Abstract
This study reports on the use of PCR to directly detect and distinguish Campylobacter species in bovine feces without enrichment. Inhibitors present in feces are a major obstacle to using PCR to detect microorganisms. The QIAamp DNA stool minikit was found to be an efficacious extraction method, as determined by the positive amplification of internal control DNA added to bovine feces before extraction. With nested or seminested multiplex PCR, Campylobacter coli, C. fetus, C. hyointestinalis, and C. jejuni were detected in all fecal samples inoculated at ≈104 CFU g−1, and 50 to 83% of the samples inoculated at ≈103 CFU g−1 were positive. At ≈102 CFU g−1, C. fetus, C. hyointestinalis, and C. jejuni (17 to 50% of the samples) but not C. coli were detected by PCR. From uninoculated bovine feces, a total of 198 arbitrarily selected isolates of Campylobacter were recovered on four commonly used isolation media incubated at three temperatures. The most frequently isolated taxa were C. jejuni (152 isolates) and C. lanienae (42 isolates), but isolates of C. fetus subsp. fetus, Arcobacter butzleri, and A. skirrowii also were recovered (≤2 isolates per taxon). Considerable variability was observed in the frequency of isolation of campylobacters among the four media and three incubation temperatures tested. With genus-specific primers, Campylobacter DNA was detected in 75% of the fecal samples, representing an 8% increase in sensitivity relative to that obtained with microbiological isolation across the four media and three incubation temperatures tested. With nested primers, C. jejuni and C. lanienae were detected in 25 and 67% of the samples, respectively. In no instance was DNA from either C. coli, C. fetus, or C. hyointestinalis detected in uninoculated bovine feces. PCR was more sensitive than isolation on microbiological media for detecting C. lanienae (17%) but not C. jejuni. Campylobacters are a diverse and fastidious group of bacteria, and the development of direct PCR not only will increase the understanding of Campylobacter species diversity and their frequency of occurrence in feces but also will enhance the knowledge of their role in the gastrointestinal tract of livestock and of the factors that influence shedding.
Campylobacter species are recognized as one the most frequent causes of acute diarrheal disease in humans throughout the world; as much as 1% of the population is thought to be infected with Campylobacter species every year in North America (Centers for Disease Control and Prevention-U.S. Department of Agriculture-Food and Drug Administration Collaborating Sites Foodborne Disease Active Survey Network [Foodnet]). Campylobacter infections also can cause enteritis and abortions in cattle (37). Alberta, Canada, possesses a very large beef cattle population (>5 million head) in the southern region of the province, but relatively limited research has investigated the prevalence of Campylobacter species associated with beef cattle. The prevalence of Campylobacter infections in humans in this region is considerably higher than the national average (Health Canada Website, http://cythera.ic.gc.ca/dsol/ndis/ndex_e.html), but a definitive link to beef cattle as a source of Campylobacter infections has not been established. Numerous selective media are used for the isolation of campylobacters; almost all contain several antibiotics as inhibitory agents (8, 17). By use of enrichment and/or isolation of campylobacters on semiselective media, a number of studies have reported the association of Campylobacter species with cattle (1, 5, 14, 15, 20, 34, 38, 44, 45). Campylobacters are very fastidious, and antimicrobial agents incorporated into media used to selectively isolate Campylobacter jejuni and C. coli have been shown to inhibit the growth of other Campylobacter species, such as C. upsaliensis, C. hyointestinalis, and C. fetus (1, 4, 12, 25, 37). As a result, microbiological methods do not provide a true measure of the frequency and diversity of Campylobacter species associated with livestock and their feces.
The application of PCR may provide a more accurate description of the prevalence of Campylobacter species associated with livestock. However, the presence of inhibitors in fecal materials is a major obstacle limiting the usefulness of PCR for detecting microorganisms in feces. A number of inhibitors are present in human feces; these include bile salts, hemoglobin degradation products, and complex polysaccharides (33, 46). In addition, polyphenolic substances from plant tissues are very inhibitory to PCR (23). A variety of strategies for removing PCR inhibitors from human stool samples have been reported. For example, Lawson et al. (25) used polyvinylpyrrolidone to reduce the inhibitory effects of polyphenolic substances in the PCR detection of C. upsaliensis and C. helveticus in human feces. Largely because of high labor costs and time constraints in processing numerous clinical samples, a number of commercial kits have been specifically developed for extracting DNA from human feces; their utility for detecting bacteria (32), including C. jejuni and C. coli (6), has been demonstrated. The primary objective of this study was to develop a PCR-based method for detecting Campylobacter species directly in bovine feces without relying on an enrichment step. Specific objectives were to (i) develop primers and an internal control for amplifying Campylobacter DNA from feces, (ii) measure the sensitivity of PCR-based detection of four species of Campylobacter added to bovine feces, and (iii) compare PCR with conventional isolation for detecting campylobacters in bovine feces.
MATERIALS AND METHODS
Progression of experiments.
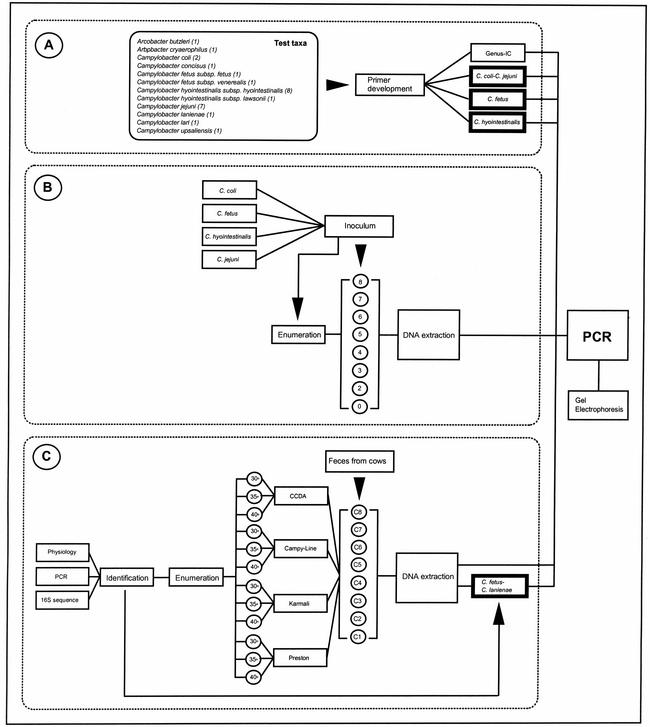
The experiments conducted for each objective are presented in Fig. 1. For the first objective (Fig. 1A), internal control DNA and primers for detecting Campylobacter species directly in feces were developed. For the second objective (Fig. 1B), the sensitivity of PCR for amplifying Campylobacter species directly from bovine feces inoculated with four taxa was compared to that of dilution plating. The efficacy of a commercial DNA extraction kit for removing PCR inhibitors also was ascertained by using internal control DNA designed for use with Campylobacter genus-specific primers. For the third objective (Fig. 1C), the detection of Campylobacter species in uninoculated bovine feces by PCR was compared to that by dilution plating. Due to the high frequency of isolation of C. lanienae, nested primers were developed for this taxon and subsequently used in a multiplex PCR with primers for C. fetus.
FIG. 1.
Flow diagram of the experiments conducted in each objective. (A) In the first objective, an internal control (IC) and primers for the genus Campylobacter, C. coli, C. jejuni, C. fetus, and C. hyointestinalis were developed. The presentation of two taxon names in the same box (on the right) represents multiplex reactions. Boxes with thick walls represent nested PCR. Names in the box labeled “Test taxa” represent taxa that were used to determine the specificities of PCRs; numbers in parentheses following the taxon names represent the numbers of isolates tested. (B) In the second objective, bovine feces were inoculated with various concentrations of C. coli, C. fetus, C. hyointestinalis, and C. jejuni (target densities in log10 CFU per gram are presented in circles); the control treatment (“0”) was not inoculated with campylobacters. CFUs were enumerated in the inoculated and control feces by using dilution plating. Concurrently, DNA was extracted from all fecal treatments and subjected to PCR with the primers shown in objective A, and amplicons were electrophoresed. (C) In the third objective, uninoculated bovine feces were collected from eight dairy cows (C1 to C8). Campylobacters were isolated and enumerated on four media maintained at three temperatures. Representative colonies were selected, established in pure cultures, and identified by using physiological characters, PCR with primers for specific taxa and, if required, extraction of DNA and sequencing of a portion of the 16S rRNA gene. During the course of this experiment, C. lanienae was frequently isolated on Karmali medium at 40°C. As a result, a nested primer set was developed for this taxon, and the C. lanienae primers were used in a multiplex PCR with primers for C. fetus. DNA was extracted from fecal samples obtained from the eight cows and subjected to PCR, and amplicons were electrophoresed. The experiments in the second and third objectives were conducted three times on separate occasions.
Objective 1. (i) Test taxa and culture conditions.
Primers for detecting DNAs of the genus Campylobacter, C. coli, C. fetus, C. hyointestinalis, and C. jejuni directly in bovine feces were developed (Table 1). We selected these taxa because of their association with bovine species. To increase sensitivity and specificity, primers for C. coli, C. fetus, C. hyointestinalis, and C. jejuni were nested or seminested. Furthermore, C. coli and C. jejuni were tested by multiplex PCR.
TABLE 1.
Primer sequences for the amplification of Campylobacter species DNA from bovine feces
| PCR target and genea | Primer | Tm(°C) | Sequence (5′ to 3′) | Size (bp) | Reference or source |
|---|---|---|---|---|---|
| Campylobacter genus and internal control | |||||
| 16S rRNA | C412F | 58 | GGATGACACTTTTCGGAGC | 816 | Linton et al. (29) |
| C1228R | CATTGTAGCACGTGTGTC | Linton et al. (29)b | |||
| Campylobacter coli and C. jejuni | |||||
| 16S rRNA | MD 16S1Upper | 58 | ATCTAATGGCTTAACCATTAAAC | 857 | Denis et al. (9); CCCJ609F modified from Linton et al. (30) |
| MD16S2Lower | GGACGGTAACTAGTTTAGTATT | Denis et al. (9); CCCJ1442R modified from Linton et al. (30) | |||
| Campylobacter coli and C. jejuni primary multiplex | |||||
| ceuE (C. coli) | COL3Upper | 58 | ATTTGAAAATTGCTCCAACTATG | 462 | Gonzales et al. (16) |
| MDCOL2Lower | TGATTTTATTATTTGTAGCAGCG | Denis et al. (10); COL2 modified from Gonzalez et al. (16) | |||
| mapA (C. jejuni) | MDmapA1 Upper | CTATTTTATTTTTGAGTGCTTGTG | 589 | Denis et al. (10) | |
| MDmapA2Lower | GCTTTATTTGCCATTTGTTTTATTA | Denis et al. (10) | |||
| Campylobacter coli and C. jejuni nested multiplex | |||||
| ceuE (C. coli) | CCceuEN3F | 58 | AAGCGTTGCAAAACTTTATGG | 330 | New primerc |
| CCceuEN3R | CCTTGTGCGCGTTCTTTATT | New primerc | |||
| mapA (C. jejuni) | CJmapAN3F | TGGTGGTTTTGAAGCAAAGA | 413 | New primerc | |
| CJmapAN3R | GCTTGGTGCGGATTGTAAA | New primerc | |||
| Campylobacter fetus and C. lanienae primary multiplex | |||||
| 23S rRNA (C. fetus) | FET1 | 56 | CTCATAATTTAATTGCACTCATA | 784 | Bastyns et al. (3) |
| HYOFET23SR | GCTTCGCATAGCTAACAT | New primer | |||
| 16S rRNA (C. lanienae) | CLAN76F | GTAAGAGCTTGCTCTTATGAG | 920 | Logan et al. (31) | |
| CLANL521021R | TCGTATCTCTACAAGGTTCTTAd | New primer; CLAN1021R modified from Logan et al. (31) | |||
| Campylobacter fetus and C. lanienae nested multiplex | |||||
| 23S rRNA (C. fetus) | FETNF | 56 | CGATAATTGATGTGAGAATCATC | 473 | New primer |
| HYOFET23SR2 | GGGAGTAAATCTTAATACAAAGTTAGG | New primer | |||
| 16S rRNA (C. lanienae) | CLANNF | TAGTTGGTGAGGTAATGGCTC | 360 | New primer | |
| CLANNR | GCAGTTTAATGGTTGAGCCA | New primer | |||
| Campylobacter hyointestinalis primary | |||||
| 23S rRNA | HYO1F | 54 | ATAATCTAGGTGAGAATCCTAGd | 611 | New primer; HYO1 modified from Bastyns et al. (3) |
| HYOFET23SR | See HYOFET23SR above | New primer | |||
| Campylobacter hyointestinalis seminested | |||||
| 23S rRNA | HYO1F | 54 | See HYO1F above | 468 | See above |
| HYOFET23SR2 | See HYOFET23SR2 | New primer |
Primary PCRs were also used to identify Campylobacter species isolated from feces.
There was a typographical error in Linton et al. (29): primer 1288 should have read primer 1228.
Developed with Primer3 software.
Bold type indicates regions of the primer that were modified.
The following reference strains were used to test the primers that we developed: Arcobacter butzleri (ATCC 49616 [American Type Culture Collection]), A. cryaerophilus (ATCC 49942), C. coli (HC 1111 [Health Canada] and ATCC 49941), C. concisus (ATCC 33237), C. fetus subsp. fetus (ATCC 25936), C. fetus subsp. venerealis (ATCC 19438), C. hyointestinalis subsp. hyointestinalis (ATCC 35217), C. hyointestinalis subsp. lawsonii (NCTC 12901 [National Collection of Type Cultures and Pathogenic Fungi]), C. jejuni (HC 1102, HC 1104, HC 1115, HC 1108, ATCC 29428, ATCC 49943, and ATCC 33291), C. lanienae (NCTC 13004), C. lari (ATCC 35221), and C. upsaliensis (LCDC 5424 [Laboratory Centre for Disease Control]). To obtain biomass, all isolates were grown on Campylobacter blood-free selective agar base (modified Campylobacter charcoal differential agar [CCDA]) (Oxoid, Nepean, Ontario, Canada) without antibiotic supplements or brucella agar (Difco, Detroit, Mich.) at 37°C in anaerobic gas jars (Oxoid). Microaerophilic conditions were generated with a CampyPak Plus microaerophilic system with a palladium catalyst (BBL, Becton Dickinson, Sparks, Md.).
(ii) Genomic DNA extraction and PCR methods.
DNA was extracted from the reference strains by using a DNeasy kit (Qiagen Inc., Mississauga, Ontario, Canada) according to the manufacturer's protocol. The conditions for primary amplification were 1 cycle at 95°C for 15 min; 25 cycles of 30 s at 94°C, 90 s at the annealing temperature (Tm), and 60 s at 72°C; and extension for 10 min at 72°C. For multiplex reactions, mixtures consisted of a total volume of 20 μl containing reaction buffer, 0.2 mM deoxynucleoside triphosphates, 2 mM MgCl2, 0.5 μM each primer (Sigma-Genosys, Oakville, Ontario, Canada), 0.2 μg of bovine serum albumin (Promega, Madison, Wis.), and 1 U of HotStar Taq polymerase (Qiagen). Each PCR was performed with a total of 2 μl of a 10−5 dilution of genomic DNA (≈500 ng μl−1). For nested and seminested amplifications, the reaction conditions were the same, with the exception that 35 cycles were used, 1 μl of the reaction mixture from the primary amplification step was used as a template, and bovine serum albumin was not included in the reaction mixture. All PCR products (10 μl) were electrophoresed in a Tris-borate-EDTA-2% agarose gel (Invitrogen Corp., Burlington, Ontario, Canada), visualized by staining with ethidium bromide, and viewed under UV light. A 100-bp ladder (Promega) was used to size products. The Tms used and estimated product sizes are shown in Table 1.
Objective 2. (i) Internal control construction.
An internal control designed to amplify under the same PCR conditions as those described for the Campylobacter genus-specific primer set was constructed by deleting a fragment of the C. jejuni (ATCC 49943) 16S rRNA gene by the strategy of Denis et al. (10). The deletion was achieved by PCR amplification of the 16S rRNA gene with mutagenic primer C1228RIC (5′-CATTGTAGCACGTGTGTCTCCCCAGGCGGTACACTTAATG-3′; the underlined sequence corresponds to C1228R). PCR amplification of the template with primers C412F and C1228RIC yielded a 475-bp product instead of an 816-bp product containing both the C412F and the C1228R primer sites. The 475-bp product was cloned into the pGEM-T EASY vector (Promega) and transformed into Escherichia coli JM109 cells. Transformed colonies were screened for the presence of an insert by PCR amplification with primers C412F and C1228. Plasmid DNA was extracted with a QIAprep spin miniprep kit (Qiagen). The plasmid-IC was linearized by digestion with NcoI enzyme (Promega). The enzyme was removed by two extractions with Strataclean resin (Stratagene) according to the manufacturer's protocol. The concentration of linearized plasmid-IC was adjusted to 700 copies μl−1 in 10 mM Tris-Cl (pH 8.5) buffer, and plasmid-IC was stored at −20°C until used.
(ii) Extraction of DNA from bovine feces.
A QIAamp DNA stool minikit (Qiagen) was used to extract DNA from 200 ± 5 mg of bovine feces according to the manufacturer's protocol for the isolation of DNA from stools for pathogen detection. Briefly, the procedure involved lysis of the bacterial cells within the fecal material in ASL buffer, adsorption of impurities to InhibitEX reagent, and purification of the DNA on a spin column. Prior to extraction, the internal control was added to each sample at the rate of 10 μl per 200 mg of feces. Extracted DNA was stored at −20°C until processed.
(iii) Inoculation.
Feces were collected from cattle in Lethbridge, Alberta, Canada. Fecal samples that were determined to be negative for Campylobacter DNA or that produced a weak amplicon with the Campylobacter genus-specific 16S rRNA gene primers were selected. Fecal samples that were free of Campylobacter DNA were infrequently obtained, and this situation necessitated the use of contaminated feces, albeit feces containing small amounts of campylobacters. Subsamples of fecal samples were stored at −20°C until required. Fecal samples were thawed once.
Isolates used to inoculate feces included C. coli ATCC 49941, C. fetus ATCC 25936, C. hyointestinalis ATCC 35217, and C. jejuni ATCC 49943 (i.e., taxon treatment). All bacteria were grown for 24 h at 37°C. In replicate 1, C. coli, C. fetus, and C. jejuni were grown on brucella agar, whereas C. hyointestinalis was grown on CCDA. In replicates 2 and 3, both C. fetus and C. hyointestinalis were grown on CCDA, and the other species were grown on brucella agar. In all instances, biomass production media were not amended with antibiotics. To obtain biomass, cells were scraped from the media and suspended in sterile brucella broth (Difco). The turbidity (A600) of the suspension was adjusted to 0.5 (this represented a cell density of approximately 2 × 109 CFU ml−1), and six 10-fold serial dilutions were prepared in brucella broth (i.e., inocula), representing a range of cell densities of approximately 103 to 109 CFU ml−1 (i.e., density treatment). To enumerate cell densities in the suspensions, dilution spread plate counts were made on either brucella agar (C. coli and C. jejuni) or CCDA (C. fetus and C. hyointestinalis). Feces were inoculated with each taxon treatment at the rate of 2 ml per 18 g (wet weight) of feces for each density; sterile brucella broth was used for the control. Immediately after addition of the inocula, feces were thoroughly mixed with a metal spatula. The experiment was conducted on three separate occasions (i.e., replicates). For each replicate, two samples were prepared (i.e., subsamples).
(iv) Detection of Campylobacter species by dilution plating.
To enumerate campylobacters in the inoculated feces by conventional microbiological plating, 2.5 g of feces was placed in 22.5 ml of phosphate-buffered saline (130 mM sodium chloride, 10 mM sodium phosphate buffer [pH 7.2]) in a 50-ml Falcon tube (DiaMed, Missassauga, Ontario, Canada), and the suspension was vortexed at the maximum setting for 1 min. The suspension was diluted in a 10-fold dilution series, and 100 μl was spread on CCDA containing selective supplement SR115E (Oxoid). Cultures were incubated microaerophilically at 37°C as described above. Colonies were enumerated at the dilution yielding 20 to 200 CFU after 48 h, and CFUs per gram of feces (fresh and dry weights) were calculated. To determine fecal dry weights, two aliquots of feces (approximately 20 g each) were dried at 50°C for 72 h. We observed no differences in water content (dry matter content ranged from 19.1 to 19.8%) among the fecal samples, and so the CFUs are presented per gram of fresh weight. A mean value (CFU per gram) for the two subsamples for each taxon treatment and density treatment was used to calculate the overall mean and SEM for the three replicates.
(v) Direct detection of Campylobacter species by PCR.
DNA was extracted from the two subsamples for each taxon treatment and density treatment with the DNA stool minikit as described above. Amplification of C. coli, C. fetus, C. hyointestinalis, and C. jejuni DNAs was achieved with the primary and nested or seminested PCR primers listed in Table 1. In all instances, the same reaction and amplification conditions as those described above were used, with the exception that 2 μl of fecal DNA was used as a template. The mean proportion of positive samples for the two subsamples for each taxon treatment and density treatment was used to calculate the overall mean and SEM (n = 3).
Objective 3. (i) Collection of feces.
Feces were aseptically collected from eight Holstein dairy cows on three separate occasions (i.e., replicates) at ca. 2-week intervals. All of the cattle used were early-lactation cows fed a total mixed ration consisting of barley, alfalfa, and corn silage. Fecal samples were obtained following the morning feeding.
(ii) Detection of Campylobacter species by plating.
On each collection date, 2.5 g of feces from each cow was suspended in 22.5 ml of phosphate-buffered saline by vortexing as described above, and 100 μl of each suspension was spread on each of four test media. The media consisted of (i) Campy-Line agar (Dalynn Biologicals) (28); (ii) Karmali agar (Oxoid) (22) with selective supplement CM935 (Oxoid); (iii) CCDA with selective supplement SR115E; and (iv) Preston agar, comprised of CM689 campylobacter base (Oxoid), 50 ml of lysed horse blood (SR48; Oxoid) liter−1, and selective supplement SR117E (Oxoid). Cultures were grown microaerophilically at 30, 35, and 40°C as described above. At 48- and 72-h intervals, CFUs of Campylobacter species were enumerated based on colony morphology and microscopic appearance. Arbitrarily selected colonies deemed to be campylobacters were transferred to the same medium from which they had been isolated and were streaked for purity. Colonies were selected based on colony appearance and frequency of occurrence. All strains were stored in brucella broth amended with 30% glycerol at −20 and −80°C until identified. In most instances, groupings based on colony appearance and morphology were determined to be monotaxic (based on the identification of representative strains), and the mean CFU per gram and SEM (n = 3) were calculated for each taxon.
(iii) Identification of Campylobacter species.
Presumptive identification of isolates recovered from bovine feces was accomplished with both physiological and molecular characters. All isolates were subjected to colony PCR with the Campylobacter genus-specific, C. coli-C. jejuni, C. fetus-C. lanienae, and C. hyointestinalis primary primer sets (Table 1). If required, Campylobacter isolates also were tested with primers for C. helveticus, C. lari, C. mucosalis, C. sputorum, and C. upsaliensis (Table 2). We also tested the C. hyointestinalis primer set of Linton et al. (29) (Table 2) but abandoned this primer set for a set described in Table 1. The same reaction and amplification conditions as those described above were used, with the exception that the DNA template consisted of 1 μl of a suspension containing 24- to 48-h-old cells of the isolate to be identified. For each isolate, cells from an individual colony were uniformly suspended in 100 μl of sterile brucella broth in 96-well microtiter plates with sterile toothpicks or pipette tips. Positive controls consisted of cells from reference strains, and the negative control consisted of brucella broth alone.
TABLE 2.
Primer sequences used to identify Campylobacter species isolated from bovine feces
| PCR target and gene | Primer | Tm (°C) | Sequence (5′ to 3′) | Size (bp) | Reference or source |
|---|---|---|---|---|---|
| Arcobacter butzleri 16S rDNA | BUTZ | 61 | CCTGGACTTGACATAGTAAGAATGA | 401 | Houf et al. (19) |
| ARCO | CGTATTCACCGTAGCATAGC | ||||
| Arcobacter cryaerophilus 23S rDNA | CRY1 | 61 | TGCTGGAGCGGATAGAAGTA | 257 | Houf et al. (19) |
| CRY2 | AACAACCTACGTCCTTCGAC | ||||
| Arcobacter skirrowii 16S rDNA | SKIR | 61 | GGCGATTTACTGGAACACA | 641 | Houf et al. (19) |
| ARCO | See ARCO above | ||||
| Campylobacter helveticus 16S rDNA | CHCU146F | 60 | GGGACAACACTTAGAAATGAG | 1225-1375 | Linton et al. (29) |
| CH1371R | CCGTGACATGGCTGATTCAC | ||||
| Campylobacter hyointestinalis 16S rDNA | CFCH57F | 65 | GCAAGTCGAACGGAGTATTA | 1,287 | Linton et al. (29) |
| CH1344R | GCGATTCCGGCTTCATGCTC | ||||
| Campylobacter lari 16S rDNA | CL594F | 64 | CAAGTCTCTTGTGAAATCCAAC | 561 | Linton et al. (29) |
| CL1155R | ATTTAGAGTGCTCACCCGAAG | ||||
| Campylobacter mucosalis 23S rDNA | MUC1 | 58 | ATGAGTAGCGATAATTGGG | 306 | Bastyns et al. (3) |
| MUC2 | ACAGTATCAAGGATTCGTC | ||||
| Campylobacter sputorum 23S rDNA | SPUT1 | 58 | ATAAGTACCGAAGTCGTAGG | 588 | Bastyns et al. (3) |
| SPUT2 | TCTAGGGCTTTAACACCC | ||||
| Campylobacter upsaliensis 16S rDNA | CHCU146F | 60 | See CHCU146F above | 878 | Linton et al. (29) |
| CU1024R | CACTTCCGTATCTCTACAGA | ||||
| Universal prokaryote 16S rRNA | UNI27F | 50 | AGAGTTTGATCCTGGCTCAG | Va | Lane (24) |
| UNI1492R | TACGG(C/T)TACCTTGTTACGACT | ||||
| Universal prokaryote 16S rRNA | UNI338F | 50 | ACTCCTACGGGAGGCAG | V | Lane (24) |
| UNI1100R | AGGGTTGCGCTCGTTG |
V, amplicon size was variable.
The abilities of strains to produce catalase and H2S, to reduce nitrate and nitrite, and to hydrolyze hippurate and indoxyl acetate and their sensitivities to nalidixic acid (30 μg; BBL) and cephalothin (30 μg; BBL) were determined as described by Logan et al. (31). In addition, C. fetus strains were tested for their ability to grow on brucella agar containing 1% glycine. For Arcobacter strains, the ability to grow in atmospheric oxygen, on brucella agar containing 1% glycine, and on MacConkey agar was determined. For C. jejuni and C. lanienae, six and seven arbitrarily selected strains were subjected to physiological tests, respectively.
For isolates presumptively identified as C. fetus, C. lanienae, A. butzleri, or A. skirrowii, the complete or partial 16S rRNA gene was sequenced. The 16S rRNA gene was amplified by PCR with eubacterial primers UNI27F and UNI1492R (Table 2). For all amplifications, the same PCR mixtures and amplification conditions as those described above for colony PCR were used. To obtain a partial sequence for the 16S rRNA gene, primers UNI338F and UNI1100R were used (Table 2) with an ABI PRISM Big Dye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif.). Prior to sequencing, excess dye was removed with a Qiagen DyeEx spin kit. Sequences were obtained with an ABI PRISM 377 automated DNA sequencer. Contigs were constructed by using Staden (Medical Research Council, Laboratory of Molecular Biology, Cambridge, England), and all sequences were compared directly with the National Center for Biotechnology Information (NCBI) GenBank nonredundant nucleotide database by using BLASTN.
The nucleotide sequences for a presumptively identified isolate of C. lanienae (L52) were aligned with data retrieved from GenBank by using the multialignment program CLUSTAL W (40), and the alignments were refined visually by using GeneDoc (www.psc.edu/biomed/genedoc). Sequences (GenBank accession numbers) for C. concisus (L04322), C. curvus (L04313), C. fetus subsp. fetus (M65012), C. fetus subsp. venerealis (M65011), C. gracilis (L04320), C. hyointestinalis subsp. hyointestinalis (AF097681, AF097689 [type], and AF097691), C. hyointestinalis (AF219235 and M65010), C. hyointestinalis subsp. lawsonii (AF097683 and AF097685 [type]), C. lanienae (AB076675, AB076677, AF043423, and AF043425[type]), C. mucosalis (L06978), C. rectus (L04317), C. sputorum (L04319), and C. jejuni subsp. jejuni (L04315) were included in the analyses.
The sequence data were analyzed by using programs contained within PHYLIP (13). Phylogenetic estimates were based on methods for determining neighbor-joining distance, maximum parsimony, and maximum likelihood. Divergence (or distance) for each pair of sequences was calculated by using DNADIST with the Kimura two-parameter model. The NEIGHBOR program was used for estimating phylogenies from the distance matrices. DNAPARS was executed to perform maximum-parsimony analysis, and DNAML was used for maximum-likelihood analysis. Support for the internal branches within the resulting trees was obtained by bootstrap analysis. A total of 1,000 bootstrap replicates for the 16S ribosomal DNA (rDNA) data were generated by using SEQBOOT, majority-rule consensus trees were constructed by using the CONSENSE program, and the trees were visualized by using TreeView (http: //taxonomy.zoology.gla.ac.uk/rod/rod.html).
(iv) Direct detection of Campylobacter species by PCR.
For each cow on each occasion, an aliquot of feces was inoculated with the internal control, DNA was extracted with the DNA stool minikit, the PCR mixtures and amplification conditions were set up, and the PCR products were visualized as described above. Due to the frequent isolation of C. lanienae on Karmali agar, primers based on the 16S rRNA gene were developed for this taxon (Table 1) and used in a multiplex PCR with primers for C. fetus. The mean proportion of positive samples and SEM (n = 3) were calculated for each taxon.
Nucleotide sequence accession number.
The partial 16S rRNA gene sequence for C. lanienae L52 has been deposited in GenBank under accession number AY288304.
RESULTS
Objective 1. (i) Genus Campylobacter.
The genus-specific primer set based on the 16S rRNA gene (29) provided amplicon products for all of the taxa of campylobacters tested. In addition, we observed weak products for A. butzleri and A. cryaerophilus.
(ii) C. coli and C. jejuni.
The primary primer sets for C. coli and C. jejuni were based on previously published results, and they were found to be highly specific. The nested primers that we designed for C. jejuni (i.e., mapA) were also highly specific, but the nested primers for C. coli (i.e., ceuU) did not confer specificity on their own. However, they also were found to be highly specific when used in conjunction with the primary primer set.
(iii) C. fetus.
The C. fetus nested primer set based on the 23S rRNA gene was found to be highly specific for the species but did not distinguish between C. fetus subsp. fetus and C. fetus subsp. venerealis. The nested primer set based on the 16S rRNA gene of C. fetus was also highly specific for C. fetus relative to other Campylobacter species in pure cultures. However, this primer set was subsequently observed to provide an amplicon of the correct size (552 bp) for all bovine fecal samples. The sequence of this PCR product exhibited relatively poor similarity with the C. fetus 16S rRNA sequences deposited at NCBI (GenBank accession numbers AF219234, AF219233, and AJ306569), and this primer set was abandoned in favor of the primer set targeting the 23S rRNA gene.
(iv) C. hyointestinalis.
One of 16S rRNA primers for C. hyointestinalis used by Linton et al. (29) binds in a polymorphic region (18), and we observed that these primers did not provide a PCR product for some of the isolates of C. hyointestinalis obtained from bovine feces at Lethbridge in a previous study (Fig. 2). To increase the comprehensiveness of detection, we modified a forward primer based on the 23S rRNA gene (HYO1F) from that of Bastyns et al. (3). We designed a new reverse primer (HYOFET23SR) to replace the reverse primer (69ar) used by Bastyns et al. (3); many of the sequences for the 23S rRNA gene of C. hyointestinalis did not include the binding region for 69ar. For the secondary reaction, a seminested primer set (HYO1F and HYOFET23SR2) was used. Both the primary and the seminested primer sets were found to be highly specific for all of the test strains of C. hyointestinalis subsp. hyointestinalis. Neither primer set provided an amplicon for C. hyointestinalis subsp. lawsonii (Fig. 2).
FIG. 2.
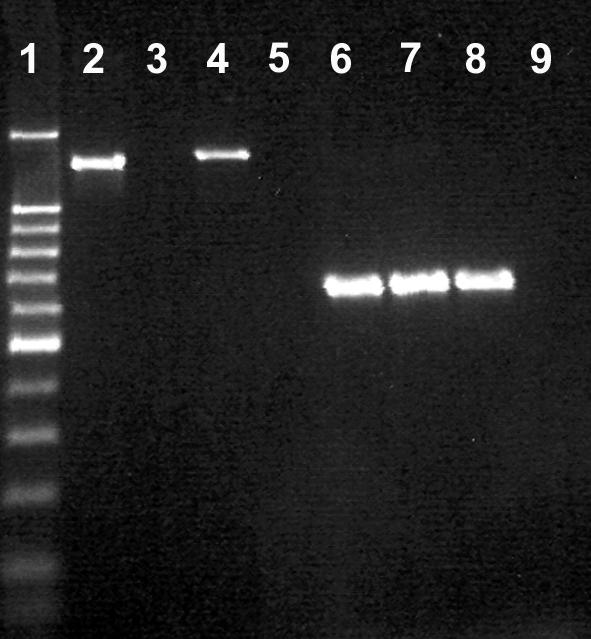
Comparison of the 16S (29) and 23S rDNA primers developed in the current study for detecting C. hyointestinalis. Lane 1, 100-bp molecular weight marker (the dark band was at 500 bp); lane 2, 16S primers with C. hyointestinalis subsp. hyointestinalis HBF; lane 3, 16S primers with C. hyointestinalis subsp. hyointestinalis HBE; lane 4, 16S primers with C. hyointestinalis subsp. hyointestinalis ATCC 35217; lane 5, 16S primers with C. hyointestinalis subsp. lawsonii NCTC 12901; lane 6, 23S primers with C. hyointestinalis subsp. hyointestinalis HBF; lane 7, 23S primers with C. hyointestinalis subsp. hyointestinalis HBE; lane 8, 23S primers with C. hyointestinalis subsp. hyointestinalis ATCC 35217; lane 9, 23S primers with C. hyointestinalis subsp. lawsonii NCTC 12901. An amplicon was not observed in either of the negative control reactions, and so these treatments were removed.
Objective 2. (i) DNA extraction.
Genomic DNA was obtained from all fecal samples by using the DNA stool minikit. The concentration of Campylobacter DNA was observed to affect the intensity of the internal control amplicon (Fig. 3). However, in the minimum detection experiment (i.e., feces inoculated with various concentrations of C. coli, C. jejuni, C. fetus, and C. hyointestinalis), the 475-bp internal control amplicon was always observed in fecal samples for which no or a weak PCR product (816 bp) was detected by the Campylobacter genus-specific primer set (Fig. 4).
FIG. 3.
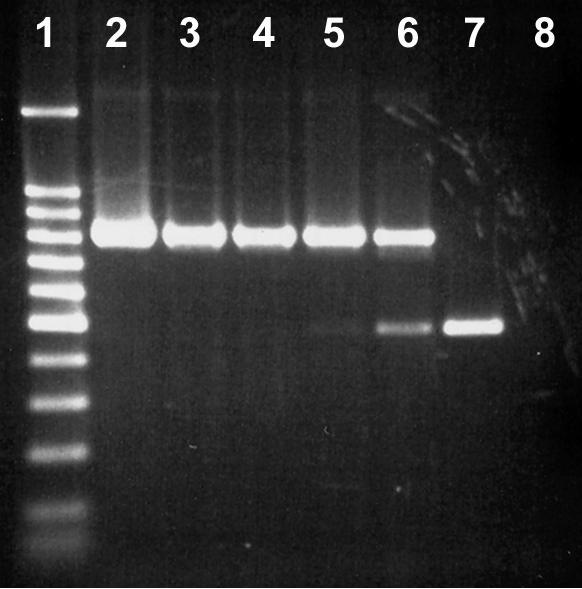
Impact of C. jejuni L102 genomic DNA concentration on the expression of the internal control amplicon. Lane 1, 100-bp molecular weight marker (the dark band was at 500 bp); lane 2, 10−1 dilution; lane 3, 10−2 dilution; lane 4, 10−3 dilution; lane 5, 10−4 dilution; lane 6, 10−5 dilution; lane 7, no template; lane 8, no internal control.
FIG. 4.
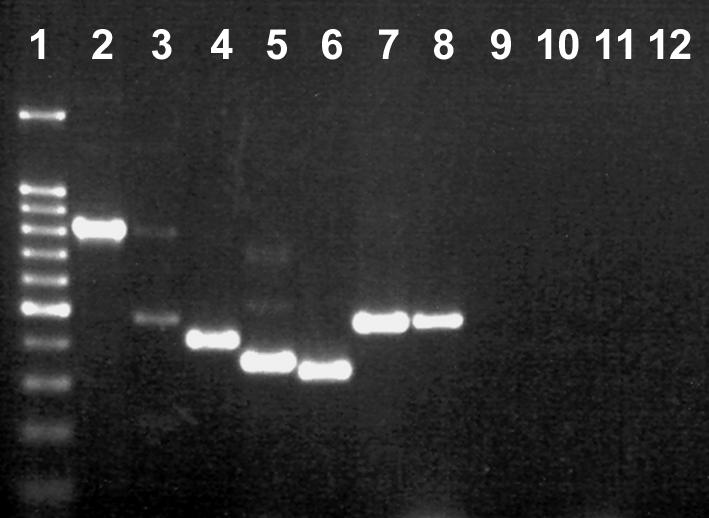
Multiplex PCR for detection of Campylobacter species in bovine feces. Lane 1, 100-bp molecular weight marker (the dark band was at 500 bp); lane 2, DNA from uninoculated feces amplified with the Campylobacter genus-specific primer set; lane 3, DNA from uninoculated feces amplified with the Campylobacter genus-specific primer set (note the weak genus amplicon and the internal control amplicon at 465 bp); lane 4, DNA from uninoculated feces amplified with the C. jejuni-C. coli multiplex nested primer set (note the C. jejuni amplicon at 413 bp); lane 5, DNA from uninoculated feces amplified with the C. fetus-C. lanienae nested primer set (note the C. lanienae amplicon at 360 bp); lane 6, DNA from feces inoculated with C. coli ATCC 49941 at a density of ≈103 CFU g−1 and amplified with the C. jejuni-C. coli multiplex nested primer set (note the C. coli amplicon at 330 bp); lane 7, DNA from feces inoculated with C. hyointestinalis subsp. hyointestinalis ATCC 35217 at a density of ≈102 CFU g−1 and amplified with the C. hyointestinalis seminested primer set (note the C. hyointestinalis amplicon at 468 bp); lane 8, DNA from feces inoculated with C. fetus ATCC 25936 at a density of ≈102 CFU g−1 and amplified with the C. fetus-C. lanienae nested primer set (note the C. fetus amplicon at 473 bp); lane 9, negative control for the internal control and Campylobacter genus-specific primers (no template added); lane 10, negative control for C. jejuni-C. coli multiplex primers; lane 11, negative control for C. fetus-C. lanienae multiplex primers; lane 12, negative control for C. hyointestinalis subsp. hyointestinalis primers.
(ii) Detection of campylobacters in bovine feces.
No amplification products were detected for the controls (uninoculated feces) for C. coli, C. fetus, C. jejuni, and C. hyointestinalis (Table 3). For all four taxa, all of the samples of feces inoculated at ≈104 CFU g−1 were positive (range of 3.67 ± 0.037 to 4.28 ± 0.012 log CFU g−1 [mean and standard error of the mean]). At ≈103 CFU g−1, 83% ± 17%, 50% ± 29%, 67% ± 17%, and 67% ± 33% of the samples of feces were positive for C. coli, C. fetus, C. jejuni, and C. hyointestinalis, respectively (range of 2.95 ± 0.051 to 3.38 ± 0.091 log CFU g−1). C. coli was not detected at a density of ≈102 CFU g−1. In contrast, 17% ± 17%, 17% ± 17%, and 50% ± 29% of the samples of feces inoculated with C. fetus, C. jejuni, and C. hyointestinalis, respectively, provided an amplification product at this population density (range of 2.13 ± 0.069 to 2.49 ± 0.114 log CFU g−1).
TABLE 3.
Microbiological isolation and direct PCR detection of Campylobacter populations in inoculated bovine feces
| Taxon and treatmenta | Mean ± SEM log CFU g−1 (fresh wt) (n = 3)b | Mean ± SEM proportion of samples found positive by PCR for:
|
|||
|---|---|---|---|---|---|
| Genusc | C. coli and C. jejunid | C. fetus and C. lanienaee | C. hyointestinalisf | ||
| Campylobacter coli | |||||
| A | 8.19 ± 0.043 | 1.0 | 1.0 | —g | — |
| B | 7.21 ± 0.038 | 1.0 | 1.0 | — | — |
| C | 6.27 ± 0.061 | 1.0 | 1.0 | — | — |
| D | 5.28 ± 0.056 | 1.0 | 1.0 | — | — |
| E | 4.17 ± 0.084 | 1.0 | 1.0 | — | — |
| F | 3.28 ± 0.087 | 0.50 ± 0.29 | 0.83 ± 0.17 | — | — |
| G | 1.98 ± 0.490 | 0.67 ± 0.17 | 0.0 | — | — |
| N | 0.0 | 0.50 ± 0.29 | 0.0 | — | — |
| Campylobacter fetus | |||||
| A | 7.97 ± 0.030 | 1.0 | — | 1.0 | — |
| B | 7.09 ± 0.065 | 1.0 | — | 1.0 | — |
| C | 6.04 ± 0.037 | 1.0 | — | 1.0 | — |
| D | 5.08 ± 0.015 | 1.0 | — | 1.0 | — |
| E | 4.07 ± 0.004 | 1.0 | — | 1.0 | — |
| F | 3.13 ± 0.103 | 1.0 | — | 0.50 ± 0.29 | — |
| G | 2.48 ± 0.059 | 0.33 ± 0.33 | — | 0.17 ± 0.17 | — |
| N | 0.0 | 0.33 ± 0.17 | — | 0.0 | — |
| Campylobacter hyointestinalis | |||||
| A | 7.73 ± 0.080 | 1.0 | — | — | 1.0 |
| B | 6.52 ± 0.129 | 1.0 | — | — | 1.0 |
| C | 5.58 ± 0.157 | 1.0 | — | — | 1.0 |
| D | 4.79 ± 0.013 | 1.0 | — | — | 1.0 |
| E | 3.67 ± 0.037 | 1.0 | — | — | 1.0 |
| F | 2.95 ± 0.051 | 1.0 | — | — | 0.67 ± 0.33 |
| G | 2.13 ± 0.069 | 0.50 ± 0.29 | — | — | 0.50 ± 0.29 |
| N | 0.0 | 0.67 ± 0.33 | — | — | 0.0 |
| Campylobacter jejuni | |||||
| A | 8.14 ± 0.071 | 1.0 | 1.0 | — | — |
| B | 7.24 ± 0.029 | 1.0 | 1.0 | — | — |
| C | 6.21 ± 0.053 | 1.0 | 1.0 | — | — |
| D | 5.28 ± 0.030 | 1.0 | 1.0 | — | — |
| E | 4.28 ± 0.012 | 1.0 | 1.0 | — | — |
| F | 3.38 ± 0.091 | 0.83 ± 0.17 | 0.67 ± 0.17 | — | — |
| G | 2.49 ± 0.114 | 0.67 ± 0.17 | 0.16 ± 0.17 | — | — |
| N | 0.0 | 0.83 ± 0.17 | 0.0 | — | — |
Treatments consisted of inoculation of feces with different concentrations of the target bacterium (10-fold dilutions); treatment N was uninoculated (control).
Feces content ranged from 19.1% ± 0.80% to 19.8% ± 0.36% dry matter.
Nonnested Campylobacter genus-specific primer set targeting the 16S rRNA gene.
Nested multiplex primer sets for C. coli and C. jejuni targeting the ceuE and mapA genes, respectively.
Nested multiplex primer sets for C. fetus and C. lanienae targeting the 23S and 16S rRNA genes, respectively.
Seminested primer set for C. hyointestinalis targeting the 23S rRNA gene.
—, not determined.
Amplification of Campylobacter DNA (with the nonnested genus-specific primer set) was observed for all fecal samples inoculated with ≈104 CFU g−1 (Table 3). At a density of ≈102 to 103 CFU g−1, Campylobacter DNA was detected in 33% ± 33% to 100% of the samples. Although DNAs of C. coli, C. fetus, C. hyointestinalis, and C. jejuni were not detected in uninoculated feces, 33% ± 17% to 83% ± 17% of these samples were observed to be positive for Campylobacter DNA.
Objective 3. (i) Detection of Campylobacter species by plating.
Considerable experience and diligence were frequently required to detect Campylobacter colonies on the test media. A total of 198 arbitrarily selected Campylobacter isolates were recovered from uninoculated bovine feces. Considerable variability was observed in the frequencies of isolation of C. jejuni and of presumptively identified C. lanienae isolates with the four media and incubation temperatures tested (Table 4). At 30°C and, to a lesser extent, at 35°C, considerable fungal contamination was encountered, making enumeration of and isolation of possible campylobacters at 72 h even more difficult; the most prevalent fungus observed was Geotrichum candidum (11). We did not observe any campylobacters at 30°C. In contrast, considerable numbers of C. jejuni isolates were recovered on all four media at 35 and 40°C. All isolates, including the C. jejuni reference strain, amplified with the mapA (C. jejuni) primer set and arbitrarily selected isolates (L36, L42, L69, L75, L92, and L97) all were able to produce catalase, did not produce H2S, reduced nitrate but not nitrite, hydrolyzed hippurate and indoxyl acetate, were sensitive to nalidixic acid, and were resistant to cephalothin (Table 5). The Campy-Line medium was less effective at recovering C. jejuni than the other three media tested at 35°C. Of the seven fecal samples that were positive for C. jejuni at 35°C (isolated on at least one medium), two (29%), seven (100%), six (86%), and five (71%) samples were positive on Campy-Line, Karmali, CCDA, and Preston media, respectively. At 40°C, the media produced very similar results, and the efficiency of isolation ranged from 78 to 89%.
TABLE 4.
Isolation of campylobacters from bovine fecesa
| Taxon and medium | Temp (°C)b | Mean log10 CFU g−1 from the following cow:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 593 | 790 | 791 | 842 | 850 | 870 | 985 | 986 | ||
| Campylobacter jejuni | |||||||||
| Campy-Line | 35 | 0.0 (0) | 1.23 (1) | 1.24 (1) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Karmali | 35 | 0.0 (0) | 2.82 (2) | 1.21 (1) | 1.49 (2) | 0.0 (0) | 2.14 (2) | 0.0 (0) | 0.0 (0) |
| CCDA | 35 | 0.0 (0) | 2.72 (2) | 1.26 (1) | 0.67 (1) | 0.0 (0) | 2.35 (2) | 0.0 (0) | 0.0 (0) |
| Preston | 35 | 0.0 (0) | 2.87 (2) | 1.39 (1) | 0.0 (0) | 0.0 (0) | 2.33 (2) | 0.0 (0) | 0.0 (0) |
| Campy-Line | 40 | 0.0 (0) | 2.84 (2) | 1.97 (2) | 0.67 (1) | 0.0 (0) | 1.77 (2) | 0.0 (0) | 0.0 (0) |
| Karmali | 40 | 0.0 (0) | 2.93 (2) | 2.12 (2) | 0.93 (1) | 0.0 (0) | 3.11 (3) | 0.0 (0) | 0.0 (0) |
| CCDA | 40 | 0.0 (0) | 2.82 (2) | 1.37 (1) | 0.83 (1) | 0.0 (0) | 3.56 (3) | 0.0 (0) | 0.0 (0) |
| Preston | 40 | 0.0 (0) | 2.98 (2) | 1.35 (1) | 0.77 (1) | 0.0 (0) | 3.70 (3) | 0.67 (1) | 0.0 (0) |
| Campylobacter lanienae | |||||||||
| Campy-Line | 35 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Karmali | 35 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| CCDA | 35 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Preston | 35 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campy-Line | 40 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Karmali | 40 | 4.83 (3) | 1.43 (1) | 2.98 (2) | 0.0 (0) | 2.61 (2) | 3.23 (2) | 3.16 (2) | 0.0 (0) |
| CCDA | 40 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Preston | 40 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
One isolate of C. fetus from cow 870 was recovered on CCDA at 40°C. Two isolates of A. skirrowii from cow 842 were recovered on Karmali agar at 40°C. One isolate of A. butzleri from cow 842 was recovered on Karmali agar at 35°C. Three replicates were conducted at ca. 2-week intervals; numbers in parentheses indicate the number of positive replicates.
No isolates of Campylobacter or Arcobacter were recovered at 30°C.
TABLE 5.
Results of physiological and molecular typing of arbitrarily selected isolates of Arcobacter and Campylobacter recovered from bovine feces
| Source and taxon (isolate) | Resulta for:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalase | H2S | Nitrate | Nitrite | Hippurate | Indoxyl acetate | Nalidixic acid | Cephalothin | Glycine (1%) | Mac- Conkey agar | O2 | PCRb | 16Sc | |
| Bovine feces | |||||||||||||
| A. butzleri (L111) | w | − | + | − | − | + | R | R | + | + | + | A. butzleri | + |
| A. skirrowii (L109) | + | − | + | − | − | + | S | S | + | − | + | A. skirrowii | + |
| A. skirrowii (L110) | + | − | + | − | − | + | S | S | + | − | + | A. skirrowii | + |
| C. fetus subsp. fetus (L49) | + | − | + | − | − | − | R | S | + | ND | ND | C. fetus | + |
| C. jejuni (L36) | + | − | + | − | + | + | S | R | ND | ND | ND | C. jejuni | |
| C. jejuni (L42) | + | − | + | − | + | + | S | R | ND | ND | ND | C. jejuni | |
| C. jejuni (L69) | + | − | + | − | + | + | S | R | ND | ND | ND | C. jejuni | |
| C. jejuni (L75) | + | − | + | − | + | + | S | R | ND | ND | ND | C. jejuni | |
| C. jejuni (L92) | + | − | + | − | + | + | S | R | ND | ND | ND | C. jejuni | |
| C. jejuni (L97) | + | − | + | − | + | + | S | R | ND | ND | ND | C. jejuni | |
| C. lanienae (L52) | + | − | + | −d | − | − | R | S | ND | ND | ND | C. lanienae | + |
| C. lanienae (L54) | + | − | + | −d | − | − | R | S | ND | ND | ND | C. lanienae | + |
| C. lanienae (L83) | + | − | + | −d | − | − | R | S | ND | ND | ND | C. lanienae | |
| C. lanienae (L85) | + | − | + | −d | − | − | R | S | ND | ND | ND | C. lanienae | |
| C. lanienae (L71) | + | − | + | −d | − | − | R | S | ND | ND | ND | C. lanienae | |
| C. lanienae (L90) | + | − | + | −d | − | − | R | S | ND | ND | ND | C. lanienae | |
| C. lanienae (L94) | + | − | + | −d | − | − | R | S | ND | ND | ND | C. lanienae | |
| Reference strains | |||||||||||||
| A. butzleri ATCC 49616 | w | − | + | − | − | + | S | Se | + | + | + | C. butzleri | |
| C. fetus subsp. fetus ATCC 25936 | + | − | + | + | − | − | R | S | + | ND | ND | C. fetus | |
| C. fetus subsp. venerealis ATCC 19438 | + | − | + | + | − | − | R | S | + | ND | ND | C. fetus | |
| C. hyointestinalis ATCC 35217 | + | + | + | +f | − | − | R | S | ND | ND | ND | C. hyointestinalis | |
| C. jejuni ATCC 49943 | + | − | + | − | + | + | S | R | ND | ND | ND | C. jejuni | |
| C. lanienae NCTC 13004 | + | − | + | −d | − | − | R | R | ND | ND | ND | C. lanienae | |
w, weak; +, positive; −, negative; R, resistant; S, susceptible; ND, not determined.
Identification based on species-specific primers.
Identification confirmed by using the sequence of the 16S rRNA gene.
Logan et al. (31) reported that C. lanienae was able to reduce nitrite.
Vandamme et al. (42) reported that 17% of A. butzleri isolates were susceptible to cephalothin.
Vandamme and De Ley (41) reported that C. hyointestinalis was not able to reduce nitrite.
Considerable numbers of presumptively identified C. lanienae isolates also were recovered from test cattle, but only on Karmali medium at 40°C (Table 4). All of these isolates and the C. lanienae reference strain were amplified with the primer set for the C. lanienae 16S rRNA gene (Fig. 5) but not with primer sets for other Campylobacter species. In addition, the reference strain of C. hyointestinalis subsp. lawsonii (NCTC 12901) but not that of C. hyointestinalis subsp. hyointestinalis (ATCC 35217) was amplified by the primer set for the C. lanienae 16S rRNA gene (Fig. 5). Seven arbitrarily selected isolates of C. lanienae (L52, L54, L83, L85, L71, L90, and L94) from bovine feces produced catalase, did not produce H2S, reduced nitrate but not nitrite, did not hydrolyze hippurate or indoxyl acetate, were resistant to nalidixic acid, and were sensitive to cephalothin (Table 5). The reference strain of C. lanienae exhibited the same physiological profile as the isolates from bovine feces, with the exception that it was resistant to cephalothin. Contrary to our findings, Logan et al. (31) reported that C. lanienae was able to reduce nitrite.
FIG. 5.
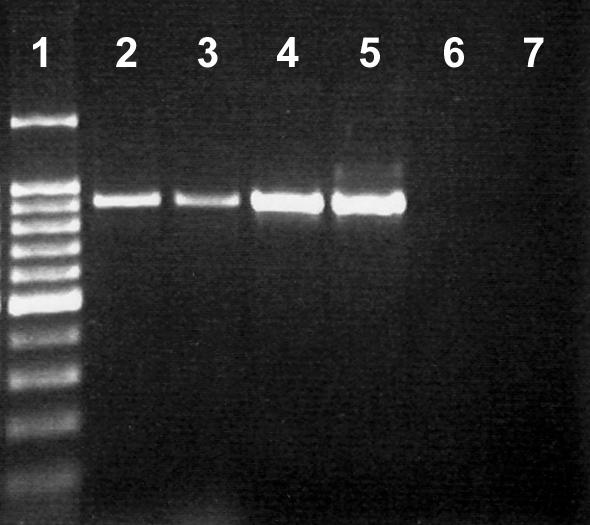
Amplification of C. lanienae isolates with the 16S rRNA gene primers (31). Lane 1, 100-bp molecular weight marker (the dark band was at 500 bp); lane 2, C. lanienae L52; lane 3, C. lanienae L54; lane 4, C. lanienae NCTC 13004; lane 5, C. hyointestinalis subsp. lawsonii NCTC 12901; lane 6, C. hyointestinalis subsp. hyointestinalis ATCC 35217; lane 7, negative control.
A comparison of two isolates from bovine feces (L52 and L54) indicated that they were 100% similar for the partial 16S rRNA gene. A comparison with other C. lanienae strains deposited at NCBI (GenBank accession numbers AB076675, AB076677, AF043423, and AF043425) indicated 98% identity. However, we detected three polymorphic bases that occurred in the annealing region of the reverse primer of Logan et al. (31), accounting for the weaker amplification products that we observed for the C. lanienae isolates from bovine feces (Fig. 5).
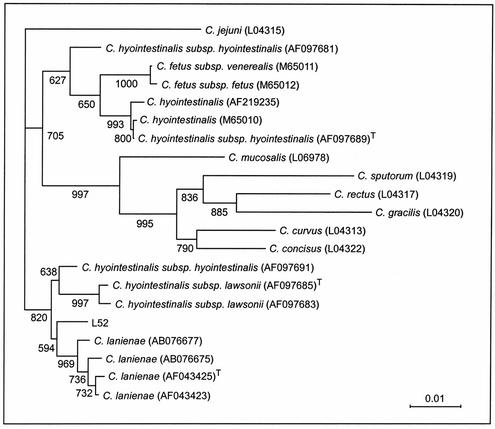
Multiple-sequence analysis was used to produce neighbor-joining, maximum-parsimony, and maximum-likelihood trees based on the 16S rRNA gene sequence of C. lanienae. The trees produced by the neighbor-joining, maximum-parsimony, and maximum-likelihood methods grouped L52 with DNA sequences obtained for other C. lanienae strains, but there was only moderate bootstrap support (42 to 59%) for this clade (Fig. 6). For all of the phylogenetic models tested, the C. lanienae clade was disparate from the C. hyointestinalis strains. However, the closest relationship was with the C. hyointestinalis subsp. lawsonii clade, which included one strain of C. hyointestinalis subsp. hyointestinalis (GenBank accession number AF097691). The other C. hyointestinalis strains included in the analysis formed a clade, but a clear relationship between the two subspecies of C. hyointestinalis was not resolved.
FIG. 6.
Dendrogram based on a majority-rule consensus tree obtained from analyzing the partial 16S rRNA gene data set with the NEIGHBOR program (neighbor-joining option) and showing DNA sequence relatedness for campylobacters. The outgroup used in the analysis was C. jejuni, and isolates indicated by “T” represent type specimens. The bar represents 0.01 nucleotide substitution per base, and numbers at selected nodes indicate support obtained by bootstrap analysis (1,000 replicates) for the internal branches within the resulting trees.
One isolate of C. fetus (L49) from cow 870 was recovered on CCDA at 40°C. This isolate, along with a C. fetus reference strain, was amplified with the C. fetus 23S rRNA primer set but not with primer sets for other species (Fig. 4). The two reference strains and the isolate of C. fetus from bovine feces produced catalase, did not produce H2S, reduced nitrate but not nitrite, did not hydrolyze hippurate or indoxyl acetate, were resistant to nalidixic acid, and were sensitive to cephalothin (Table 5). Like the reference strain of C. fetus subsp. fetus, L49 exhibited positive growth on brucella agar containing 1% glycine. Analysis of the 16S rRNA gene sequence for L49 indicated 98 to 99% identity with C. fetus subsp. venerealis (GenBank accession number AF482990) and C. fetus subsp. fetus (GenBank accession numbers AF219234, AF219233, and AJ306569) sequences deposited at NCBI.
Three isolates from cow 842 that were recovered on Karmali agar provided relatively weak amplicon bands with the Campylobacter genus-specific primer set and did not provide an amplification product with any of the Campylobacter species-specific primer sets. Multiplex PCR indicated that isolates L109 and L110 were A. skirrowii and that isolate L111 was A. butzleri (Fig. 7); the two presumptively identified A. skirrowii isolates were recovered on Karmali agar at 40°C, and the A. butzleri isolate was recovered on Karmali agar at 35°C. Analysis of the 16S rRNA genes for these three isolates indicated that L109 and L110 possessed 96% identity with A. skirrowii and that L111 possessed 99% identity with A. butzleri.
FIG. 7.
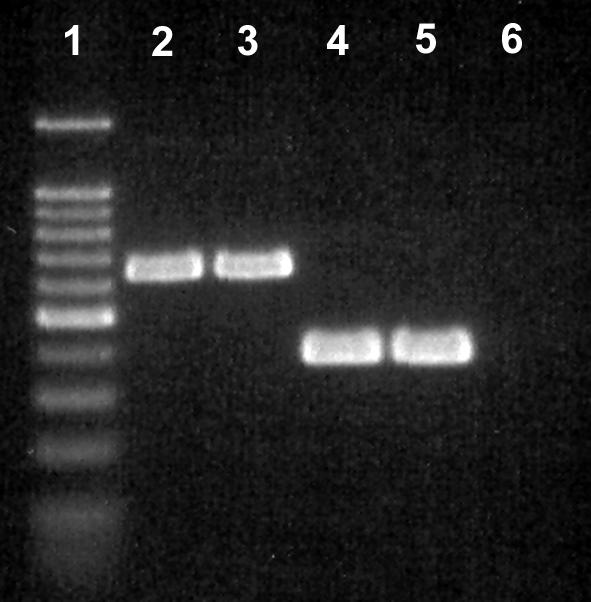
Multiplex PCR of 16S and 23S rRNA genes for distinguishing Arcobacter species (19). Lane 1, 100-bp molecular weight marker ladder (the dark band was at 500 bp); lane 2, A. skirrowii L109 from bovine feces; lane 3, A. skirrowii L110 from bovine feces; lane 4, A. butzleri L111 from bovine feces; lane 5, A. butzleri ATCC 49616; lane 6, negative control.
(ii) Direct detection of Campylobacter species by PCR.
Campylobacter species DNA was detected in 75% (n = 18) of the fecal samples tested, but none of the fecal samples obtained from cow 986 was positive (Table 6). In no instance was DNA from either C. coli, C. fetus, or C. hyointestinalis detected in bovine feces. In contrast, 25% (n = 4) of the fecal samples were positive for C. jejuni, and 67% (n = 16) of the samples were positive for C. lanienae. Relative to the efficacy of isolation on microbiological media (combined media and incubation temperatures), PCR was more sensitive for total campylobacters and C. lanienae in 8 and 17% of the fecal samples, respectively. Conversely, microbiological isolation was more sensitive than PCR for C. jejuni in 13% of the samples.
TABLE 6.
Microbiological isolation and PCR detection of campylobacters in bovine feces by replicatea
| Taxon and method | Positive results from the following cow:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 593 | 790 | 791 | 842 | 850 | 870 | 985 | 986 | |
| Genus Campylobacter | ||||||||
| Campy-Line | 0-0-0 | 1-1-0 | 0-1-1 | 0-1-0 | 0-0-0 | 1-1-0 | 0-0-0 | 0-0-0 |
| Karmali | 1-1-1 | 1-1-0 | 0-1-1 | 1-1-1 | 0-1-0 | 1-1-1 | 0-1-1 | 0-0-0 |
| CCDA | 0-0-0 | 1-1-0 | 0-1-0 | 1-1-0 | 0-0-0 | 1-1-1 | 0-0-0 | 0-0-0 |
| Preston | 0-0-0 | 1-1-0 | 0-1-0 | 0-1-0 | 0-0-0 | 1-1-1 | 0-1-0 | 0-0-0 |
| PCR—Campylobacter | 1-1-1 | 1-1-1 | 0-1-1 | 0-1-0 | 1-1-1 | 1-1-1 | 1-1-1 | 0-0-0 |
| PCR—internal control | 0-0-0 | 0-0-0 | 1-0-0 | 1-0-1 | 0-0-0 | 0-0-0 | 0-0-0 | 1-1-1 |
| Campylobacter jejuni | ||||||||
| Campy-Line | 0-0-0 | 1-1-0 | 0-1-1 | 0-1-0 | 0-0-0 | 1-1-0 | 0-0-0 | 0-0-0 |
| Karmali | 0-0-0 | 1-1-0 | 0-1-1 | 1-1-0 | 0-0-0 | 1-1-1 | 0-0-0 | 0-0-0 |
| CCDA | 0-0-0 | 1-1-0 | 0-1-0 | 1-1-0 | 0-0-0 | 1-1-1 | 0-0-0 | 0-0-0 |
| Preston | 0-0-0 | 1-1-0 | 0-1-0 | 0-1-0 | 0-0-0 | 1-1-1 | 0-1-0 | 0-0-0 |
| PCR—C. jejuni | 0-0-0 | 1-1-0 | 0-1-0 | 0-0-0 | 0-0-0 | 1-1-1 | 0-0-0 | 0-0-0 |
| Campylobacter lanienae | ||||||||
| Campy-Line | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 |
| Karmali | 1-1-1 | 0-1-0 | 0-1-1 | 0-0-0 | 0-1-1 | 1-1-0 | 0-1-1 | 0-0-0 |
| CCDA | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 |
| Preston | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 | 0-0-0 |
| PCR—C. lanienae | 1-1-1 | 0-1-1 | 0-1-1 | 0-0-0 | 1-1-1 | 1-1-1 | 1-1-1 | 0-0-0 |
No C. coli, C. fetus, or C. hyointestinalis was detected by PCR. For all media, results are compiled across the temperature treatment. Data represent a positive result for each of three replicates conducted over time (i.e., replicate 1 [R1]-R2-R3), where 1 is positive and 0 is negative. Bold type indicates a discrepancy between the results of the isolation and PCR detection methods. In all instances when there was a positive result for isolation and a negative result for PCR, very small numbers of CFU were recovered (<6).
DISCUSSION
We isolated C. jejuni, C. fetus subsp. fetus, C. lanienae, A. butzleri, and A. skirrowii from the feces of dairy cows. Interestingly, we detected multiple species (≥2) in feces from five of eight cows. To our knowledge, this is the first report of C. lanienae associated with a livestock source; this bacterium was first isolated from humans working in abattoirs in Switzerland that had been exposed to cattle carcasses, but its pathogenicity or virulence in humans has not been ascertained (31). Although we found some disparity, the isolates that we obtained from cattle feces clustered with known strains of C. lanienae based on the 16S rRNA gene. Interestingly, C. hyointestinalis subsp. lawsonii was observed to be more closely related to C. lanienae than it was to many strains of C. hyointestinalis subsp. hyointestinalis based on 16S rDNA sequences. The relatedness of these two taxa was further supported by our observation of positive amplification of C. hyointestinalis subsp. lawsonii but not C. hyointestinalis subsp. hyointestinalis with the C. lanienae primer set. Logan et al. (31) did not include C. hyointestinalis subsp. lawsonii in their description of C. lanienae as a new species, and Harrington and On (18) did not include C. lanienae in their phylogenetic analysis of C. hyointestinalis strains. The type specimen of C. hyointestinalis subsp. lawsonii (GenBank accession number AF097685) possesses 97% rDNA sequence identity with the type specimen of C. lanienae (GenBank accession number AF043425), and 95% identity with the type specimen of C. hyointestinalis subsp. hyointestinalis (GenBank accession number AF09769). Furthermore, considerable variation in 16S rDNA sequences was observed within the seven strains of C. hyointestinalis examined in the current study, and the phylogenetics of this species may require revision. For example, should C. lanienae be lowered to a subspecies of C. hyointestinalis, or should C. hyointestinalis subsp. lawsonii be elevated to species status?
Extreme care had to be taken to detect colonies of Campylobacter on the test media. Furthermore, considerable variability was observed in the frequency of isolation of species among the four media and incubation temperatures tested. The primary taxa that we recovered were C. jejuni and C. lanienae, but C. fetus subsp. fetus, A. butzleri, and A. skirrowii also were isolated infrequently. The media that we used were primarily developed for selectively isolating C. jejuni and C. coli, and the ineffectiveness of these media for the isolation of other taxa of campylobacters has been well documented by others (1, 4, 12, 37). C. hyointestinalis and C. coli are often isolated from bovine feces at low frequencies (1), and we have previously isolated these taxa on CCDA in Alberta, Canada. However, neither of these bacteria was recovered in the current study, a result which was likely a function of the small number of animals sampled and not of the isolation media that we used. We included 30 and 35°C incubation treatments in an attempt to facilitate the isolation of nonthermophilic taxa, such as C. fetus. We only recovered one isolate of C. fetus subsp. fetus on CCDA at 40°C. This finding contrasts with the much higher frequency of isolation of C. fetus subsp. fetus from bovine fecal samples by others. For example, Atabay and Corry (1) isolated this bacterium in feces from 11% of the dairy cattle they tested. Unfortunately, the effect of temperature on isolation frequency was confounded by the extensive growth of fungal contaminants. The most prevalent fungus that we observed on all four media was G. candidum, an arthroconidial fungus that is able to grow in low-oxygen environments (11). This fungus possesses a cosmopolitan distribution, it is commonly associated with feces, and it is a human pathogen causing geotrichoses affecting oral, bronchial, and bronchopulmonary epithelia and/or skin. In the current study, we relied on direct plating of feces and did not use enrichment methods which have been shown to be effective in selectively isolating a number of Campylobacter taxa from bovine feces, including C. sputorum (1). While enrichment methods may facilitate the isolation of some taxa, the enrichment medium used also is selective and will discriminate against some taxa.
As a result of the fastidiousness and diversity of campylobacters and arcobacters, it is clear that no one medium will provide an accurate measure of their occurrence. Furthermore, inexperienced personnel can easily overlook Campylobacter colonies, and species may be missed, particularly rarely occurring taxa that do not produce distinct colonies. For this reason, the use of PCR-based detection methods is very attractive. However, a variety of PCR inhibitors occur in feces, and the presence of these inhibitory compounds may inhibit the PCR and provide false-negative results (2, 7, 47). The QIAamp DNA stool minikit possesses a polysaccharide mixture that is used to remove PCR inhibitors of fecal origin, and it was found to provide the best performance in PCR for human feces compared to three other commercial kits (32). To determine whether PCR inhibitors have been removed, a number of researchers have designed internal primers and added them to feces prior to DNA extraction (33, 35, 39). We designed our internal control based on the strategy of Denis et al. (10), but our internal control was designed to amplify with the Campylobacter genus-specific primer set. In all instances, we observed either amplification of the internal control or Campylobacter DNA from extracted bovine feces, indicating that the DNA stool minikit sufficiently removed PCR inhibitors present in the feces.
Considerable variation in the consistency (e.g., water content and presence of mucus) and composition (e.g., fiber versus grain) of the cattle feces used in the current study were observed, and these and other variables may have an impact on the efficacy of inhibitor removal. We used the amount of feces recommended by the manufacturer (200 mg [fresh weight]). However, very little is known about the spatial distribution of bacteria in feces, and if campylobacters are aggregated, the small amount of feces sampled may provide an erroneous measure of Campylobacter species prevalence. It may be possible to increase the amount of feces sampled, but increased sample size may overload the purification step. Complex polysaccharides are commonly encountered inhibitors of PCR found in feces (33). Furthermore, polyphenolic compounds of plant origin can be very inhibitory to PCR (23), and the high plant content of cattle feces would be expected to contribute large quantities of both complex polysaccharides and polyphenolic compounds, a factor which may limit the biomass of feces that can be efficaciously processed by the QIAamp DNA stool minikit. This possibility remains to be determined.
The quality of the extracted DNA is only one aspect of achieving highly specific and sensitive detection of a target bacterium. The amount of DNA present is also important, and nested PCR has been used with various degrees of success to increase the sensitivity of detection, particularly for genes with one or a small number of copies (21, 32, 36, 43). We observed that the nested and seminested primers were highly sensitive (102 to 103 CFU g−1) in detecting C. coli, C. fetus, C. hyointestinalis subsp. hyointestinalis, and C. jejuni. This sensitivity is substantially higher (≈100 times) than that of nonnested PCRs used by others (25, 27). Given that only 200 mg of bovine feces was processed, we obtained a detection sensitivity of approximately 20 cells. In addition to the direct effects of nested PCR, the nested step may also increase sensitivity by diluting out inhibitors; some amplification would be expected to occur in the primary step even in the presence of PCR inhibitors, providing a sufficient template for the secondary reaction (32). Interestingly, we observed variability in detection sensitivities among the three sample times. Reasons for the variability among the replicates are uncertain, but the variability does not appear to be due to PCR inhibitors.
We observed that the nonnested 16S rDNA gene primers designed to detect campylobacters in feces were more sensitive in general than the nested primers for specific taxa. Even in uninoculated feces (i.e., minimum detection experiment), positive amplification of Campylobacter DNA was detected (33 to 83% of the samples tested). Analysis of the sequences of these amplicons demonstrated high similarity with Campylobacter (data not shown). A number of Campylobacter species not targeted in the current study (i.e., with nested or seminested primers) have been reported from bovine feces. For example, Giacoboni et al. (15) isolated C. lari, Atabay and Corry (1) recovered C. sputorum, and we recovered C. lanienae from bovine feces. Furthermore, we isolated A. butlzeri and A. skirrowii from bovine feces, and the genus-specific primers were observed to amplify DNA of these taxa, contrary to previous findings (29). Therefore, the apparently higher sensitivity of the genus-specific primers than of the species-specific primers may not be due to increased sensitivity but rather to the presence of additional Campylobacter species in the feces. In contrast to the minimum detection experiment, we included a primer set specific for C. lanienae in the direct PCR of campylobacters in the bovine feces experiment. The combined amplification frequency for C. jejuni and C. lanienae corresponded closely with that of the genus-specific primer set in this experiment. Nevertheless, the development and testing of nested primers for additional species of campylobacters and arcobacters in bovine feces are necessary, and we are currently pursuing this goal.
Relative to microbiological isolation, PCR was found to be more sensitive for detecting total campylobacters and C. lanienae but not C. jejuni. The media that we tested are designed to isolate C. jejuni and C. coli (1, 4, 12, 37), and this factor provides an explanation for the high isolation sensitivity that we observed for C. jejuni. Given the diversity of campylobacters associated with cows, the advantages of PCR over conventional isolation methods that we observed agree with the conclusions of Lawson et al. (26), who found that direct PCR of human feces provided unique data about mixed infections by non-C. coli and non-C. jejuni campylobacters.
We observed that the QIAamp DNA stool minikit effectively removed any inhibitors in bovine feces, as indicated by the amplification of an internal control designed to amplify under the same conditions as a 16S rDNA primer set for campylobacters and arcobacters. Using nested and seminested PCRs, we were able to detect C. coli, C. fetus, C. hyointestinalis, and C. jejuni at densities of 102 to 103 CFU g−1 of feces (fresh weight). The PCR-based detection method was found to be substantially more effective in detecting C. lanienae than conventional culturing methods, and this is the first report of the detection of C. lanienae outside of humans, to our knowledge. Given the logistical advantages and high resolution for detecting campylobacters in bovine feces, PCR-based methods will facilitate the understanding of the interaction of this fastidious group of pathogens with cattle.
Acknowledgments
We thank Kathaleen House (Agriculture and Agri-Food Canada Research Centre, Lethbridge, Alberta, Canada [AAFC]) for technical assistance in the sequencing of 16S rDNA, Tom Graham (Animal Diseases Research Institute, Health Canada, Lethbridge, Alberta, Canada) for providing microbiological media, Brian Barron (AAFC) for providing access to the dairy cattle, Hilma Busz (AAFC) and Roger Johnson (Health Canada, Guelph, Ontario, Canada) for providing isolates of Arcobacter and/or Campylobacter, Jay Yanke (AAFC) for lyophilizing isolates of Campylobacter, Kaarina Benkel (AAFC) for conducting the sequencing of 16S rDNA, Georg Hausner (University of Manitoba, Winnipeg, Manitoba, Canada) for assistance with the phylogenetic analyses, and Ron Teather (AAFC) and two anonymous reviewers for comments on the manuscript.
This study was supported by a grant from the Canada-Alberta Beef Industry Development Fund (CABIDF).
Footnotes
Contribution 02097 from the Agriculture and Agri-Food Canada Research Centre, Lethbridge, Alberta, Canada.
REFERENCES
- 1.Atabay, H. I., and J. E. L. Corry. 1998. The isolation and prevalence of campylobacters from dairy cattle using a variety of methods. J. Appl. Microbiol. 84:733-740. [DOI] [PubMed] [Google Scholar]
- 2.Ballagi-Pordany, A., and S. Belak. 1996. The use of mimics as internal standards to avoid false negatives in diagnostic PCR. Mol. Cell. Probes 10:159-164. [DOI] [PubMed] [Google Scholar]
- 3.Bastyns, K., S. Chapelle, P. Vandamme, H. Goossens, and R. de Wachter. 1994. Species-specific detection of campylobacters important in veterinary medicine by PCR amplification of 23S rDNA areas. Syst. Appl. Microbiol. 17:563-568. [Google Scholar]
- 4.Baylis, C. L., S. MacPhee, K. W. Martin, T. J. Humphrey, and R. P. Betts. 2000. Comparison of three enrichment media for the isolation of Campylobacter spp. from foods. J. Appl. Microbiol. 89:884-891. [DOI] [PubMed] [Google Scholar]
- 5.Busato, A., D. Hofer, T. Lentze, C. Gaillard, and A. Burnens. 1999. Prevalence and infection risks of zoonotic enteropathogenic bacteria in Swiss cow-calf farms. Vet. Microbiol. 69:251-263. [DOI] [PubMed] [Google Scholar]
- 6.Collins, E., M. Glennon, S. Hanley, A. M. Murray, M. Cormican, T. Smith, and M. Maher. 2001. Evaluation of PCR-DNA probe colorimetric membrane assay for identification of Campylobacter spp. in human stool specimens. J. Clin. Microbiol. 39:4163-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cone, R., A. Hobson, and M. Huang. 1992. Coamplified positive control detects inhibition of polymerase chain reactions. J. Clin. Microbiol. 30:3185-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corry, J. E., D. E. Post, P. Colin, and M. J. Laisney. 1995. Culture media for the isolation of campylobacters. Int. J. Food Microbiol. 26:43-76. [DOI] [PubMed] [Google Scholar]
- 9.Denis, M., J. Refrégier-Petton, M.-J. Laisney, G. Ermel, and G. Salvat. 2001. Campylobacter contamination in French chicken production from farm to consumers. Use of a PCR assay for detection and identification of Campylobacter jejuni and Camp. coli. J. Appl. Microbiol. 91:255-267. [DOI] [PubMed] [Google Scholar]
- 10.Denis, M., C. Soumet, K. Rivoal, G. Ermel, D. Blivet, G. Salvat, and P. Colin. 1999. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 29:406-410. [DOI] [PubMed] [Google Scholar]
- 11.Domsch, K. H., W. Gams, and T. H. Anderson. 1980. Compendium of soil fungi. Academic Press Ltd., London, United Kingdom.
- 12.Engberg, J., S. L. W. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1995. PHYLIP (phylogeny inference package), version 3.572.c. University of Washington, Seattle.
- 14.Garcia, M. M., H. Lior, R. B. Stewart, G. M. Ruckerbauer, J. R. R. Trudel, and A. Skljarevski. 1985. Isolation, characterization, and serotyping of Campylobacter jejuni and Campylobacter coli from slaughter cattle. Appl. Environ. Microbiol. 49:667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giacoboni, G. I., K. Itoh, K. Hirayama, E. Takahashi, and T. Mitsuoka. 1993. Comparison of fecal Campylobacter in calves and cattle of different ages and areas in Japan. J. Vet. Med. Sci. 55:555-559. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez, I., K. A. Grant, P. T. Richardson, S. F. Park, and M. D. Collins. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goossens, H., and J. P. Butzler. 1992. Isolation and identification of Campylobacter spp., p. 93-109. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins. (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, D.C.
- 18.Harrington, C. S., and S. L. W. On. 1999. Extensive 16S rRNA gene sequence diversity in Campylobacter hyointestinalis strains: taxonomic and applied implications. Int. J. Syst. Bacteriol. 49:1171-1175. [DOI] [PubMed] [Google Scholar]
- 19.Houf, K., A. Tutenel, L. De Zutter, J. Van Hoof, and P. Vandamme. 2000. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 193:89-94. [DOI] [PubMed] [Google Scholar]
- 20.Humphrey, T. J., and P. Beckett. 1987. Campylobacter jejuni in dairy cows and raw milk. Epidemiol. Infect. 98:263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, C., C. Li, T. Ha, D. A. Ferguson, D. S. Chi, J. J. Laffan, and E. Thomas. 1998. Identification of H. pylori in saliva by a nested PCR assay derived from a newly cloned DNA probe. Dig. Dis. Sci. 43:1211-1218. [DOI] [PubMed] [Google Scholar]
- 22.Karmali, M. A., A. E. Simor, M. Roscoe, P. C. Fleming, S. S. Smith, and J. Lane. 1986. Evaluation of a blood-free, charcoal-based, selective medium for the isolation of Campylobacter organisms from feces. J. Clin. Microbiol. 23:456-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonjul, P. K., W. F. Brandt, J. M. Farrant, and G. G. Lindsey. 1999. Inclusion of polyvinylpyrrolidone in the polymerase chain reaction reverses the inhibitory effects of polyphenolic contamination of RNA. Nucleic Acids Res. 27:915-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley & Sons, Chichester, United Kingdom.
- 25.Lawson, A. J., D. Linton, J. Stanley, and R. J. Owen. 1997. PCR detection and speciation of Campylobacter upsaliensis and Campylobacter helveticus in human faeces, and comparison with cultural methods. J. Appl. Microbiol. 83:375-380. [DOI] [PubMed] [Google Scholar]
- 26.Lawson, A. J., J. M. J. Logan, G. L. O'Neill, M. Desai, and J. Stanley. 1999. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR- enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37:3860-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawson, A. J., M. S. Shafi, K. Pathak, and J. Stanley. 1998. Detection of campylobacter in gastroenteritis: comparison of direct PCR assay of faecal samples with selective culture. Epidemiol. Infect. 121:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Line, J. E. 2001. Development of a selective differential agar for isolation and enumeration of Campylobacter spp. J. Food Prot. 64:1711-1715. [DOI] [PubMed] [Google Scholar]
- 29.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 30.Linton, D., A. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logan, J. M., A. Burnens, D. Linton, A. J. Lawson, and J. Stanley. 2000. Campylobacter lanienae sp. nov., a new species isolated from workers in an abattoir. Int. J. Syst. E vol. Microbiol. 50:865-872. [DOI] [PubMed] [Google Scholar]
- 32.McOrist, A. L., M. Jackson, and A. R. Bird. 2002. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J. Microbiol. Methods 50:131-139. [DOI] [PubMed] [Google Scholar]
- 33.Monteiro, L., D. Bonnemaison, A. Vekris, K. G. Petry, J. Bonnett, R. Vidal, J. Cabrita, and F. Mégraud. 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono, K., H. Masaki, and Y. Tokumaru. 1995. Isolation of Campylobacter spp. from slaughtered cattle and swine on blood-free selective media. J. Vet. Med. Sci. 57:1085-1087. [DOI] [PubMed] [Google Scholar]
- 35.Rijpens, N., L. Herman, F. Vereecken, G. Jannes, J. De Smedt, and L. De Zutter. 1999. Rapid detection of stressed Salmonella spp. in dairy and egg products using immunomagnetic separation and PCR. Int. J. Food Microbiol. 46:37-44. [DOI] [PubMed] [Google Scholar]
- 36.Saruta, K., T. Matsunaga, M. Kono, S. Hoshina, S. Kanemoto, O. Sakai, and K. Machida. 1997. Simultaneous detection of Streptococcus pneumoniae and Haemophilus influenzae by nested PCR amplification from cerebrospinal fluid samples. FEMS Immunol. Med. Microbiol. 19:151-157. [DOI] [PubMed] [Google Scholar]
- 37.Skirrow, M. B. 1994. Diseases due to Campylobacter, Helicobacter and related bacteria. J. Comp. Pathol. 111:113-149. [DOI] [PubMed] [Google Scholar]
- 38.Stanley, K. N., J. S. Wallance, J. E. Currie, P. J. Diggle, and K. Jones. 1998. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85:472-480. [DOI] [PubMed] [Google Scholar]
- 39.Staub, J. A., C. Hertel, D. Mäde, and W. P. Hammes. 1999. Development and application of a heterologous internal standard for polymerase-chain-reaction-based detection of Campylobacter jejuni and Campylobacter coli in foods. Eur. Food Res. Technol. 209:180-184. [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vandamme, P., and J. De Ley. 1991. Proposal for a new family, Campylobacteraceae. Int. J. Syst. Bacteriol. 41:451-455. [Google Scholar]
- 42.Vandamme, P., M. Vancanneyt, B. Pot, L. Mels, B. Hoste, D. Dewettinck, L. Vlaes, C. van den Borre, R. Higgins, J. Hommez, K. Kersters, J.-P. Butzler, and H. Goossens. 1992. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42:344-356. [DOI] [PubMed] [Google Scholar]
- 43.Verdin, E., C. Saillard, A. Labbe, J. M. Bove, and M. Kobisch. 2000. A nested PCR assay for detection of Mycoplasma hyopneumoniae in tracheobronchiolar washings from pigs. Vet. Microbiol. 76:31-40. [DOI] [PubMed] [Google Scholar]
- 44.Warner, D. P., J. H. Bryner, and G. W. Beran. 1986. Epidemiologic study of campylobacteriosis in Iowa cattle and the possible role of unpasteurized milk as a vehicle of infection. Am. J. Vet. Res. 47:254-258. [PubMed] [Google Scholar]
- 45.Wesley, I. V., S. J. Wells, K. M. Harmon, A. Green, L. Schroeder-Tucker, M. Glover, and I. Siddique. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl. Environ. Microbiol. 66:1194-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widjojoatmodjo, M. N., A. C. Fluit, R. Torensma, G. P. H. T. Verdonk, and J. Verhoef. 1992. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J. Clin. Microbiol. 30:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, C. J. 1997. Inhibition and facilitation of nucleic acid amplification. J. Appl. Microbiol. 26:9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]