Abstract
Genetic relationships among 62 Vibrio vulnificus strains of different geographical and host origins were analyzed by multilocus enzyme electrophoresis (MLEE), random amplification of polymorphic DNA (RAPD), and sequence analyses of the recA and glnA genes. Out of 15 genetic loci analyzed by MLEE, 11 were polymorphic. Cluster analysis identified 43 distinct electrophoretic types (ETs) separating the V. vulnificus population into two divisions (divisions Ι and ΙΙ). One ET (ET 35) included all indole-negative isolates from diseased eels worldwide (biotype 2). A second ET (ET 2) marked all of the strains from Israel isolated from patients who handled St. Peter's fish (biotype 3). RAPD analysis of the 62 V. vulnificus isolates identified 26 different profiles separated into two divisions as well. In general, this subdivision was comparable (but not identical) to that observed by MLEE. Phylogenetic analysis of 543 bp of the recA gene and of 402 bp of the glnA gene also separated the V. vulnificus population into two major divisions in a manner similar to that by MLEE and RAPD. Sequence data again indicated the overall subdivision of the V. vulnificus population into different biotypes. In particular, indole-negative eel-pathogenic isolates (biotype 2) on one hand and the Israeli isolates (biotype 3) on the other tended to cluster together in both gene trees. None of the methods showed an association between distinct clones and human clinical manifestations. Furthermore, except for the Israeli strains, only minor clusters comprising geographically related isolates were observed. In conclusion, all three approaches (MLEE, RAPD, and DNA sequencing) generated comparable but not always equivalent results. The significance of the two divisions (divisions Ι and ΙΙ) still remains to be clarified, and a reevaluation of the definition of the biotypes is also needed.
Vibrio vulnificus is a common worldwide inhabitant of estuarine and marine waters. It can be isolated under a wide range of salinity and temperature conditions from oysters, clams, mussels, and fish, as well as from sediment and plankton (5). This gram-negative bacterium is an opportunistic human pathogen, causing disease mostly in persons with underlying chronic disorders, especially those with a high iron level in their blood (23, 24). In immunocompromised individuals, V. vulnificus may be fatal. Ingestion of V. vulnificus-infected shellfish can lead to gastroenteritis and primary septicemia, while contact with existing skin lesions can result in edema, erythema, and necrotizing wound infections.
Based on phenotypic characteristics and host range criteria, V. vulnificus isolates have been grouped into three different biotypes. Biotype 1 strains have been associated with pathogenicity in humans, have a positive indole reaction, and are serologically heterogeneous. Biotype 2 strains are pathogenic for both humans and eels, typically have a negative indole reaction, and are characterized by a homogeneous lipopolysaccharide (LPS) O antigen (3, 8). This subdivision has been reinforced by several genetic assays, which showed that, in contrast to biotype 1 organisms, biotype 2 strains are genetically homogeneous and harbor high-molecular-weight plasmids (8, 9). Several studies have documented the emergence of a few atypical strains, which are apparently pathogenic for eels but have a positive indole reaction and a different genotype than indole-negative eel-pathogenic strains (2, 8). Other atypical isolates include a few indole-negative strains isolated from the environment and from a human wound infection, not from diseased eels (8). The existence of atypical isolates, including indole-positive strains isolated from diseased eels, has led to the suggestion to replace the designation biotype 2 by serovar E (8). Serovar E is a homogeneous LPS O serogroup to which have been assigned all former biotype 2 strains as well as the atypical strains, on the basis of the serological characteristics of their LPS side chains, reacting with specific antisera against biotype 2 strains E22 and NCIMB 2137 (8). The high-molecular-weight portion of their LPS O side chains seems to protect serovar E isolates against the bactericidal action of the eel serum complement (1, 18).
Recently, a third V. vulnificus variant has been found in Israel (called biotype 3 by Bisharat et al. 10). The organisms were isolated from patients who handled St. Peter's fish (Tilapia spp.).
Classification methods used in the past have been insufficient to reveal the phylogenetic relationships between the three V. vulnificus variants and to measure the genetic distances separating them. To achieve this and to obtain the best possible evaluation of the V. vulnificus population structure, we used three different molecular methods: multilocus enzyme electrophoresis (MLEE) (26), random amplification of polymorphic DNA (RAPD) (28), and sequence typing of two genes, recA and glnA. The recA gene encodes RecA, a protein involved in homologous recombination, DNA repair, and the SOS response (21). The glnA gene encodes a glutamine synthetase, an enzyme involved in nitrogen metabolism and ammonia assimilation in eukaryotes as well as in prokaryotes (17). This study is the first to analyze the V. vulnificus population structure based on the comparative use of a variety of population genetic methods.
MATERIALS AND METHODS
Bacterial strains.
Sixty-two V. vulnificus strains originating from different countries (Denmark, Israel, Spain, and the United States), as well as a Vibrio diazotrophicus strain and a Listonella anguillarum strain, were studied (Table 1). The latter two species were added as outgroups to allow the tree rooting in the phylogenetic analysis based on gene sequencing. This collection comprised clinical and environmental isolates as well as isolates from healthy and diseased eels. The indole reaction was verified by a standard method.
TABLE 1.
Origin and characteristics of the isolates
| Isolate | Indole reaction | Biotype | Serovar Ea | Sample origin | Geographic originb | Source (reference)c |
|---|---|---|---|---|---|---|
| 523 | + | 2 | + | Diseased eel | Unknown | E. G. Biosca, CECT |
| 58 | + | 3 | NAd | Human clinicale | Israel | Israel (13) |
| 162 | + | 3 | NA | Human clinicale | Israel | Israel (13) |
| 1033 | + | 3 | NA | Human clinicale | Israel | Israel (13) |
| 11028 | + | 3 | NA | Human clinicale | Israel | Israel (13) |
| 530 | + | 1 | − | Diseased eel | Belgium | E. G. Biosca, CECT |
| 9060-96 | + | NA | NA | Human clinicalf | Texas, GB | B. Swaminathan, CDC |
| 9083-96 | + | NA | NA | Human clinicalf | Louisiana, SL | B. Swaminathan, CDC |
| 9048-96 | + | NA | NA | Human clinicalg | United States, NA | B. Swaminathan, CDC |
| 96-9-114s | + | 1 | NA | Sediment | Denmark | L. Høi, Denmark |
| EDL-174 | + | NA | NA | Blood | Georgiah | J. Powell, Maryland |
| TW1 | + | 1 | − | Fish farm tank water | Spain | E. G. Biosca, CECT |
| UNCC913 | + | NA | NA | Environment | Michiganh | J. Powell, Maryland |
| 9038-96 | + | NA | NA | Human clinicalf | Louisiana, GB | B. Swaminathan, CDC |
| 9070-96 | + | NA | NA | Human clinicalf | Louisiana, GB | B. Swaminathan, CDC |
| 9081-96 | + | NA | NA | Human clinicalf | United States, NA | B. Swaminathan, CDC |
| 96-8-129AG | + | 1 | − | Wild eel | Denmark | L. Høi, Denmark |
| 96-10-9M | + | 1 | NA | Blue mussel | Denmark | L. Høi, Denmark |
| 95-6-1R | + | 1 | − | Tiger shrimp | Asia | L. Høi, Denmark |
| 9031-96 | + | NA | NA | Human clinicalf | Florida, AB | B. Swaminathan, CDC |
| 9046-96 | + | NA | NA | Human clinicalg | United States, NA | B. Swaminathan, CDC |
| 9076-96 | + | NA | NA | Human clinicalf | Louisiana, SL | B. Swaminathan, CDC |
| 9003-97 | + | NA | NA | Human clinicalf | Louisiana, BB | B. Swaminathan, CDC |
| 9074-96 | + | NA | NA | Human clinicalf | Texas, GB; CB | B. Swaminathan, CDC |
| 9067-96 | + | NA | NA | Human clinicalf | Texas, GB | B. Swaminathan, CDC |
| MO6-24 | + | NA | − | Blood | Californiah | J. Powell, Maryland |
| V1015H | + | NA | NA | Blood | Louisianah | J. Powell, Maryland |
| 2809-78 | + | NA | NA | Blood | South Carolinah | J. Powell, Maryland |
| 85A5667 | + | NA | NA | Blood | Californiah | J. Powell, Maryland |
| 529 | + | 1 | − | Blood | United States | CECT |
| CVD773d | + | NA | NA | Environment | Marylandh | J. Powell, Maryland |
| 5C1326 | + | NA | NA | Blood | Marylandh | J. Powell, Maryland |
| 52785 | + | NA | NA | Blood | New Yorkh | J. Powell, Maryland |
| E109 | + | 1 | − | Healthy eel | Spain | E. G. Biosca, CECT |
| 96-7-137 | + | 2 | NA | Diseased eel | Denmark | L. Høi, Denmark |
| 94-8-110 | + | 1 | − | Blood | Denmark | L. Høi, Denmark |
| 94-8-111 | + | NA | NA | Wound infection | Denmark | L. Høi, Denmark |
| BO62312 | + | NA | NA | Blood | Marylandh | J. Powell, Maryland |
| 96-9-113v | + | 1 | NA | Tank water | Denmark | L. Høi, Denmark |
| 94-8-112 | − | 2 | + | Wound infection | Denmark | L. Høi, Denmark |
| 898 | − | 2 | NA | Diseased eel | Japan | M. Nishiluchi, CECT |
| ATCC 33149 | − | 2 | + | Diseased eel | Japan | R. Aznar and C. R. Arias, Spain |
| NCIMB 2136 | − | 2 | + | Diseased eel | Japan | R. Aznar and C. R. Arias, Spain |
| NCIMB 2138 | − | 2 | + | Diseased eel | Japan | R. Aznar and C. R. Arias, Spain |
| E86 | − | 2 | + | Diseased eel | Spain | R. Aznar and C. R. Arias, Spain |
| E22 | − | 2 | + | Diseased eel | Spain | E. G. Biosca, CECT |
| E39 | − | 2 | + | Diseased eel | Spain | E. G. Biosca, CECT |
| E58 | − | 2 | + | Diseased eel | Spain | E. G. Biosca, CECT |
| E116 | − | 2 | + | Diseased eel | Spain | E. G. Biosca, CECT |
| CIP 8190 | − | NA | NA | Blood | France | C. Kingombe |
| 90-2-11 | − | 2 | + | Diseased eel | Denmark | L. Høi, Denmark |
| 94-9-123 | − | 2 | NA | Environment | Denmark | L. Høi, Denmark |
| 9005-97 | + | NA | NA | Human clinicalf | Louisiana, GC | B. Swaminathan, CDC |
| 9075-96 | + | NA | NA | Human clinicalf | Florida, AB | B. Swaminathan, CDC |
| 94-9-115 | + | 1 | − | Clinical | Denmark | L. Høi, Denmark |
| M63 | + | 1 | − | Healthy eel | Spain | R. Aznar and C. R. Arias, Spain |
| M79 | + | 1 | − | Healthy eel | Spain | R. Aznar and C. R. Arias, Spain |
| M89 | + | 1 | − | Healthy eel | Spain | R. Aznar and C. R. Arias, Spain |
| M90 | + | 1 | − | Healthy eel | Spain | R. Aznar and C. R. Arias, Spain |
| 94-9-114 | + | 1 | − | Blood | Denmark | L. Høi, Denmark |
| 6353 | + | NA | NA | Blood | Marylandh | J. Powell, Maryland |
| V345-83 | + | NA | NA | Environment | Louisianah | J. Powell, Maryland |
| 627i | + | Spain | E. G. Biosca, CECT | |||
| 522j | − | Spain | E. G. Biosca, CECT |
Serovar E was determined either by Western blotting with an antiserum against V. vulnificus biotype 2 strain E22 (8) or by slide agglutination with an antiserum against V. vulnificus biotype 2 strain E39 (2). +, serovar E; −, not serovar E. For strains obtained from Denmark, minus means that the isolates do not belong to any known serotype and plus means serovar O4, which, according to Høi et al. (20), corresponds to serovar E.
Specific origins within the United States of the raw shellfish responsible for disease in patients: GB, Galveston Bay; CB, California Bay; SL, Sister Lake; GC, Grand Caillou; AB, Apach Bay; BB, Black Bay; NA, data not available.
CECT, Colección Española de Cultivos Tipo; CDC, Centers for Disease Control and Prevention.
NA, data not available.
Human infected Tilapia spp.
Human clinical cases associated with oyster consumption.
Human clinical origin associated with shellfish consumption.
Only the U.S. state is available.
V. diazotrophicus from Strongylocentrotus droebachiensis.
L. anguillarum from Gadus morhua.
Culture conditions and specimen storage.
Strains were grown on enriched Columbia blood agar plates. Cultures were incubated for 24 h at 25°C under atmospheric conditions. Bacteria were harvested, suspended in skim milk (Difco Laboratories, Detroit, Mich.), and stored at −80°C.
Enzyme extraction.
For the MLEE analysis, bacteria grown on two plates were harvested in 1.5 ml of buffer solution (10 mM Tris, 1 mM EDTA, 0.5 mM NADP [pH 6.8]). The enzyme extraction was performed as described by Boerlin and Piffaretti (11) with slight modifications.
Enzyme electrophoresis.
Bacterial lysates were thawed and subjected to gel electrophoresis under nondenaturing conditions in 10% starch gels (Connaught Laboratories; Fisher Scientific, Nepean, Ontario, Canada) as described by Selander et al. (26). Of 28 different enzymes tested with different electrophoretic buffers, 15 could be reliably used: nucleoside phosphorylase, catalase (CAT), serine-methionine peptidase, phenylalanine-proline peptidase (FP), and phosphogluconate dehydrogenase with buffer system F (Tris-maleate, pH 8.2); glucose 6-phosphate dehydrogenase, malate dehydrogenase, phosphoglucose isomerase, and isocitrate dehydrogenase with buffer system G (potassium phosphate, pH 6.7 and 7); indophenol oxidase (IPO), glyceraldehyde-phosphate dehydrogenase (GP1), and glutamic-pyruvic transaminase with buffer system E (Tris maleate, pH 7.4); and malic enzyme (ME), threonine dehydrogenase (THD), and alanine dehydrogenase with buffer system A (Tris-citrate, pH 8). Enzyme staining was performed as described by Selander et al. (26). Specific staining procedures for CAT were performed by the method of Harris and Hopkinson (16).
DNA extraction.
A few bacterial colonies were suspended in 500 μl of sterile water. DNA for PCR was extracted by using a commercial ion-exchange resin (InstaGene matrix; Bio-Rad Laboratories, Richmond, Calif.) according to the manufacturer's instructions.
For RAPD, colony suspensions were heated to 98°C for 10 min, and after centrifugation (12,000 × g for 10 min), 1 μl of the supernatant was further diluted in 100 μl of sterile water.
RAPD analysis.
RAPD was performed according to the protocol of Aznar et al. (4), with slight modifications.
PCR and sequencing reactions.
PCR was performed with 5 μl of the DNA extract, a 0.5 μM concentration of each primer, and 1 U of Taq polymerase (Boehringer Mannheim, Germany) in a total reaction volume of 50 μl with the buffer provided by the manufacturer.
Primers recA-1 (5′-GACGAGAATAAACAGAAGGC-3′) and recA-2 (5′-TCGCCGTTATAGCTGTACC-3′), amplifying a 543-bp fragment of the recA gene, were designed on the basis of the DNA sequence alignment of two Vibrio cholerae sequences (GenBank accession numbers U10162 and L42384), Vibrio anguillarum (GenBank accession number M80522), and Aeromonas salmonicida (GenBank accession number U83688). A 35-cycle PCR was performed with these primers and the following thermal profile: 94°C for 60 s, 58°C for 60 s, and 72°C for 90 s.
Primers glnA-1 (5′-TGACCCACGCTCTATCGC-3′) and glnA-2 (5′-GCGTGTGCAACGTTGTG-3′), amplifying a 402-bp fragment of the glnA gene, were designed on the basis of the DNA sequence alignment of the glnA sequence of V. cholerae (GenBank accession number AF013513) and Vibrio alginolyticus (GenBank accession number L08499). These primers were used in a 35-cycle PCR with the following thermal profile: 94°C for 60 s, 52°C for 60 s, and 72°C for 60 s.
Templates for direct sequencing were prepared by a simple purification of the PCR products with the QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. Cycle sequencing reactions were performed in both directions with the above-described primers (recA-1-recA-2 and glnA-1-glnA-2) in total volumes of 15 μl with an ABI Prism dRhodamine dye terminator Cycle Sequencing Ready Reaction Kit (dRhodamine terminator; Perkin-Elmer Applied Biosystems International Inc., Foster City, Calif.) on an ABI Prism 310 genetic analyzer (Perkin-Elmer).
Data analysis.
Statistical analysis of the MLEE data was performed with a computer program designed by Whittam et al. (26, 29) and as described previously (14). A dendrogram was constructed with the average-linkage method from a matrix of coefficients of pairwise genetic distances (26). Due to the low level of mutation, an unweighted tree-building method was preferred, because it does not emphasize single mutations.
Sequence data were analyzed and assembled by DNASTAR (1994 release; DNAstar Inc., Madison, Wis.). Genetic distances and sequence statistics (base composition, codon usage, and transition/transversion ratios) were determined with MEGA (22). Phylogenetic trees were constructed by the neighbor-joining method (25), and the robustness of each node was tested by bootstrap analysis (18). Estimates of the number of nucleotide substitutions per site for the recA and glnA genes were determined by the method of Tamura and Nei (27).
RAPD profiles were analyzed and compared by using the program GelCompar 4.1 (Comparative Analysis of Electrophoresis Patterns; Applied Maths, Kortrijk, Belgium). Trees were constructed by the unweighted pair group method using arithmetic averages.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported in this paper are AF311473 to AF311600.
RESULTS
Genetic diversity observed by MLEE.
Eleven of the 15 enzymes analyzed in 62 V. vulnificus isolates were polymorphic, and four (IPO, GP1, THD, and ME) were monomorphic. The number of alleles per locus varied from 1 to 8 (FP), with an average of 3.1 (Table 2). The level of genetic diversity among electrophoretic types (ETs) at each locus ranged from 0 for the monomorphic enzymes to 0.793 for CAT, with a mean value per locus of 0.281 (Table 2). A total of 43 ETs were identified for the 62 V. vulnificus isolates.
TABLE 2.
Number of alleles and genetic diversity per enzyme locus for 62 V. vulnificus isolates
| Enzyme locusa | No. of alleles | Genetic diversity per locus (h) |
|---|---|---|
| NSP | 2 | 0.047 |
| PGI | 2 | 0.047 |
| 6PG | 7 | 0.688 |
| SM | 3 | 0.534 |
| ME | 1 | 0 |
| G6P | 4 | 0.557 |
| MDH | 2 | 0.047 |
| IPO | 1 | 0 |
| FP | 8 | 0.722 |
| CAT | 6 | 0.793 |
| ALD | 2 | 0.091 |
| IDH | 4 | 0.643 |
| THD | 1 | 0 |
| GP1 | 1 | 0 |
| GPT | 2 | 0.047 |
| Mean | 3.067 | 0.281 |
NSP, nucleoside phosphorylase; PGI, phosphoglucose isomerase; 6PG, phosphogluconate dehydrogenase; SM, serine-methionine peptidase; G6P, glucose 6-phosphate dehydrogenase; MDH, malate dehydrogenase; ALD, alanine dehydrogenase; IDH, isocitrate dehydrogenase; GPT, glutamic-pyrovic transaminase.
V. diazotrophicus and L. anguillarum could be distinguished from each other and from V. vulnificus by species-specific alleles at 11 loci (Fig. 1). Furthermore, each of the three different species exhibited a distinct electromorph for IPO, GP1, THD, and ME. Within V. vulnificus, these enzymes were monomorphic.
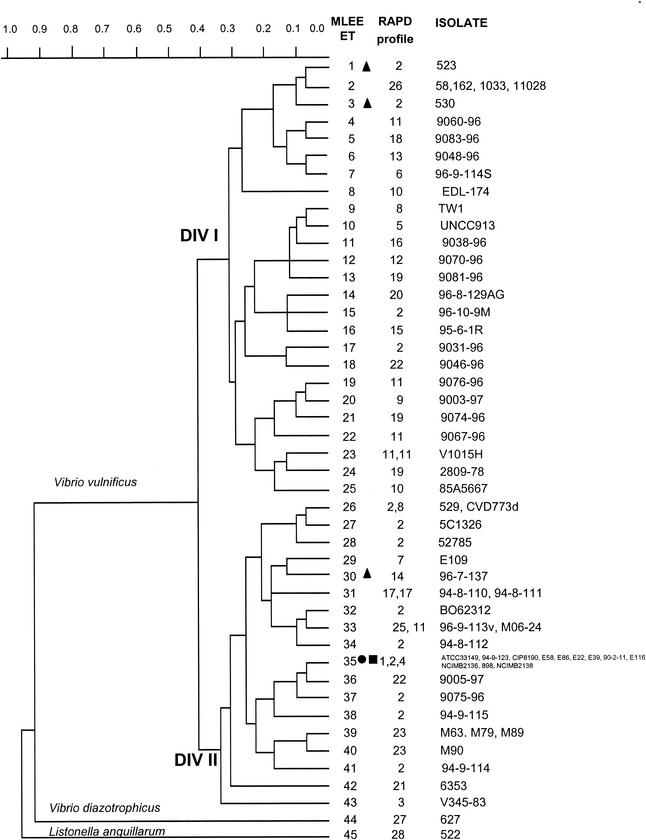
FIG. 1.
Genetic relationships among 62 V. vulnificus strains, 1 L. anguillarum strain, and 1 V. diazotrophicus strain, based on allelic variations at 15 enzyme loci. The dendrogram was generated by using the average-linkage method of clustering and a matrix of pairwise coefficients of genetic distance. •, V. vulnificus indole-negative strains isolated from diseased eels; ▪, V. vulnificus indole-negative strains not isolated from diseased eels; ▴, V. vulnificus indole-positive strains isolated from diseased eels.
Genetic relationships among MLEE ETs and source of the isolates.
Cluster analysis of the 43 ETs of V. vulnificus showed no significant clone associated with the clinical or environmental origin of the isolates (Fig. 1; Table 1). The 43 ETs were separated into two major divisions (divisions Ι and ΙΙ) by a distance of 0.41. ET 35 marked 10 indole-negative eel-pathogenic isolates of different geographic origins (Denmark, Japan, or Spain) (Table 1). In addition, this ET included two indole-negative isolates which did not originate from diseased eels (94-8-112 from the environment in Denmark and CIP 8190 from a French patient). In contrast, three indole-positive strains isolated from diseased eels (523, 530, and 96-7-137) belonged to ETs 1, 3, and 30, respectively. Not all strains isolated from diseased eels clustered in the same division; isolates marked by ETs 1 and 3 were found in division Ι, while those marked by ETs 30 and 35 were found in division ΙΙ.
In a few instances, isolates of the same geographic origin had the same genotype: isolates 58, 162, 1033, and 11028 from Israel (ET 2); isolates 94-8-110 and 94-8-111 from Denmark (ET 31); isolates M63, M79, and M89 from Spain (ET 39); and isolates 529 and CVD773d (ET 26) from the United States.
V. diazotrophicus (ET 44) and L. anguillarum (ET 45) were separated from each other by a genetic distance of 0.913 and were separated from V. vulnificus by a distance of 0.963. This result confirms that they are different species (Fig. 1).
Genetic relationships inferred by RAPD.
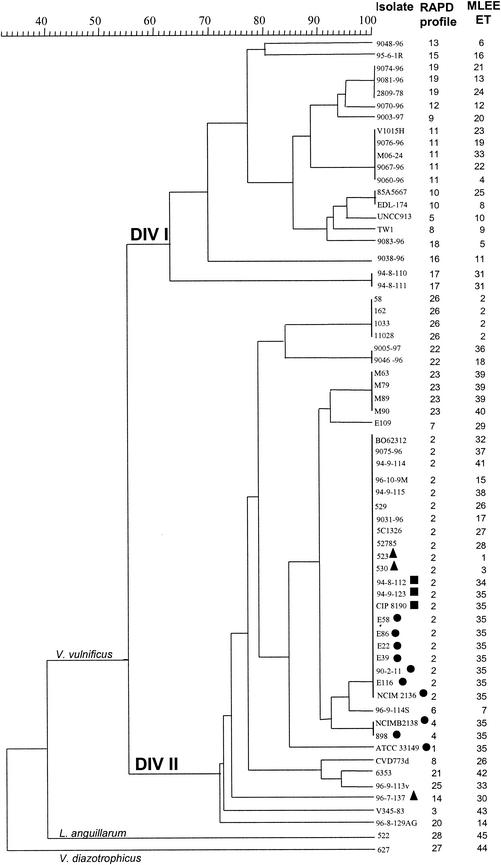
The RAPD technique, based on primer M13 (4), was applied to all 62 V. vulnificus strains, L. anguillarum, and V. diazotrophicus. A total of 28 different RAPD profiles were obtained. RAPD separated the V. vulnificus population into two divisions (divisions Ι and ΙΙ). One cluster within division ΙΙ (profiles 2, 4, and 6) included all of the strains originating from diseased eels as well as isolates not associated with eel pathogenicity and exhibiting a positive indole reaction (Fig. 2). Another cluster within division ΙΙ comprised the strains isolated in Israel. These isolates had identical RAPD profiles (profile 26) and were well differentiated from the other V. vulnificus strains. Additional minor clusters associated with geographic origin were observed: profile 23 represented four Spanish isolates, profile 17 represented two Danish isolates, and profiles 10, 11, and 19 represented two, five, and three isolates from the United States, respectively. As with MLEE, RAPD allowed a clear distinction between V. vulnificus, V. diazotrophicus (profile 27), and L. anguillarum (profile 28) (Fig. 2).
FIG. 2.
Genetic relationships among 62 V. vulnificus strains, 1 L. anguillarum strain, and one V. diazotrophicus strain, based on RAPD electrophoretic patterns obtained by PCR with the universal primer M13. Symbols are as described for Fig. 1.
Genetic diversity observed by recA and glnA sequences.
Alignment of 543 bp of the recA gene revealed 35 nucleotide substitutions, 28 of which were shared by more than one sequence. Two nucleotide substitutions resulted in amino acid substitutions (strain 5C1326, Val154Met; strain ATCC 33149, Thr171Ile). Tamura-Nei genetic distance values (27) among V. vulnificus strains ranged from 0 to 0.046.
Alignment of 402 bp of the glnA gene revealed 32 nucleotide substitutions, 25 of which were shared by more than one sequence. Only one amino acid substitution was present (strain TW1,Gly78Arg). Tamura-Nei genetic distances ranged from 0 to 0.036.
Genetic relationships inferred by recA and glnA DNA sequence analysis.
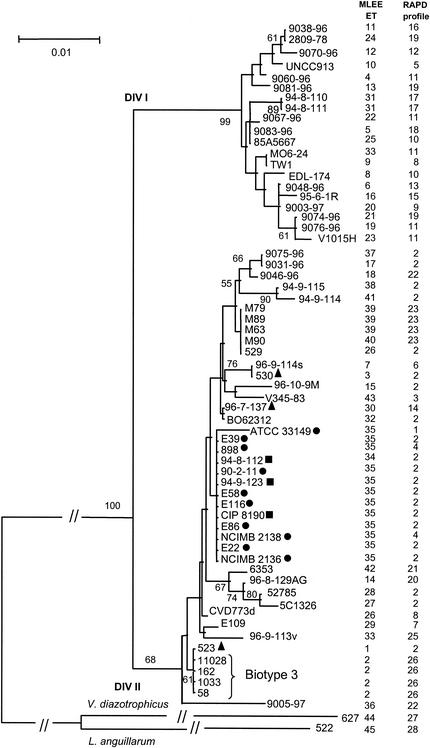
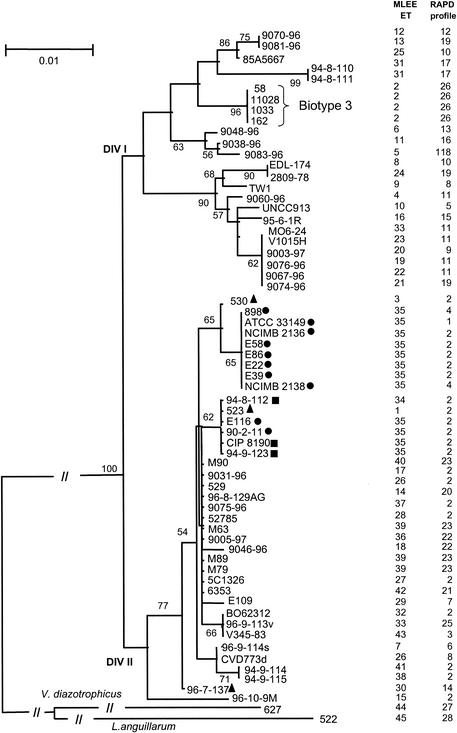
Phylogenetic analysis of the recA (Fig. 3) and glnA (Fig. 4) gene sequences also separated the V. vulnificus population into two major heterogeneous divisions (division Ι and ΙΙ), which do not seem to be correlated with a particular phenotypic trait. In any case, this subdivision was supported in both trees by a bootstrap confidence level of 100%. In general, there is a low level of nucleotide substitution in both genes, within (0.1 to 2%), as well as between (3 to 4%),the two divisions. No differences in nucleotide composition and in codon usage could be identified between the divisions. Moreover, no division-specific nucleotide substitutions existed, and the positions of single strains or clones were variable in the two gene trees examined. This was particularly evident with the Israeli isolates (biotype 3), which form a cluster in division ΙΙ of the recA tree but belong to division Ι in the glnA tree (Fig. 3 and 4).
FIG. 3.
Neighbor-joining tree based on the sequence alignment of a 543-bp fragment of the recA gene, using Tamura-Nei genetic distances (27). Only bootstrap values higher than 50% are shown. Symbols are as described for Fig. 1.
FIG. 4.
Neighbor-joining tree based on the sequence alignment of a fragment of 402 bp of the glnA gene, using Tamura-Nei genetic distances (27). Only bootstrap values higher than 50% are shown. Symbols are as described for Fig. 1.
Even if most of the clusters within the two major divisions are not well supported by bootstrap values, our data indicate an overall subdivision of the V. vulnificus population into different biogroups. In particular, besides the cluster of the Israeli strains (biotype 3), another group could be identified. Indole-negative eel-pathogenic isolates (biotype 2) form a cluster in the recA gene tree (but are divided into two groups within division ΙΙ in the glnA tree). Indole-negative isolates not coming from diseased eels are always found within the biotype 2 strains in both trees. Biotype 1 strains are distributed throughout the recA and glnA phylogenies, which represents evidence of the heterogeneous nature of these isolates.
Indole-positive eel-pathogenic strains do not form a monophyletic group in either of the gene trees, and the positions of these strains are variable within division ΙΙ. In general, the indole-positive eel-pathogenic strains do not cluster together with indole-negative eel-pathogenic isolates (the only exception is represented by the indole-positive strain 523 in the glnA tree [Fig. 3]).
DISCUSSION
Similarity and discrepancies between the divisions obtained by MLEE, RAPD, and sequence typing.
MLEE, RAPD, and sequence typing separated the V. vulnificus population into two distinct divisions. In general, the divisions obtained by these methods were comparable, and 80% of the isolates were included in the same genetic groups. A few important exceptions were observed. Strains 523 (ET 1, profile 2) and 530 (ET 3, profile 2), isolated from diseased eels, clustered in MLEE division Ι. In contrast, RAPD analysis and sequence typing indicated that the isolates were members of division ΙΙ. Another significant exception was the clone which comprised the Israeli isolates (ET 2, profile 26): in the dendrogram obtained by MLEE and glnA sequencing this clone was found in division Ι, while the RAPD analysis and the recA phylogeny positioned this clone in division ΙΙ. The inconstancies observed between the four trees are troubling and deserve a critical discussion. MLEE is a well-established method for estimating phylogenetic relationships between genotypes at the intraspecific level. It is based on the analysis of various loci encoding housekeeping enzymes and thus considered selectively neutral. Provided that a sufficient number of loci are analyzed (generally 15 to 20), a global representation of the genome evolution is deduced. A drawback is that variability is the consequence of the changes of the electrostatic charges on the target proteins, and hence a significant fraction of the mutations are not detected (e.g., third-codon-position mutations). In the case of V. vulnificus, we considered 15 loci, 4 of which were monomorphic, and estimated the mean genetic diversity at 0.281 per locus, a value regarded as low compared to those for other bacterial species such as V. cholerae (0.463 [6]), Escherichia coli (0.52 [29]), or Bacteroides fragilis (0.393 [14]). Thus, a few mutations are sufficient to associate a V. vulnificus clone with division Ι instead of division ΙΙ.
Sequencing of genes encoding housekeeping enzymes also allows the inferring of phylogenetic relationships among bacteria. Obviously, this method, named multilocus sequence typing, should also consider a sufficient number of loci in order to avoid estimating the evolution of single genes and not of the whole genome. This requirement is well supported by the comparison to the MLEE-deduced dendrograms of the trees generated by sequencing the recA and glnA genes. Again, the level of discrepancies is increased by low variability in single genes. For the V. vulnificus recA and glnA genes, we found polymorphism values of 6.45 and 7.96%, respectively. For comparison, the values we determined for B. fragilis were 14.8% in the recA gene and 16.3% in the glnA gene (15).
Dendrograms generated by RAPD are even more difficult to interpret and compare to MLEE or multilocus sequence typing trees, since in the case of the former, variability between strains depends not only on the evolution of the sequences corresponding to the primers used but also on genome rearrangements, which are due mainly to the presence of insertion sequences or short sequence repeats.
In conclusion, our data emphasize that comparison of trees obtained by different methods should always be done with caution. In our case and under our conditions, we estimate that the MLEE data obtained by the analysis of 15 housekeeping enzyme loci are probably the most representative of the structure of the V. vulnificus population.
One major clone represented most eel-pathogenic isolates.
As a consequence of the isolation of various atypical strains, a debate has emerged concerning whether or not eel-pathogenic isolates constitute a distinct biogroup or serogroup (biotype 2 or serovar E) (8, 18, 19). The atypical strains have phenotypic and genetic characteristics different from those of the majority of eel-pathogenic isolates (8, 18). In general, despite low variability, all of the approaches (MLEE, RAPD, and sequence typing) indicated that indole-negative eel-pathogenic strains isolated from different geographic regions tend to cluster as a separate genotype (ET 35) (Fig. 1 to 4). Hence, our data provide additional evidence for the existence of a distinct genetic subgroup associated with disease in eels. Indole-positive eel-pathogenic strains (523, 530, and 96-7-137, marked by MLEE ETs 1, 3, and 30, respectively) do not form a monophyletic group, and the individual positions of these strains are variable in each phylogeny constructed. In addition, the indole-positive eel-pathogenic strains rarely cluster together with the indole-negative eel-pathogenic isolates. Therefore, the designation serovar E is not well supported by the phylogenetic data presented in this study. The possibility that both eel-pathogenic and nonpathogenic strains coexist in the same animal and that isolates marked by ETs 1 (strain 523), 3 (strain 530), and 30 (strain 96-7-137), although grown from diseased eels, may be nonpathogenic has not received support. Indeed, a virulence test on healthy eels confirmed the pathogenicity of isolate 530 (8). Alternatively, isolates of ETs 1, 3, and 30 might have acquired, by horizontal transfer from ET 35 strains, a set of genes conferring virulence to the recipient host. The majority of biotype 2 strains (ET 35), but not biotype 1 isolates, harbor high-molecular-weight plasmids (7, 8). Interestingly, isolate 96-7-137 has been shown to have a plasmid profile similar to those of pathogenic strains (12).
Geographic distribution of genotypes.
All of the methods we used indicated that the Israeli isolates (ET 2) were clearly distinct from all of the other V. vulnificus isolates (Fig. 1 to 4), supporting the existence of a new biotype in Israel (biotype 3 [10]). This clone might have emerged and evolved independently due to geographic isolation. Alternatively, and more likely, the association with Tilapia spp. (all patients were infected while cleaning this fish) might suggest that ET 2 has evolved as a distinct genotype due to niche separation. The isolation of eel-pathogenic strains of the same genotype (ET 35) from diverse geographic regions provides further support for the view that interaction with a particular host has influenced the evolution of V. vulnificus.
With the exception of the Israeli clone, only a few minor ETs representing more than one isolate of the same geographical origin were observed. Thus, geographical isolation does not seem to play a major role in the evolution of V. vulnificus.
Interestingly, isolates of ET 35 appear to be absent from North America. This is likely explained by the absence of eel farming in the United States. The expansion of ET 35 might be hindered in the absence of large eel monocultures. Alternatively, for some unknown reasons, ET 35 strains do not survive in the North America environment.
In conclusion, existing biotype and biogroup designations did not always correlate with the phylogenies generated by MLEE, recA and glnA gene sequence analysis, and RAPD analysis. Strains from biotype 1 are distributed throughout the phylogenetic trees, and in general indole-negative strains are separated from indole-positive isolates. From a phylogenetic point of view, the designation biotype 2 should not be limited to the indole-negative isolates originating from diseased eels. In addition, the designation serovar E, which presently includes biotype 2 strains as well as other eel-pathogenic isolates, is not supported by our data. Finally, the Israeli isolates (biotype 3) form a cluster in all trees. A reevaluation of the present criteria defining biogroups, taking into consideration new phenotypic, serological, and genetic data, is greatly needed.
Acknowledgments
We thank V. Agmon, C. R. Arias, C. Kingombe, J. Kopelowitz, J. Powell, and B. Swaminathan for supplying bacterial strains. We are grateful to C. Kingombe for helpful discussions. We thank S. Reid for reading the manuscript and for helpful discussions.
This research was supported by grants 31-45914.95 and 31-64976.01 from the Swiss National Science Foundation to J.-C.P.
REFERENCES
- 1.Amaro, C., L.-I. Hor, E. Marco-Noales, T. Bosque, B. Fouz, and E. Alcaide. 1999. Isolation of Vibrio vulnificus serovar E from aquatic habitats in Taiwan. Appl. Environ. Microbiol. 65:1352-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, C. R., L. Verdonck, J. Swings, R. Aznar, and E. Garay. 1997. A polyphasic approach to study the intraspecific diversity amongst Vibrio vulnificus isolates. Syst. Appl. Microbiol. 20:622-633. [Google Scholar]
- 3.Arias, C. R., M. J. Pujalte, E. Garay, and R. Aznar. 1998. Genetic relatedness among environmental, clinical, and diseased-eel Vibrio vulnificus isolates from different geographic regions by ribotyping and randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 64:3403-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aznar, R., W. Ludwig, and K. H. Schleifer. 1993. Ribotyping and randomly amplified polymorphic DNA analysis of V. vulnificus biotypes. Syst. Appl. Microbiol. 16:303-309. [Google Scholar]
- 5.Aznar, R., W. Ludwig, R. I. Amann, and K. H. Schleifer. 1994. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of V. vulnificus with rRNA-targeted nucleotide probes. Int. J. Syst. Bacteriol. 44:330-337. [DOI] [PubMed] [Google Scholar]
- 6.Beltran, P., G. Delgado, A. Navarro, F. Trujillo, R. K. Selander, and A. Cravioto. 1999. Genetic diversity and population structure of Vibrio cholerae. J. Clin. Microbiol. 37:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biosca, E. G., H. Llorens, E. Garay, and C. Amaro. 1993. Presence of a capsule in Vibrio vulnificus biotype 2 and its relationship to virulence for eels. Infect. Immun. 61:1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biosca, E. G., C. Amaro, J. L. Larsen, and K. Pedersen. 1997. Phenotypic and genotypic characterization of Vibrio vulnificus: proposal for the substitution of the subspecific taxon biotype for serovar. Appl. Environ. Microbiol. 63:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biosca, E. G., R. M. Collado, J. D. Oliver, and C. Amaro. 1999. Comparative study of biological properties and electrophoretic characteristics of lipopolysaccharide from eel-virulent and eel-avirulent Vibrio vulnificus strains. Appl. Environ. Microbiol. 65:856-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. L. Swerdlow, J. J. Farmer, et al. 1999. Clinical, epidemiological, and microbiological features of V. vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 11.Boerlin, P., and J.-C. Piffaretti. 1995. Multilocus enzyme electrophoresis. Methods Mol. Biol. 46:63-68. [DOI] [PubMed] [Google Scholar]
- 12.Dalsgaard, I., L. Høi, R. J. Siebeling, and A. Dalsgaard. 1999. Indole-positive Vibrio vulnificus isolated from disease outbreaks on a Danish eel farm. Dis. Aquat. Org. 35:187-194. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 14.Gutacker, M., C. Valsangiacomo, and J.-C. Piffaretti. 2000. Identification of two genetic groups in Bacteroides fragilis by multilocus enzyme electrophoresis: distribution of antibiotic resistance (cfiA, cepA) and enterotoxin (bft) encoding genes. Microbiology 146:1241-1254. [DOI] [PubMed] [Google Scholar]
- 15.Gutacker, M., C. Valsangiacomo, M. Bernasconi, and J.-C. Piffaretti. 2002. RecA and glnA sequences separate the Bacteroides fragilis population into two genetic divisions associated with the antibiotic resistance genotypes cepA and cfiA. J. Med. Microbiol. 51:123-130. [DOI] [PubMed] [Google Scholar]
- 16.Harris, H., and D. A. Hopkinson. 1978. Handbook of enzyme electrophoresis in human genetics. North-Holland Publishing Co., Amsterdam, The Netherlands.
- 17.Hill, R. T., J. R. Parker, H. J. Goodman, D. T. Jones, and D. R. Woods. 1989. Molecular analysis of a novel glutamine synthetase of the anaerobe Bacteroides fragilis. J. Gen. Microbiol. 135:3271-3279. [DOI] [PubMed] [Google Scholar]
- 18.Høi, L., A. Dalsgaard, J. L. Larsen, J. M. Warner, and J. D. Oliver. 1997. Comparison of ribotyping and randomly amplified polymorphic DNA PCR for characterization of Vibrio vulnificus. Appl. Environ. Microbiol. 63:1674-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Høi, L., I. Dalsgaard, A. DePaola, R. J. Siebeling, and A. Dalsgaard. 1998. Heterogeneity among isolates of Vibrio vulnificus recovered from eels (Anguilla anguilla) in Denmark. Appl. Environ. Microbiol. 64:4676-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Høi, L., J. L. Larsen, I. Dalsgaard, and A. Dalsgaard. 1998. Occurrence of Vibrio vulnificus biotypes in Danish marine environments. Appl. Environ. Microbiol. 64:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlin, S., G. M. Weinstock, and V. Brendel. 1995. Bacterial classifications derived from recA protein sequence comparisons. J. Bacteriol. 177:6881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 23.Linkous, D. A., and J. D. Oliver. 1999. Pathogenesis of V. vulnificus. FEMS Microbiol. Lett. 174:207-214. [DOI] [PubMed] [Google Scholar]
- 24.Oliver, J. D., R. A. Warner, and D. R. Cleland. 1983. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl. Environ. Microbiol. 45:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 26.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 28.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittam, T. S., H. Ochman, and R. K. Selander. 1983. Multilocus genetic structure in natural populations of Escherichia coli. Proc. Natl. Acad. Sci. USA 80:1751-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]