis in a 2% agarose gel. Amplified PCR fragments were cloned into the pCR2.1 TA vector (Invitrogen, Carlsbad, Calif.). The constructed plasmids were introduced into E. coli INV-αF′ competent cells (Invitrogen). Both directions of the insert fragments were sequenced, and all sequencing was completed on an ABI model 3700 instrument at the Biological Resource Center, Cornell University. The DNA sequences were aligned by using the MegAlign program, and all sequence comparisons were computed as percent identity by using Clustal (DNASTAR, Madison, Wis.).
TABLE 2.
Primers used in this study to amplify and analyze portions of a 26.8-kb region of naphthalene catabolic plasmids pDTG1 and pCg1a
| Primer name | Sequence (5′→3′) | Corresponding region of 26.886 kb |
|---|---|---|
| Primers used for cloning and sequencing | ||
| mpfA-Fb | CCC GGT ACC CAA ACC CTG CAA AGC GTA ATC | 21145-21125 |
| mpfA-Rb | CCC GTC GAC GGC CAT GCT CTT CAA ACC ACT | 20972-20992 |
| orf25-F | ATG CAG AGC CTA GAC AAA AGA G | 21521-21500 |
| mpfB-R | TCA ACG AGG CGC ATA ACT TCT | 20474-20494 |
| parA-Fb | GGG GGT ACC CAT ATA TAG GGT TTA CTT TTC | 22102-22082 |
| mpfA-R2b | CCC GTC GAC GTC CAA CTT TGG AAG TTT | 21205-21222 |
| tra-Fb | CGC GAA TTC GTC AAA GTC TCA ATA CAC AAC ACG | 2683-2706 |
| traB-Rb | CGC GGT ACC AAT CGA CTT CTC TTT TAT GC | 3316-3297 |
| Primers used for RFLP analysis | ||
| 493F | CGC GTG CCG AAG TGG TC | 493-509 |
| 1272R | TGG TCG GGC AGA TGA TGG AAG ATT | 1272-1249 |
| 1629F | TTG TTG AAG ACC ACC AGA ATC C | 1629-1650 |
| 2437R | TGA GTC AAA GGC AGC GAA AAT ACG | 2437-2414 |
| 4122F | GGA CTC CGA GGG CTA CTT | 4122-4138 |
| 5108R | TCC GAC GAT TAA CTA TTT GAC | 5108-5088 |
| 6779F | CGA CAG AAG AAC CAG GAA CTA AAA | 6779-6802 |
| 7510R | TGT AAG ACC TTG GGC GCT GTG GAC | 7510-7487 |
| 9422F | TCT TTG GCC ATG TCC TAA TCC TCT | 9422-9445 |
| 10591R | GCT CGC GCT ATG GTT TCT AAG TTG | 10591-10568 |
| 12475F | ACG CGG CCA GGT TTA CGA ATA CA | 12475-12497 |
| 13114R | GCG CGG GTG CCT CCA ACT CC | 13114-13095 |
| 17460F | GTG TTG ACC AGT TCC TCG TAA T | 17460-17481 |
| 18239R | TCA TGC AGA TCC GCC GTA AAA | 18239-18219 |
| 19699F | GCT CAG CTC GCA GTA TTC CT | 19699-19718 |
| 20391R | GAG TTG GTG GTC AGC GAG ATG | 20391-20371 |
Nucleotide positions 1 and 26886 correspond to the extreme left and right, respectively, of Fig. 2. The mpf gene cluster (Fig. 2) is bounded by positions 8794 and 21232.
Restriction enzyme sites, used for cloning purposes, are underlined; when restriction enzyme sites were inserted into a primer, three additional nucleotides were added to the 5′ end.
Construction of polar mutation in the mpf region.
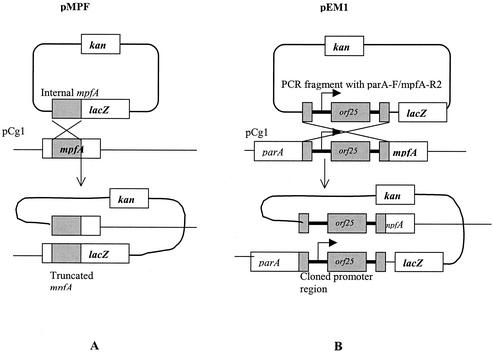
The suicide translational fusion vector pVIK110 (27) was used for constructing a polar mutation of mpf regions (Fig. 1A). Primer pair mpfA-F/mpfA-R was designed to target an internal region of a putative mpfA gene. The amplified 192-bp fragment of the internal mpfA fragment gene was cloned into the KpnI-SalI cloning sites of pVIK110, creating the pMPF plasmid (full sequencing of the modification of pVIK110 verified that the sought genetic construct was achieved [Fig. 1A]), which was introduced into E. coli S-17-1 λpir. Then conjugation was performed by filter mating with E. coli S-17-1 λpir (pMPF) and P. putida Cg1 as donor and recipient bacteria, respectively. The transconjugants (P. putida Cg1pMPF) were selected on MSB-P medium (MSB plus 0.1% pyruvate) containing kanamycin at room temperature (22°C). Confirmation of transconjugants was conducted by PCR using primers specific to pCg1 or pVIK110. The expected PCR fragment was amplified only in the transconjugant (data not shown).
FIG. 1.
Construction of pMPF and pEM1 via Campbell-type homologous recombination with the pCg1 plasmid. The internal regions are depicted as a gray box. pMPF was designed to create a polar mutation (knockout) of mpfA. pEM1 reported transcriptional activity of the mpfA operon.
RT-PCR.
Reverse transcriptase PCR (RT-PCR) was performed on total RNA extracted from P. putida Cg1. Primers (orf25-F, mpfA-R, and mpfB-R) used for RT-PCR are listed in Table 2. Total RNA was extracted according to the manufacturer's procedure (RNeasy mini kit; Qiagen). The extracted total RNA was treated with DNase I (Gibco BRL) for 15 min at 37°C. DNase I was inactivated with boiling at 70°C for 10 min. Reverse transcription and PCR were performed by using Superscript II RT (Gibco BRL) as follows: the 50-μl volume reaction mixture in each tube contained each primer (1 μM), 2× reaction buffer (Gibco BRL), and 1 μl of RT/Taq polymerase mixture (Gibco BRL). A cycling regimen of 50°C for 20 min and 94°C for 2 min (one cycle) for cDNA synthesis and of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min (35 cycles), and 72°C for 5 min (one cycle) for PCR was used. To confirm that cDNA synthesis occurred and that RNA preparation was free of genomic DNA, negative control RT-PCR was performed only with Taq polymerase.
Construction of mpfA-lacZ fusions.
Two independent reporter fusions were constructed and used, i.e., pEM1 and pAH7. The suicide transcriptional fusion vector pVIK112 (27) was used for constructing an mpfA-lacZ transcriptional fusion reporter system (Fig. 1B). The PCR fragment containing parA-F and mpfA-R2 (Table 2) was cloned into TA cloning vector pCR2.1. EcoRI-digested fragments were cloned into the EcoRI site of pVIK112, creating plasmid pEM1. In addition, the suicide translational fusion vector pVIK110 was used for constructing a translational fusion reporter system. For this, the PCR fragment with parA-F and mpfA-R2 was cloned into pVIK110, creating pAH7 (analogous to pEM1 in Fig. 1B), after digestion with KpnI and SalI restriction enzymes. The orientation of the fragments was confirmed by sequencing the cloned region in the pEM1 and pAH7 plasmids. Then the same cloning and transformation procedures described for creating the polar mutation in the mpf region (above) were applied in constructing P. putida Cg1pEM1 (P. putida Cg1 containing mpfA-lacZ transcriptional fusion on pCg1) and P. putida Cg1pAH7 (P. putida Cg1 containing mpfA-lacZ translational fusion on pCg1).
β-Galactosidase activity assay.
β-Galactosidase activity was measured by using a microtiter plate modification (49) of the original method of Miller (35). β-Galactosidase activities were measured by using o-nitrophenyl β-d-galactopyranoside (10 mg of it per ml in 0.1 M KPO4, pH 7.0) as a substrate. MSB-N (MSB plus 0.1% naphthalene crystals) and MSB-G (MSB plus 10 mM glucose) were used as the minimal medium.
Conjugation frequency.
P. putida Cg1pEM1 served as the donor and P. putida KT2440-Tc (39) served as the recipient in standard filter mating as described elsewhere (52). Donor cells (≈107 to 109, grown overnight in Luria-Bertani [LB] broth, in MSB-G and in MSB-N amended with 100 μg of kanamycin/ml) were harvested and washed with phosphate-buffered saline (PBS) (1). To the same tube, ≈107 to 109 recipient cells grown overnight in LB broth were added and harvested by centrifugation. For solid medium conjugation, cells were resuspended in 50 μl of PBS, collected on a sterile filter (0.2-μm pore size, 25-mm diameter; Millipore, Bedford, Mass.), and placed on an LB or MSB-G plate. The plate contents were incubated for 1 to 24 h, agar side down, at room temperature. After mating, filters were removed and vortexed in 1 ml of PBS for 30 s to disperse the cells. For liquid medium conjugation, cells were resuspended with 50 μl of LB or MSB-G broth and were incubated at room temperature. After mating for 1 to 24 h, serial dilutions were prepared and transconjugants were enumerated on LB plates amended with kanamycin and tetracycline. Donor cells only were enumerated on LB plates amended with kanamycin, and recipient cells only were enumerated on LB plates amended with tetracycline. Donors were also plated on LB amended with kanamycin and tetracycline to account for spontaneous tetracycline resistance, while recipients were also plated onto LB amended with kanamycin and tetracycline to account for potential experimental errors. No spontaneous mutants of donor or recipient cells appeared on the selective media during the entire conjugation assays.
Nucleotide sequence accession number.
The nucleotide sequences of the entire pDTG1 plasmid of P. putida NCIB 9816-4 have been deposited by G. Zylstra (Rutgers University) in GenBank under accession no AF491307. The nucleotide-numbering system in this paper differs from that of the GenBank submission. Nucleotides 1 to 28886 described here correspond to nucleotides 61380 to 83042 and 1 to 5223 in GenBank. The nucleotide sequences of pCg1 covering traA (634 bp) and orf25-mpfB (1,629 bp) have been deposited in GenBank under accession numbers AY249146 and AY249147.
RESULTS
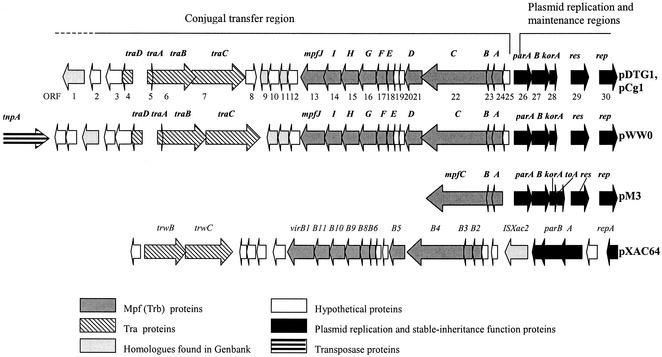
Identification and sequence analysis of mpf regions from plasmid pCg1.
The map of a 26.8-kb portion of pDTG1/pCg1 shows that it has tra and trb (referred to henceforth as mpf) regions (Fig. 2). To confirm and extend prior measures (43, 52) of homology between pDTG1 and pCg1, we conducted RFLP analysis of PCR-amplified tra and mpf regions by using primers listed in Table 2 (primers 493F/1272R [780-bp product], 1629F/2437R [809-bp product], 4122F/5108R [987-bp product], 6779F/7510R [732-bp product], 9422F/10591R [1,170-bp product], 12475F/13114R [639-bp product], 17460F/18239R [780-bp product]; and 19699F/20391R [693-bp product]). Over a ∼20-kb region both plasmids showed indistinguishable patterns (data not shown). Sequence comparison of the fragments amplified with primer pairs (tra-F/traB-R, parA-F/mpfA-R2, and orf25-F/mpfB-R; ∼2.5 kb) confirmed that in these regions pCg1 was 100% identical to pDTG1.
FIG. 2.
Comparison of the gene order in mating pair formation (mpf) and tra gene clusters in ∼30-kb regions of five related plasmids.
As shown in Fig. 2 and Table 3, there is considerable similarity in both the order and sequence of many open reading frames in the putative transfer regions in pCg1, pDTG1, pWW0 (TOL plasmid in P. putida; IncP-9 incompatibility group [15]), and pM3 (IncP-9 incompatibility group; found in 16 isolates of P. putida species originating from farmland soil in Belarus, from industrial site soil in Belarus and Azerbaijan, and from sewage from a pharmaceutical plant in Belarus [16]). Nine of the 10 genes in the mpf cluster in pDTG1/pCg1 have their closest homologue (65 to 87% identity) in pWW0. The closest match to the remaining gene (mpfA) was in pM3 (73% identity). Two other matches between pDTG1/pCg1 and pM3 homologues were also high (70 and 77% identity). Figure 2 further reveals that there are four tra-like genes shared by pWW0 and pDTG1/pCg1. The oriT region of pWW0 was identified between the traD and traA genes (15). The putative oriT site of pDTG1/pCg1 is also present between traD and traA genes, and its sequence is 100% identical to the oriT site of pWW0.
TABLE 3.
Summary of computer-based nucleotide sequence analyses of open reading frames in the conjugal transfer regions in pDTG1/pCg1 plasmids
| ORFa | Geneb | aaf length | Mol mass (kDa) | pI | BLAST resultc | Degree of relatedness (aa pDTG1/aa match, % aa identity)c | Motif found in GenBank |
|---|---|---|---|---|---|---|---|
| orf1 | hyp | 261 | 28.8 | 6.22 | Hyp of pWW0 | 261/263, 77 | |
| Mpr (Zinc metalloproteinase) of IncN plasmid R64 | 261/263, 41 | ||||||
| orf2 | hyp | 128 | 13.8 | 6.67 | Hyp of pWW0 | 128/130, 67 | |
| orf3 | hyp | 239 | 26.5 | 5.33 | Hyp of pWW0 | 239/239, 73 | |
| orf4 | traD | 151 | 16.7 | 8.90 | TraD of pWW0 | 151/151, 60 | |
| orf5 | traA | 72 | 7.9 | 4.48 | TraA of pWW0 | 72/127, 56 | |
| orf6 | traB | 518 | 58.3 | 6.97 | TraB of pWW0 | 518/516, 86 | |
| TrwB of pXAC64 | 518/524, 65 | ||||||
| orf7 | traC | 981 | 109.0 | 9.46 | TraC of pWW0 | 981/978, 80 | Viral helicase I |
| TrwC of pXAC64 | 981/991, 63 | ||||||
| orf8 | hyp | 193 | 21.9 | 10.03 | Hyp (ORF7) of pWW0 | 193/244, 40 | |
| orf9 | nuc | 212 | 22.9 | 9.09 | Putative nuclease (ORF188) of pWW0 | 212/210, 60 | |
| orf10 | hyp | 160 | 17.7 | 6.51 | Putative nuclease (ORF188) of pWW0 | 160/210, 23 | |
| Hyp (XACb0057) of pXAC64 | 160/395, 23 | ||||||
| orf11 | nuc | 201 | 21.6 | 8.39 | Putative nuclease (ORF189) of pWW0 | 201/185, 73 | |
| Endonuclease of IncN | 201/177, 48 | ||||||
| Plasmid R64 | |||||||
| orf12 | hyp | 109 | 11.7 | 8.47 | Hyp of pXAC64 | 109/111, 68 | |
| orf13 | mpfJ | 311 | 33.6 | 7.21 | MpfJ of pWW0 | 311/315, 65 | Transglycosylase (SLT domaind) |
| VirB1 of pXAC64 | 311/292, 35 | ||||||
| orf14 | mpfI | 342 | 38.3 | 6.13 | Mpf of pWW0 | 342/343, 81 | GSPIIe (bacterial type II secretion) |
| VirB11 of pXAC64 | 342/340, 44 | ||||||
| orf15 | mpfH | 427 | 45.5 | 5.54 | MpfH of pWW0 | 427/421, 65 | |
| VirB10 of p XAC64 | 427/406, 40 | ||||||
| orf16 | mpfG | 263 | 29.1 | 9.04 | MpfG of pWW0 | 263/261, 87 | |
| VirB9 of pXAC64 | 263/260, 52 | ||||||
| orf17 | mpfF | 221 | 25.4 | 8.90 | MpfF of pWW0 | 221/226, 66 | |
| VirB8 of pXAC64 | 221/224, 36 | ||||||
| orf18 | mpfE | 287 | 31.1 | 5.30 | MpfE of pWW0 | 287/287, 73 | |
| VirB6 of pXAC64 | 287/288, 43 | ||||||
| orf19 | hyp | 134 | 14.2 | 8.28 | Hyp of pWW0 | 134/130, 67 | |
| orf20 | hyp | 70 | 7.9 | 9.60 | Hyp of pWW0 | 70/70, 73 | |
| orf21 | mpfD | 229 | 24.9 | 5.46 | MpfD of pWW0 | 229/229, 84 | |
| VirB5 of pXAC64 | 229/220, 55 | ||||||
| orf22 | mpfC | 894 | 101.7 | 5.79 | MpfC of pWW0 | 894/894, 75 | |
| TrbE of pM3 | 894/339, 70 | ||||||
| VirB4 of pXAC64 | 894/877, 56 | ||||||
| orf23 | mpfB | 97 | 10.8 | 9.84 | MpfB of pWW0 | 97/98, 77 | |
| TrbD of pM3 | 97/98, 77 | ||||||
| VirB3 of pXAC64 | 97/99, 47 | ||||||
| orf24 | mpfA | 153 | 16.8 | 10.01 | TrbC of pM3 | 153/153, 73 | |
| MpfA of pWW0 | 153/153, 71 | ||||||
| VirB2 of pXAC64 | 153/125, 27 | ||||||
| orf25 | hyp | 90 | 10.0 | 6.55 | Hyp of pWW0 | 90/115, 81 | |
| Hyp of pXAC64 | 90/118, 43 |
See Fig. 2 for position of open reading frames (ORFs) in the conjugal transfer regions.
hyp, hypothetical.
1:1 Clustal alignment between the two proteins over the entire sequences.
Soluble lytic domain.
General secretory pathway II.
aa, amino acid.
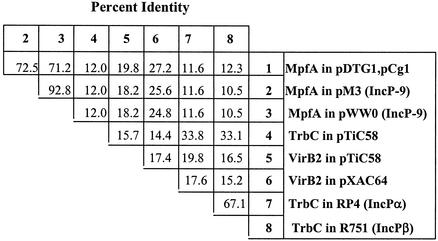
An extended comparison of mpfA-related proteins (Fig. 3) confirms close relatedness between pDTG1/pCg1 and pM3/pWW0 (∼72% identity) and distant relatedness (11.6% identity) to well-characterized trbC genes of broad-host-range enteric plasmids IncPα (RP4) and IncPβ (R751). The matrix in Fig. 3 also shows a moderate match between MpfA and VirB2 of pXAC64 of Xanthomonas axonopodis pv. citri. (8) and pTiC58 of Agrobacterium tumefaciens. Relatedness is confirmed by sequence comparison between structural genes adjacent to mpfA and corresponding genes in VirbB (pXAC64) and Trb (pM3) systems (Table 3; Fig. 2).
FIG. 3.
Similarity matrix comparing percent identity scores for complete pilin subunit proteins encoded on seven plasmids. The scores were calculated by using the MEGALIGN program (LASER-GENE package; DNASTAR Inc.).
Insertional inactivation of the mpfA gene.
The mpfA gene is predicted to encode the precursor of the pilin subunit of the conjugal pilus based on homology with other known pilin subunit genes. To test the hypothesis that mpfA of pCg1 is critical for conjugation, perhaps playing the role of other trbC-type products, we disrupted it by using Campbell-type single-crossover recombination (Fig. 1A). The insertional inactivation used was designed to cause a polar mutation and disrupt other genes downstream of mpfA as well, should expression of these be controlled by a shared promoter. A standard filter mating was performed between P. putida Cg1pMPF and P. putida Cg1CR (52). Conjugation was not detected. The number of transconjugants and the number of spontaneous rifampin-resistant colonies per donor were equal (10−7). In contrast, the positive control filter mating performed between wild-type P. putida Cg1 donor and P. putida Cg1CR recipient yielded 10−3 transconjugants per donor and 10−8 spontaneous rifampin resistance colonies per donor (22). This experiment showed that the mpf region of pCg1 plasmid is essential for conjugation.
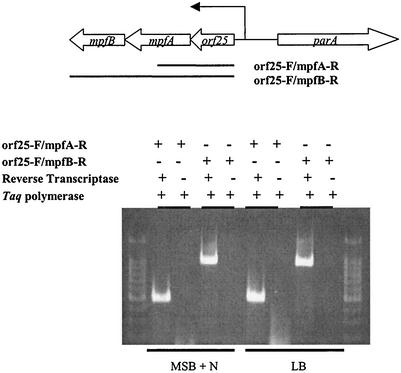
Detection of mpf gene transcripts by RT-PCR.
Computer analysis (Neural Network Promoter Prediction; http://www.fruitfly.org/seq_tools/promoter.html) showed that there is one putative promoter region upstream of mpfA and orf25 (hypothetical protein) (Fig. 2 and 4). To analyze expression of the mpf region, we extracted DNA-free total RNA from P. putida Cg1 grown in MSB liquid medium with naphthalene as the carbon source (MSB-N) or in LB broth. The extracted RNA was used as the template for RT reactions. Subsequently, the cDNA template was amplified with PCR primers yielding products corresponding to two intercistronic regions, i.e., orf25-mpfA and orf25-mpfA-mpfB (Fig. 4). Total RNA extracted from naphthalene-grown cells and LB broth-grown cells both showed amplified products of the anticipated sizes (Fig. 4). Negative control treatments that omitted RT did not yield any amplified products. This result (Fig. 4) indicated that expression of the mpf region was not linked to naphthalene utilization and that orf25, mpfA, and mpfB are cotranscribed.
FIG. 4.
RT-PCR analysis assay for detecting pCg1 plasmid gene expression (mRNA transcripts) in P. putida Cg1. Diagram at top shows construct used to probe for mpf gene expression. Middle diagram shows four pairs of treatments that verify transcript detection. Bottom gel image shows that the mpf genes were expressed in cells grown in LB and MSB-N media.
Expression of PmpfA-lacZ reporter construct in P. putida Cg1.
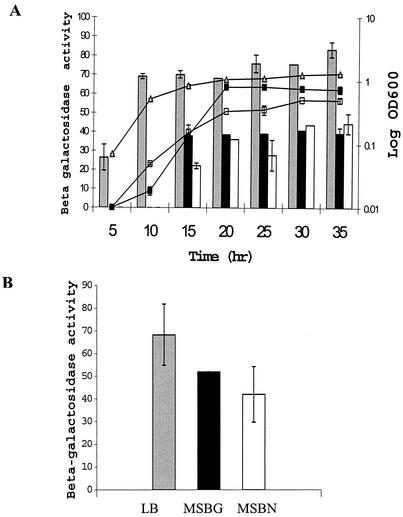
For quantification of expression of the mpfA gene, we constructed an mpfA promoter transcriptional fusion to lacZ (PmpfA-lacZ). This construct, pEM1 (Fig. 1B), contained the fragment extending from a portion of partial parA through orf25 to just downstream of the ATG start site of the mpfA gene (+28 nucleotides). We believe that this construct contained the promoter region of the mpf operon based on computer analysis and operon structure. We measured β-galactosidase activity in P. putida Cg1pEM1 (22°C) in cells harvested from both liquid (Fig. 5A) and solid (Fig. 5B) preparations of three media. Promoter activity of PmpfA-lacZ was increased when cells were grown in rich medium (LB). The level of β-galactosidase activity of cells grown in LB liquid medium was roughly two times higher than that of cells grown in MSB-N and MSB-G during the exponential and stationary phases (Fig. 5A). The β-galactosidase activity measured by using cells grown in MSB-G liquid medium was similar to the β-galactosidase activity of cells in MSB-N liquid medium (Fig. 5A). Thus, this expression was not linked to naphthalene catabolic function of the pCg1 plasmid. The expression of mpf genes in the agar media was induced to levels similar to those in liquid media (Fig. 5B). The RT-PCR assay (that produced the data in Fig. 4) and the PmpfA-lacZ reporter assays showed that the mpf genes of pCg1 were constitutively expressed even in the absence of naphthalene and were highly expressed both in liquid and on semisolid media.
FIG. 5.
Use of pEM1, containing a PmpfA-lacZ fusion, to examine the influence of medium composition and growth phase on expression of conjugal transfer genes in P. putida Cg1. Expression of mpfA during growth in three different liquid media (A) and after 1 day of growth on agar (B). Labels for media appear in panel B. Each treatment was replicated three times. Error bars show standard deviation. β-galactosidase activities are expressed in Miller units. OD600, optical density at 600 nm.
Because we recognize that the level of transcription is not the same as the level of expression, we attempted to monitor the promoter activity of the mpf region by using a translational fusion construct in P. putida Cg1pAH7. However, that construct failed to express β-galactosidase activity in any condition. The expression of β-galactosidase did not occur on semisolid or liquid media in the presence of recipient cells (22). An explanation for failure to detect β-galactosidase activity in P. putida Cg1pAH7 is not straightforward. It is possible that the constructed translational fusion suffered from unstable transcripts or impaired initiation of translation (44).
Conjugation frequency of the pCg1 plasmid of P. putida Cg1.
Because the reporter assay above indicated that the expression of mpf region is constitutive and is higher in rich (LB) media than in minimal (MSB-G or MSB-N) media, we predicted that the conjugation frequency would be higher in LB-grown cells if the expression of the mpf region is the rate-limiting factor that governs conjugation frequency. We completed conjugation assays examining the influence of medium and other key variables (mating duration and the presence of a solid surface) on conjugation frequency (Table 4). Comparison of columns 5 and 6 shows that conjugal transfer of pCg1 was consistently higher (∼100-fold) on solid media. The pattern within the first five lines of Table 4 (in which donor medium was held constant but mating time increased) indicates that conjugal transfer was roughly proportional to the duration of cell-cell contact. However, extensive incubation of donor and recipient cells beyond 6 (to 24) h did not increase the conjugation frequency. In addition, the trend in assays in which donor cells were prepared in minimal salt media (last four entries [Table 4]) indicates that a longer period was required for successful mating than for donor cells grown on LB broth. Data in Table 4 indicate that several factors, in addition to expression of the mpf region, influence conjugation frequency.
TABLE 4.
Influence of mating time, medium, and solid surface on conjugal transfer frequencies of pCg1 between P. putida Cg1 (host) and P. putida KT2440-Tc (recipient)
| Donor and recipient density in mating mix | Mating duration (h) | Donor medium | Conjugation frequencya
|
||
|---|---|---|---|---|---|
| Donor | Recipient | Solid | Liquid | ||
| 1.5 × 108 (1.30)b | 2.7 × 108 (1.50) | 1 | LB | 1.5 × 10−6 | BDc |
| 3.6 × 108 (0.98) | 3.8 × 108 (1.30) | 3 | LB | 2.7 × 10−7 | 1.3 × 10−9 |
| 3.2 × 107 (0.61) | 7.4 × 107 (0.90) | 3 | LB | 4.4 × 10−5 | 4.6 × 10−7 |
| 2.3 × 108 (1.10) | 3.8 × 108 (1.30) | 6 | LB | 1.3 × 10−4 | 3.9 × 10−6 |
| 1.9 × 108 (0.90) | 1.9 × 109 (0.70) | 24 | LB | 1.9 × 10−4 | 1.1 × 10−5 |
| 2.0 × 107 (1.10) | 2.7 × 108 (1.50) | 1 | MSB-G | BD | BD |
| 1.8 × 108 (0.99) | 5.8 × 108 (1.05) | 3 | MSB-G | BD | BD |
| 9.8 × 108 (0.95) | 6.0 × 106 (0.70) | 6 | MSB-G | 3.7 × 10−7 | BD |
| 2.4 × 108 (1.05) | 6.0 × 106 (0.70) | 6 | MSB-N | 3.9 × 10−6 | BD |
Frequencies are expressed as the number of transconjugants per donor cell (transconjugant/donor ratio).
Measurement of optical density at 600 nm is given in parentheses.
BD, below detection limit.
DISCUSSION
Among the motivations for this study was a desire to understand the mechanism by which naturally occurring microbial populations at a coal tar-contaminated field site (2, 20, 23, 33) adapt to introduced organic substrates. Previously it has been shown that Cg1-like plasmids, involved in horizontal transfer of naphthalene catabolic genes between bacteria at the study site, are homologous to the archetypal naphthalene catabolic plasmid pDTG1 from P. putida NCIB 9816-4 (despite some variability in overall plasmid sizes [e.g., pDTG1, 81 kb; and pCg1, 86 kb {52}]). This led us to investigate further how pCg1 conjugation occurs and is regulated. The entire pDTG1 plasmid has been sequenced (GenBank accession no. AF491307). In this study we have shown by using RFLP and sequencing analysis that the conjugal transfer regions in pCg1 and pDTG1 are closely related in sequence and operon structure to the putative mpf genes of broad-host-range plasmids pWW0 and pM3. The incompatibility group of pDTG1 and pCg1 has not been determined. Nonetheless, we feel it is prudent to deem these plasmids “IncP-9-like” because pWW0 and pM3 and the closely related plasmid from P. putida NCIB 9816-3 all belong to the IncP-9 group (15, 52, 56). The mpf genes of IncP-9 plasmids along with vir genes of pXAC64 share distinct sequence identity and operon structure; thus, evolutionary divergence from other plasmids is suggested.
We recognize that complex networks of transcriptional and translational regulatory systems often control plasmid conjugation (57-60). For example, in the trb operon (the genes for mating pair formation start with trbB) of plasmid RK2, it is thought that translational control on trbB transcripts is exerted by the trbA promoter (responsible for steady-state transcription of the trb operon as well as trbA). Furthermore, repression of the trbB promoter is exerted by both KorB and TrbA proteins (25, 36, 58, 60). Thus, it is likely that conjugal transfer genes in enteric IncPα plasmids (e.g., RK2 and RP4) are controlled by a network of regulatory elements (regulators and DNA sequences) that reduce the metabolic burden on the bacterial host. It is very probable that a regulatory system unlike that of the IncPα system operates in pDTG1/pCg1. Our data suggest that the mpf region of pCg1 is not controlled at the level of transcription because transcription was constitutive in the promoter-probing construct in P. putida Cg1pEM1 (Fig. 5).
It is notable that, although transcription of the mpf region of pCg1 occurs constitutively, conjugation frequency is low (Table 4). To place this frequency in perspective, the TOL plasmid of P. putida mt-2 (also thought to be naturally derepressed for conjugation) transfers at 2.5 × 10−1 transconjugants per donor h−1 (4). Consistent with data reported here on the key role of the mpfA pilin subunit in pCg1 conjugation, the thick, flexible conjugative pili encoded by the TOL plasmid (IncP-9) in P. putida mt-2 were found to be important for plasmid transfer (4). Interestingly, conjugation frequency of the TOL plasmid depends on the identity of the host cell. The number of transconjugants per donor cell dropped 1,000-fold when the host was altered from P. putida mt-2 to P. putida S388 (4). We have similar observations for conjugation of pDTG1/pCg1-like plasmids: the conjugation frequency of pCg1 in P. putida Cg1 after overnight mating on the filter is 10−3 to 10−4 (transconjugants per donor), yet conjugation is often below the detection limit (10−8) in the case of pDTG1 in P. putida NCIB 9816-4 (20, 43, 52). We conclude then (presuming that any sequence differences between pDTG1 and pCg1 are immaterial) that conjugation of pDTG1/pCg1-like plasmids is highly dependent upon the host in which the plasmids reside.
Systematic observation of relationships between solid surfaces and plasmid transfer has led to the proposal of three categories of conjugation (3, 4, 15): universal mating type (equal conjugation frequency on solid and in liquid medium; e.g., IncK, IncFII, and IncH1), surface-obligatory mating type (conjugation frequency is >2,000 times higher on a solid surface; e.g., IncP-1, IncI, IncW, and IncN), and the solid-preferred type (conjugation frequency is 45 to 450 times higher on a solid surface; e.g., IncP-7 and IncP-9). Data presented here clearly place pCg1 in the latter class (Table 4). Surface-preferred conjugation of pCg1 may be the result of IncP-9-type pili and related, stable mating pair formation on a semisolid surface, where high-density cell-to-cell contact is fostered. Solid surface-preferred conjugation is also observed in the Ti plasmids of Agrobacterium (13, 45). It has been thought that this surface, preference might result from the thick, flexible morphology of pili (12) or from biofilm formation that might increase conjugation frequency with quorum-sensing molecules (13, 45). Based on the observations above and discussion by others (4, 6, 7, 19, 37-39, 45, 46, 50), conjugation frequency is confirmed to be governed by many factors, including cell density, surfaces, growth rate, incubation time, media, pH, and moisture. Clearly the mpfA and/or cotranscribed genes in the conjugal transfer region of pCg1 are necessary but not sufficient for successful conjugation.
Acknowledgments
This research was supported by National Science Foundation grants MCB-0084175 (to E.L.M.) and MCB-0078465 (to G.J.Z.).
Constructive criticism from three anonymous reviewers was appreciated.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struthl. 1999. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bakermans, C., and E. L. Madsen. 2002. Diversity of 16S rDNA and naphthalene dioxygenase genes from coal-tar-waste-contaminated aquifer waters. Microb. Ecol. 44:95-106. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, D. E., D. E. Taylor, and D. R. Cohen. 1980. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J. Bacteriol. 143:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley, D. E., and P. A. Williams. 1982. The TOL plasmid is naturally derepressed for transfer. J. Gen. Microbiol. 128:3019-3024. [DOI] [PubMed] [Google Scholar]
- 5.Bushman, F. 2002. Lateral gene transfer. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Christensen, B. B., C. Sternberg, and S. Molin. 1996. Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (GFP) from Aequorea victoria as a marker. Gene 173:59-65. [DOI] [PubMed] [Google Scholar]
- 7.Dahlberg, C., M. Bergstrom, and M. Hermansson. 1998. In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl. Environ. Microbiol. 64:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Sluys, N. F. Almeida, L. M. Alves, A. M. Do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. De Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade Dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 9.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 10.Droge, M., A. Puhler, and W. Selbitschka. 1998. Horizontal gene transfer as a biosafety issue: a natural phenomenon of public concern. J. Biotechnol. 64:75-90. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, N. W., and I. C. Gunsalus. 1973. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J. Bacteriol. 114:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost, L. S. 1993. Conjugative pili and pilus-specific phages. Plenum, New York, N.Y.
- 13.Fuqua, W. C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuya, N., and T. Komano. 1996. Nucleotide sequence and characterization of the trbABC region of the IncI1 plasmid R64: existence of the pnd gene for plasmid maintenance within the transfer region. J. Bacteriol. 178:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greated, A., L. Lambertsen., P. A. Williams, and C. M. Thomas. 2002. Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4:856-871. [DOI] [PubMed] [Google Scholar]
- 16.Greated, A., M. Titok, R. Krasowiak, R. J. Fairclough, and C. M. Thomas. 2000. The replication and stable-inheritance functions of IncP-9 plasmid pM3. Microbiology 146:2249-2258. [DOI] [PubMed] [Google Scholar]
- 17.Haase, J., R. Lurz, A. M. Grahn, D. H. Bamford, and E. Lanka. 1995. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J. Bacteriol. 177:4779-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallier-Soulier, S., V. Ducrocq, and N. Truffaut. 1999. Conjugal transfer of a TOL-like plasmid and extension of the catabolic potential of Pseudomonas putida F1. Can. J. Microbiol. 45:898-904. [PubMed] [Google Scholar]
- 19.Hausner, M., and S. Wuertz. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65:3710-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrick, J. B., K. G. Stuart-Keil, W. C. Ghiorse, and E. L. Madsen. 1997. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl. Environ. Microbiol. 63:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohnstock, A. M., K. G. Stuart-Keil, E. E. Kull, and E. L. Madsen. 2000. Naphthalene and donor cell density influence field conjugation of naphthalene catabolism plasmids. Appl. Environ. Microbiol. 66:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hohnstock-Ashe, A. M. 2002. Ph.D. thesis. Cornell University, Ithaca, N.Y.
- 23.Hohnstock-Ashe, A. M., S. M. Plummer, R. M. Yager, P. Baveye, and E. L. Madsen. 2001. Further biogeochemical characterization of a trichloroethene-contaminated fractured dolomite aquifer: electron source and microbial communities involved in reductive dechlorination. Environ. Sci. Technol. 35:4449-4456. [DOI] [PubMed] [Google Scholar]
- 24.Jacoby, G. A., and J. A. Shapiro. 1977. Plasmids studied in Pseudomonas aeruginosa and other pseudomonads, p. 639-656. In A. I. Bukhari, J. A. Shapiro, and S. L. Adhya (ed.), DNA insertion elements, plasmids and episomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Jagura-Burdzy, G., D. P. Macartney, M. Zatyka, L. Cunliffe, D. Cooke, C. Huggins, L. Westblade, F. Khanim, and C. M. Thomas. 1999. Repression at a distance by the global regulator KorB of promiscuous IncP plasmids. Mol. Microbiol. 32:519-532. [DOI] [PubMed] [Google Scholar]
- 26.Junker, F., and A. M. Cook. 1997. Conjugative plasmids and the degradation of arylsulfonates in Comamonas testosteroni. Appl. Environ. Microbiol. 63:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalogeraki, V. S., and S. C. Winans. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69-75. [DOI] [PubMed] [Google Scholar]
- 28.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lessl, M., D. Balzer, K. Weyrauch, and E. Lanka. 1993. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J. Bacteriol. 175:6415-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lessl, M., and E. Lanka. 1994. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77:321-324. [DOI] [PubMed] [Google Scholar]
- 32.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 33.Madsen, E. L., J. L. Sinclair, and W. C. Ghiorse. 1991. In situ biodegradation: microbiological patterns in a contaminated aquifer. Science 252:830-833. [DOI] [PubMed] [Google Scholar]
- 34.Margesin, R., and F. Schinner. 1997. Heavy metal resistant Arthrobacter sp.—a tool for studying conjugational plasmid transfer between gram-negative and gram-positive bacteria. J. Basic Microbiol. 37:217-227. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Motallebi-Veshareh, M., D. Balzer, E. Lanka, G. Jagura-Burdzy, and C. M. Thomas. 1992. Conjugative transfer functions of broad-host-range plasmid RK2 are coregulated with vegetative replication. Mol. Microbiol. 6:907-920. [DOI] [PubMed] [Google Scholar]
- 37.Muela, A., M. Pocino, I. Arana, J. I. Justo, J. Iriberri, and I. Barcina. 1994. Effect of growth phase and parental cell survival in river water on plasmid transfer between Escherichia coli strains. Appl. Environ. Microbiol. 60:4273-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neilson, J. W., K. L. Josephson, I. L. Pepper, R. B. Arnold, G. D. Di Giovanni, and N. A. Sinclair. 1994. Frequency of horizontal gene transfer of a large catabolic plasmid (pJP4) in soil. Appl. Environ. Microbiol. 60:4053-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Normander, B., B. B. Christensen, S. Molin, and N. Kroer. 1998. Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris). Appl. Environ. Microbiol. 64:1902-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogunseitan, O. A. 1995. Bacterial genetic exchange in nature. Sci. Prog. 78:183-204. [PubMed] [Google Scholar]
- 41.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 42.Park, W., C. O. Jeon, and E. L. Madsen. 2002. Interaction of NahR, a LysR-type transcriptional regulator, with the alpha subunit of RNA polymerase in the naphthalene degrading bacterium, Pseudomonas putida NCIB 9816-4. FEMS Microbiol. Lett. 213:159-165. [DOI] [PubMed] [Google Scholar]
- 43.Park, W., P. Padmanabhan, S. Padmanabhan, G. J. Zylstra, and E. L. Madsen. 2002. nahR, encoding a lysR-type transcriptional regulator, is highly conserved among naphthalene degrading bacteria isolated from a coal tar waste-contaminated site and in extracted community DNA. Microbiology 148:2319-2329. [DOI] [PubMed] [Google Scholar]
- 44.Pessi, G., C. Blumer, and D. Haas. 2001. lacZ fusions report gene expression, don't they? Microbiology 147:1993-1995. [DOI] [PubMed] [Google Scholar]
- 45.Piper, K. R., and S. K. Farrand. 1999. Conjugal transfer but not quorum-dependent tra gene induction of pTiC58 requires a solid surface. Appl. Environ. Microbiol. 65:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richaume, A., J. S. Angle, and M. J. Sadowsky. 1989. Influence of soil variables on in situ plasmid transfer from Escherichia coli to Rhizobium fredii. Appl. Environ. Microbiol. 55:1730-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serdar, C. M., and D. T. Gibson. 1989. Isolation and characterization of altered plasmids in mutant strains of Pseudomonas putida NCIB 9816. Biochem. Biophys. Res. Commun. 164:764-771. [DOI] [PubMed] [Google Scholar]
- 48.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 49.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smets, B. F., B. E. Rittmann, and D. A. Stahl. 1993. The specific growth rate of Pseudomonas putida PAW1 influences the conjugal transfer rate of the TOL plasmid. Appl. Environ. Microbiol. 59:3430-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Starodub, M. E., and J. T. Trevors. 1990. Mobilization of Escherichia coli R1 silver-resistance plasmid pJT1 by Tn5-Mob into Escherichia coli C600. Biol. Met. 3:24-27. [DOI] [PubMed] [Google Scholar]
- 52.Stuart-Keil, K. G., A. M. Hohnstock, K. P. Drees, J. B. Herrick, and E. L. Madsen. 1998. Plasmids responsible for horizontal transfer of naphthalene catabolism genes between bacteria at a coal tar-contaminated site are homologous to pDTG1 from Pseudomonas putida NCIB 9816-4. Appl. Environ. Microbiol. 64:3633-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Syvanen, M., and C. I. Kado. 2002. Horizontal gene transfer, 2nd ed. Academic Press, San Diego, Calif.
- 54.White, G. P., and N. W. Dunn. 1978. Compatibility and sex specific phage plating characteristics of the TOL and NAH catabolic plasmids. Genet. Res. 32:207-213. [DOI] [PubMed] [Google Scholar]
- 55.Worning, P., L. J. Jensen, K. E. Nelson, S. Brunak, and D. W. Ussery. 2000. Structural analysis of DNA sequence: evidence for lateral gene transfer in Thermotoga maritima. Nucleic Acids Res. 28:706-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yen, K. M., and I. C. Gunsalus. 1982. Plasmid gene organization: naphthalene/salicylate oxidation. Proc. Natl. Acad. Sci. USA 79:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young, C., D. H. Bechhofer, and D. H. Figurski. 1984. Gene regulation in plasmid RK2: positive control by korA in the expression of korC. J. Bacteriol. 157:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zatyka, M., L. Bingle, A. C. Jones, and C. M. Thomas. 2001. Cooperativity between KorB and TrbA repressors of broad-host-range plasmid RK2. J. Bacteriol. 183:1022-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zatyka, M., G. Jagura-Burdzy, and C. M. Thomas. 1994. Regulation of transfer genes of promiscuous IncP alpha plasmid RK2: repression of Tra1 region transcription both by relaxosome proteins and by the Tra2 regulator TrbA. Microbiology 140:2981-2990. [DOI] [PubMed] [Google Scholar]
- 60.Zatyka, M., G. Jagura-Burdzy, and C. M. Thomas. 1997. Transcriptional and translational control of the genes for the mating pair formation apparatus of promiscuous IncP plasmids. J. Bacteriol. 179:7201-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]