Abstract
Knowledge of bacteriophage ecology in vegetable fermentations is essential for developing phage control strategies for consistent and high quality of fermented vegetable products. The ecology of phages infecting lactic acid bacteria (LAB) in commercial sauerkraut fermentations was investigated. Brine samples were taken from four commercial sauerkraut fermentation tanks over a 60- or 100-day period in 2000 and 2001. A total of 171 phage isolates, including at least 26 distinct phages, were obtained. In addition, 28 distinct host strains were isolated and identified as LAB by restriction analysis of the intergenic transcribed spacer region and 16S rRNA sequence analysis. These host strains included Leuconostoc, Weissella, and Lactobacillus species. It was found that there were two phage-host systems in the fermentations corresponding to the population shift from heterofermentative to homofermentative LAB between 3 and 7 days after the start of the fermentations. The data suggested that phages may play an important role in the microbial ecology and succession of LAB species in vegetable fermentations. Eight phage isolates, which were independently obtained two or more times, were further characterized. They belonged to the family Myoviridae or Siphoviridae and showed distinct host ranges and DNA fingerprints. Two of the phage isolates were found to be capable of infecting two Lactobacillus species. The results from this study demonstrated for the first time the complex phage ecology present in commercial sauerkraut fermentations, providing new insights into the bioprocess of vegetable fermentations.
Like most vegetable fermentations, sauerkraut fermentation is spontaneous and relies on a very small population of lactic acid bacteria (LAB), which are naturally present on fresh vegetables, for preservation. It is known that a succession of various LAB species and their metabolic activities are responsible for the quality and safety of these products (25). The process is characterized by an initial heterofermentative stage, followed by a homofermentative stage. Heterofermentative Leuconostoc mesenteroides initiates the fermentation and quickly predominates the early stage of the fermentation because it is present at an initially higher number (approximately 103 CFU/ml) and has a shorter generation time at 18°C (the typical temperature of sauerkraut fermentation) than most other epiphytic LAB (22, 23, 24, 25, 27). The quality characteristics of sauerkraut are largely dependent upon the growth of this species (25). Between 3 and 7 days after the start of the fermentation, heterofermentative Leuconostoc species are usually succeeded by the more acid-tolerant homofermentative Lactobacillus species, due to the accumulation of lactic acid to 1% (wt/vol) or more and the decrease in pH below 4.5 (21, 25). Lactobacillus plantarum completes the fermentation, with a final pH of approximately 3.5 (25, 29).
The correct sequence of LAB species is essential in achieving a stable product with the typical flavor and aroma of sauerkraut. It is generally thought that microbial succession is largely due to the initial microbial load on cabbage, salt and acid concentrations, pH, and temperature (11, 13, 25). The development of low-salt (1% or less) fermentation technology (H. P. Fleming, unpublished data) to reduce chloride waste during the production of sauerkraut may require the use of LAB starter cultures. These cultures may be used to ensure that a normal microbial succession is initiated. However, the use of starter cultures raises questions about possible interference by bacteriophages.
The presence of bacteriophages active in vegetable fermentations has only recently been studied. Yoon et al. (31) presented the first report of nine bacteriophages isolated from a commercial sauerkraut fermentation. However, the diversity and ecological role of phages in the succession of LAB in commercial sauerkraut fermentations remain largely unexplored. The objectives of this study were to investigate the diversity and ecology of phages active against LAB in commercial sauerkraut fermentations, to explore the possible role of phages in microbial succession in the fermentation, and to characterize predominant LAB phages isolated from the commercial fermentations. The information from this study provides new insights into vegetable fermentation, which should facilitate the development of controlled vegetable fermentation strategies, whereby starter cultures could be used. To our knowledge, this is the first study to examine the phage ecology in commercial vegetable fermentations.
MATERIALS AND METHODS
Commercial sauerkraut fermentation and sample collection.
Four commercial sauerkraut fermentation tanks (one in 2000 and three in 2001) were examined in this study. Each tank had a 90-ton capacity. These fermentations were carried out with 2.3% NaCl (after equilibration with the shredded cabbage) and an average temperature of 18°C. The samples were obtained between October 2000 and December 2001. Fresh shredded cabbage samples (500 g) were collected in sterile plastic bags prior to salting. Brine samples (100 ml each) were obtained from fermentation tanks on various days after the start of the fermentation (days 1, 3, 7, 9, 14, 22, 30, 60, and 100) with a stainless steel tube (1 cm in diameter). The tube was inserted to a depth of about 60 cm from the top of the fermentation tank (about 60 cm from the edge of the tank) and allowed to fill with brine through a small hole at the bottom. Each brine sample was transferred into two separate 50-ml sterile plastic tubes, cooled on ice, and immediately shipped to our laboratory by overnight mail.
Preparation of shredded cabbage samples for microbiological and chemical analyses.
For microbiological analysis, 30 g of the shredded cabbage sample was homogenized in a stomacher with 270 g of saline (0.85% NaCl) for 3 min at the maximal speed (Stomacher 400; Tekmar, Cincinnati, Ohio). The cabbage extract (1 ml) was obtained from the stomacher bag for microbiological analysis. For chemical analysis, 100 g of the shredded cabbage sample was blended with 200 g of water for 3 min in a Waring blender (Dynamic Products Corp., New Hartford, Conn.) and then stomached as described above. The cabbage extract (30 ml) was transferred from the filter side of the stomacher bag to a 50-ml sterile plastic tube and frozen at −20°C for later analysis.
The treatment of brine samples for host and phage isolations.
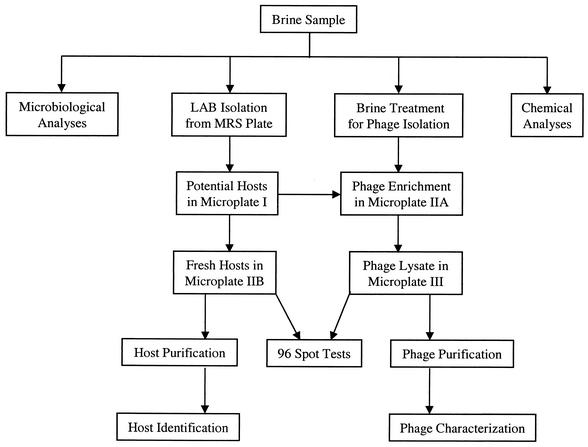
Each brine sample was divided into several portions for microbiological and chemical analyses and for isolation of phages and their hosts (Fig. 1). One milliliter of brine was saved for immediate microbiological analysis and host isolation. The remaining brine sample was centrifuged aseptically at 10,000 × g (GSA rotor, RC-5B; Sorvall, Newtown, Conn.) and 4°C for 10 min to remove solid particles. The supernatant was filtered (Whatman filter paper no. 4; W & R Balston Limited, Maidstone, England). A portion (30 ml) of the filtrate was stored at −20°C for later chemical analysis. The pH of the remaining filtrate was measured and then adjusted to approximately 6.3 with 3.0 N NaOH. After a second filtration (0.45-μm-pore-size filter), the brine was stored at 4°C for later use as a potential phage source.
FIG. 1.
Flow diagram for analyses of a brine sample and isolation of phages and their hosts in a sauerkraut fermentation studied in 2000.
Microbiological analysis.
Brine samples were plated on plate count agar (PCA; Difco Laboratories, Detroit, Mich.), violet red bile glucose (VRBG) agar (VRB agar [Difco] supplemented with 1% glucose [Sigma Chemical Co, St. Louis, Mo.]), DeMan Rogosa Sharpe (MRS) agar (Difco), and yeast and mold (YM) agar (Difco) to enumerate total aerobic microflora, Enterobacteriaceae, LAB, yeasts, and molds, respectively, using a spiral plater (Autoplate 4000; Spiral Biotech, Inc., Bethesda, Md.). A modified MRS agar (MMRS) was prepared by adding 0.02 M sodium azide prior to pouring of the plates to prevent the growth of yeasts and molds and to ensure selection for LAB. The plates were incubated at 30°C for 1 day (PCA and VRBG) or 2 days (MRS or MMRS) or at room temperature for 4 days (YM). The colonies on the plates were enumerated with an automated colony counter (Protos Plus; Bioscience International, Rockville, Md.).
Chemical analyses.
The salt (NaCl) content in brine was determined by titration with standard AgNO3 using dichlorofluorescein as an indicator (16). Sugars, alcohols, and organic acids were determined by high-performance liquid chromatography. Sugars and mannitol were separated by a Carbopac PA1 column (Dionex Corp., Sunnyvale, Calif.) with a 0.8-ml/min flow rate of 0.12 N NaOH at room temperature and detected by a pulsed amperometric detector (model PAD-2; Dionex). Cellobiose was used as an internal standard. Organic acids and ethanol were analyzed with an anion-exchange column (Aminex HPX-87H; Bio-Rad Laboratories, Richmond, Calif.) with a 0.8-ml/min flow rate of 0.03 N H2SO4 at 75°C. A UV detector (UV-6000; Thermo Separation Products Inc., San Jose, Calif.) and a differential refractometer (Waters 410; Waters, Milford, Mass.) were connected in series for detection of organic acids (at 210 nm) and ethanol, respectively. Isobutyric acid was used as an internal standard.
Isolation of phages and their hosts.
Ninety-six isolated colonies were randomly picked from MRS agar plates and used for inoculating 96 wells in a microplate (microplate I) (Fig. 1). Each well in microplate I contained 200 μl of MRS broth and cells and was overlaid with 75 μl of mineral oil after inoculation. The microplate was then incubated overnight at 30°C. Ten microliters of each of the 96 overnight cultures in microplate I was transferred into two new microplates (IIA and IIB) (Fig. 1). These cultures would serve as potential hosts for phage enrichment and isolation. Each well in microplates IIA and IIB contained 200 μl of MRS broth. Microplate IIA was incubated at 30°C for 6 h, whereas microplate IIB was stored at 4°C for later use. A filter-sterilized brine sample (50 μl) was then added to each well in microplate IIA to enrich the phages in the brine. The incubation of microplate IIA was continued overnight. Microplate IIB was incubated at 30°C to provide fresh cultures for spot tests. After 8 to 10 h of incubation, microplate IIA was placed in a microplate carrier (SH-3000 swinging bucket rotor; Sorvall) and centrifuged (RC-5B; Sorvall) at 3,300 × g and 4°C for 10 min. Ninety-six individual spot tests were performed by spotting 10 μl of supernatant from a well in microplate IIA onto the corresponding bacterial lawn that resulted from 100 μl of overnight culture from the corresponding well in microplate IIB. The 96-well plates were incubated overnight at 30°C. Primary phage-host relationships were indicated by positive spot test plates and confirmed by plaque assay after the host was colony purified. Each host isolate was assigned an identification consisting of a number indicating the day when it was isolated, followed by an alphanumeric identification of the well on microplate IIB in which the host was grown. The corresponding phage isolate was assigned the same designation with the prefix φ. For example, φ1-A3 indicated that the phage was isolated on day 1 and propagated in well A3 of microplate IIA, and its principal host was 1-A3. Theoretically, the detection limit for phages in 2000 was 20 PFU/ml of brine sample, because 50 μl of the brine sample was used for phage enrichment. All of the resulting phage isolates were purified by one round of single-plaque isolation according to the method described by Lu et al. (19). Additional spot tests were performed to determine host range and phage typing, thereby identifying distinct phages and hosts.
In 2001, the hosts isolated in 2000 were used to propagate phages in brine samples from three commercial fermentation tanks. With a few exceptions, hosts isolated from a specific day in 2000 were used mainly for phage isolation from the brine samples of the same day of the fermentation in 2001. Fresh host cultures were prepared in 1.5-ml microcentrifuge tubes instead of microplates. MRS broth (0.5 ml) supplemented with 10 mM of CaCl2, 0.5 ml of filter-sterilized brine, and 20 μl of host culture (in log phase) was added to a 1.5-ml microcentrifuge tube, which was then treated as described above. The theoretical detection limit of phage in 2001 was 2 PFU/ml of brine sample, because a larger volume (0.5 ml) of brine was used for phage enrichment than in 2000. Phages and their hosts were stored at 4°C in MRS broth or at −84°C in MRS broth supplemented with 16% glycerol.
Host identification.
All host isolates were subjected to biochemical tests for gas (CO2) production from glucose with a Durham tube in MRS broth, malolactic enzyme activity in MD medium (10), dextran (slime) production on 5% sucrose agar (2), and catalase activity using 5% H2O2. Each individual host isolate obtained in 2000 was tested against every phage isolate obtained in the same year, with spot tests to determine phage typing of the host. Based on phage typing, distinct hosts were determined. Host strains were identified by an rRNA operon PCR fingerprinting method (7). To further identify selected isolates, the 5′ end of the 16S rRNA gene was sequenced as described by Barrangou et al. (3).
Characterization of phages.
Each phage isolate obtained in 2000 was tested against all host isolates obtained from the same tank to determine the host range by spot test on MRS agar media. Based on host ranges, distinct phages were identified. Eight frequently occurring phages were selected for further characterization by their morphology, major structural protein profiles, and restriction endonuclease digestion patterns by using the methods described by Lu et al. (19). Briefly, phages were concentrated from 1 liter of phage lysate by polyethylene glycol precipitation and purified by ultracentrifugation in a CsCl step gradient (26). Electron micrographs of phages, restriction analysis of phage DNA, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of structural proteins were carried out as described by Lu et al. (19).
RESULTS
Microbiological analysis.
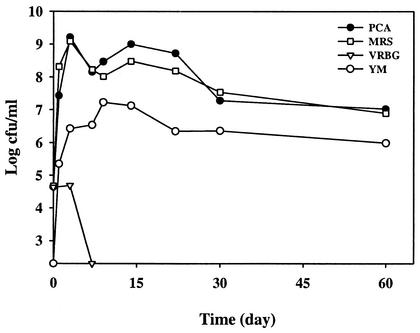
Figure 2 shows the profiles of PCA counts for total aerobes, VRBG counts for Enterobacteriaceae, MRS counts for LAB, and YM counts for yeasts and molds in a commercial sauerkraut fermentation tank over a 60-day period in 2000. The initial PCA, MRS, and VRBG counts on cabbage were around 4 × 104 CFU/g, and YM counts for yeasts and molds were below the detection limit (102 CFU/ml). The MRS count increased rapidly after day 0 and reached 109 CFU/ml on day 3. The YM count increased to 106 CFU/g. After day 7, the VRBG count was below the detection limit, whereas MRS and YM counts gradually decreased. On day 60, the MRS count was around 107, while the YM count was 106. The PCA count was very close to the MRS count during the 60-day period. Similar microbial count profiles were obtained in the three fermentation tanks studied in 2001 (data not shown).
FIG. 2.
Microbial counts in a commercial sauerkraut fermentation.
Chemical analyses.
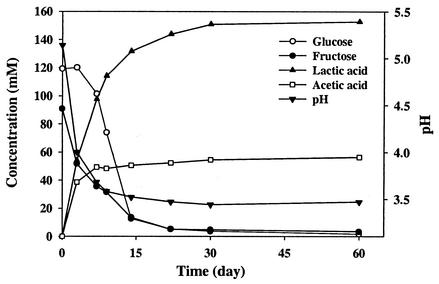
The fermentable-substrate composition of shredded cabbage used for the fermentations is shown in Table 1. The concentrations of glucose, fructose, sucrose, and malic acid in the cabbage used in 2000 were lower than those reported by Fleming et al. (15) but higher than those in the cabbage used in 2001. No sucrose was detected in the cabbage used in 2001. Sugar consumption, acid production, and pH reduction were rapid during the first 2 weeks of the fermentation in the tank studied in 2000 (Fig. 3) and slower thereafter and remained unchanged after day 30. The pH on day 60 was slightly lower than 3.5. Similar profiles were obtained with three other tanks studied in 2001 (data not shown). Malic acid was exhausted in all four tanks (data not shown). The salt content in brines from the four tanks was in the range of 2.2 to 2.3% (data not shown).
TABLE 1.
Fermentable substrate composition of shredded cabbage used in sauerkraut fermentationsa
| Year | Tank | Concn of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Glucose
|
Fructose
|
Sucrose
|
Malic acid
|
||||||
| mM | % | mM | % | mM | % | mM | % | ||
| 2000 | 1 | 119.2 | 2.15 | 90.8 | 1.64 | 4.8 | 0.17 | 5.6 | 0.08 |
| 2001 | 1 | 92.4 | 1.66 | 81.7 | 1.47 | ND | ND | 3.9 | 0.05 |
| 2001 | 2 | 90.8 | 1.63 | 83.5 | 1.50 | ND | ND | 3.5 | 0.05 |
| 2001 | 3 | 81.7 | 1.47 | 73.8 | 1.33 | ND | ND | 4.7 | 0.06 |
Percentages are by wet weight. ND, not detected.
FIG. 3.
Changes in the concentrations of substrates and products and pH in a commercial sauerkraut fermentation.
Isolation of phages and their hosts.
In 2000, 864 randomly picked bacterial isolates from MRS plates were obtained from nine brine samples over a 100-day period. These isolates were tested as potential hosts for phage enrichment and isolation. A total of 46 phage-host relationships were established by spot tests and confirmed by plaque assays (Table 2). Two phage isolates (φ1-F9 and φ14-A4) lost their infectivity for unknown reasons and were not included in Table 2. Among the remaining 44 phage isolates, 12 were obtained on day 1, 11 were from day 3, 2 were from day 7, 1 were from day 9, 9 were from day 14, 5 were from day 22, and 4 were from day 60. A number of phages were found to attack multiple hosts. No phages were isolated on day 30 or 100.
TABLE 2.
Phages and their hosts isolated from a commercial sauerkraut fermentation tank in 2000a
| Host | Isolation of phage on days 1 and 3
|
Isolation of phage on days 7, 9, 14, 22, and 60
|
||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| φ1-A3 | φ3-H2 | φ1-B8 | φ1-C5 | φ1-F7 | φ1-F10 | φ1-A4 | φ1-D6 | φ1-G7 | φ1-E10 | φ1-F8 | φ1-H3 | φ3-D5 | φ3-A4 | φ3-B5 | φ1-C4 | φ3-B1 | φ3-G9 | φ3-811 | φ3-F4 | φ3-E11 | φ3-G1 | φ3-G10 | φ7-E1 | φ14-A2 | φ14-B7 | φ14-D10 | φ14-H7 | φ22-G10 | φ9-B4 | φ14-F3 | φ14-H4 | φ14-E10 | φ22-A2 | φ22-E2 | φ22-D10 | φ22-F3 | φ60-H11 | φ7-C4 | φ60-D8 | φ14-C8 | φ14-F10 | φ60-E4 | φ60-E8 | |
| 1-A3 | + | + | ||||||||||||||||||||||||||||||||||||||||||
| 3-H2 | + | + | ||||||||||||||||||||||||||||||||||||||||||
| 1-A4 | + | + | + | + | ||||||||||||||||||||||||||||||||||||||||
| 1-D6 | + | + | + | + | ||||||||||||||||||||||||||||||||||||||||
| 1-F9 | + | + | + | + | ||||||||||||||||||||||||||||||||||||||||
| 1-G7 | + | + | + | + | ||||||||||||||||||||||||||||||||||||||||
| 1-B8 | + | + | + | + | + | |||||||||||||||||||||||||||||||||||||||
| 1-E10 | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||||
| 1-C5 | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||||
| 1-F8 | + | + | + | + | + | |||||||||||||||||||||||||||||||||||||||
| 3-B5 | + | + | + | + | + | |||||||||||||||||||||||||||||||||||||||
| 3-D5 | + | + | + | + | + | |||||||||||||||||||||||||||||||||||||||
| 1-H3 | + | + | + | + | + | |||||||||||||||||||||||||||||||||||||||
| 3-A4 | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||||
| 1-F10 | + | + | + | |||||||||||||||||||||||||||||||||||||||||
| 1-F7 | + | |||||||||||||||||||||||||||||||||||||||||||
| 1-C4 | + | + | ||||||||||||||||||||||||||||||||||||||||||
| 3-B1 | + | + | + | |||||||||||||||||||||||||||||||||||||||||
| 3-G9 | + | + | + | |||||||||||||||||||||||||||||||||||||||||
| 3-B11 | + | + | ||||||||||||||||||||||||||||||||||||||||||
| 3-F4 | + | + | ||||||||||||||||||||||||||||||||||||||||||
| 3-E11 | + | |||||||||||||||||||||||||||||||||||||||||||
| 3-G1 | + | |||||||||||||||||||||||||||||||||||||||||||
| 3-G10 | + | |||||||||||||||||||||||||||||||||||||||||||
| 7-C4 | + | + | + | + | ||||||||||||||||||||||||||||||||||||||||
| 7-E1 | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| 14-A2 | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| 14-B7 | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| 14-D10 | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| 14-H7 | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| 22-G10 | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| 14-A4 | + | + | + | |||||||||||||||||||||||||||||||||||||||||
| 9-B4 | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||||
| 14-E10 | + | |||||||||||||||||||||||||||||||||||||||||||
| 14-F3 | + | + | + | + | ||||||||||||||||||||||||||||||||||||||||
| 22-A2 | + | |||||||||||||||||||||||||||||||||||||||||||
| 14-H4 | + | |||||||||||||||||||||||||||||||||||||||||||
| 22-E2 | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| 22-D10 | + | + | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||
| 60-D8 | + | + | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||
| 14-C8 | + | + | + | + | + | + | + | + | + | |||||||||||||||||||||||||||||||||||
| 14-F10 | + | + | + | + | + | + | + | + | + | |||||||||||||||||||||||||||||||||||
| 22-F3 | + | + | + | + | + | + | + | + | + | |||||||||||||||||||||||||||||||||||
| 60-H11 | + | + | + | + | + | + | + | + | + | |||||||||||||||||||||||||||||||||||
| 60-E4 | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| 60-E8 | + | + | + | + | + | + | + | |||||||||||||||||||||||||||||||||||||
+, host is sensitive to the phage or the phage is infective for the host. In host designations, the first number indicates the isolation day and the alphanumeric label indicates the well in a microplate where the host was grown. The corresponding phage is given the same designation with the prefix φ.
Based on host range, 26 distinct phages were obtained (Table 3). Nine distinct phages were isolated more than once on the same or different days. Phage φ7-E1 was isolated six times (once each on days 7 and 22 and four times on day 14), whereas φ14-C8 appeared in the phage isolates four times (twice on day 14 and twice on day 60). Many phages were capable of infecting two or more bacterial strains (Table 3). Phages φ14-C8, φ14-E10, and φ22-A2 had very broad host ranges, capable of infecting five or six stains. Several phages had a very restricted host range and infected only a single strain.
TABLE 3.
Features of 26 distinct phages isolated from a commercial saucrkraut fermentation in 2000
| Phage | No. of times isolated | Days of isolation | Host(s) |
|---|---|---|---|
| φ1-A4 | 3 | 1, 1, 1 | 1-A4, 1-B8, 1-E10 |
| φ1-B8 | 1 | 1 | 1-B8 |
| φ1-C5 | 1 | 1 | 1-C5, 1-F8, 3-A4 |
| φ1-E10 | 1 | 1 | 1-E10, 1-A4, 1-B8, 3-A4 |
| φ1-F7 | 1 | 1 | 1-F7 |
| φ1-F8 | 3 | 1, 1, 3 | 1-F8, 3-A4 |
| φ1-F10 | 1 | 1 | 1-F10 |
| φ3-A4 | 2 | 3, 3 | 3-A4, 1-C5, 1-F8, 1-F10 |
| φ3-B1 | 2 | 1, 3 | 3-B1, 1-C4, 1-E10 |
| φ3-B11 | 2 | 3, 3 | 3-B11 |
| φ3-E11 | 1 | 3 | 3-E11 |
| φ3-G1 | 1 | 3 | 3-G1 |
| φ3-G9 | 1 | 3 | 3-G9 |
| φ3-G10 | 1 | 3 | 3-G10 |
| φ3-H2 | 2 | 1, 3 | 3-H2 |
| φ7-C4 | 1 | 7 | 7-C4, 14-C8, 22-D10 |
| φ7-E1 | 6 | 7, 14, 14, 14, 14, 22 | 7-E1 |
| φ9-B4 | 1 | 9 | 9-B4, 14-A4 |
| φ14-C8 | 4 | 14, 14, 60, 60 | 14-C8, 22-E2, 22-D10, 60-E4, 60-E8 |
| φ14-E10 | 1 | 14 | 14-E10, 7-E1, 22-G10, 14-A4, 9-B4, 14-F3 |
| φ14-F3 | 1 | 14 | 14-F3 |
| φ14-H4 | 1 | 14 | 14-H4, 14-F3 |
| φ22-A2 | 1 | 22 | 22-A2, 7-E1, 22-G10, 14-A4, 9-B4, 14-F3 |
| φ22-D10 | 3 | 22, 22, 60 | 22-D10, 7-C4, 14-C8 |
| φ22-E2 | 1 | 22 | 22-E2, 22-D10 |
| φ60-D8 | 1 | 60 | 60-D8, 14-C8, 60-E4 |
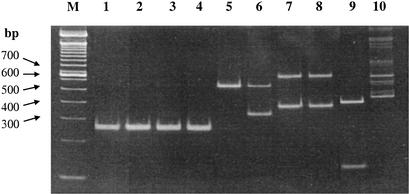
Based on phage typing, 28 distinct hosts were identified(Table 4). These hosts were further identified by their intergenic transcribed spacer (ITS) restriction digestion patterns and 16S rRNA gene sequence analyses. The biochemical characteristics and phage sensitivities of the 28 distinct hosts are listed in Table 4. These hosts were identified as 9 L. mesenteroides strains, 1 Leuconostoc pseudomesenteroides strain, 1 Leuconostoc citreum strain, 3 Leuconostoc fallax strains, 1 Weissella species (not conclusively identified), 10 L. plantarum strains, 2 Lactobacillus paraplantarum strains, and 1 Lactobacillus brevis strain (Table 4). Representative restriction profiles of ITS PCR products obtained from 10 distinct phage hosts are shown in Fig. 4. The four L. mesenteroides strains (1-A4, 1-F8, 3-A4, and 3-B1) had identical ITS restriction profiles when digested with RsaI. L. pseudomesenteroides (3-B11), L. brevis (7-E1), L. fallax (3-G10), and Weissella (3-H2) presented distinct ITS patterns. The two L. plantarum strains (14-C8 and 22-D10) had the same ITS profile. The Weissella digest showed multiple bands, presumably due to nonspecific priming of the original PCR product. We have found this pattern to be typical of this strain (data not shown).
TABLE 4.
Characteristics of 28 distinct host strains isolated from a commercial sauerkraut fermentation in 2000a
| Host | Species | Gas production | Dextran test | Malolactic activity | Phage typingb result |
|---|---|---|---|---|---|
| 1-A4 | L. mesenteroides | + | + | − | φ1-A4, φ1-E10 |
| 1-B8 | L. mesenteroides | + | + | − | φ1-B8, φ1-A4, φ1-E10 |
| 1-C4 | L. mesenteroides | + | + | + | φ1-C4, φ3-B1 |
| 1-C5 | L. mesenteroides | + | + | − | φ1-C5, φ1-F8, φ3-A4 |
| 1-E10 | L. mesenteroides | + | + | − | φ1-E10, φ1-A4, φ1-C4, φ3-B1 |
| 1-F7 | L. citreum | + | + | − | φ1-F7 |
| 1-F8 | L. mesenteroides | + | + | − | φ1-F8, φ3-A4 |
| 1-F10 | L. mesenteroides | + | + | − | φ1-F10, φ3-A4 |
| 3-A4 | L. mesenteroides | + | + | − | φ3-A4, φ1-E10, φ1-F8 |
| 3-B1 | L. mesenteroides | + | + | + | φ3-B1, φ1-C4, φ3-G9 |
| 3-B11 | L. pseudomesenteroides | + | + | + | φ3-B11 |
| 3-E11 | L. fallax | + | + | − | φ3-E11 |
| 3-G1 | L. fallax | + | + | − | φ3-G1 |
| 3-G10 | L. fallax | + | + | − | φ3-G10 |
| 3-H2 | Weissella sp. | + | + | + | φ3-H2 |
| 7-C4 | L. plantarum | − | − | + | φ7-C4, φ22-D10 |
| 7-E1 | L. brevis | + | − | + | φ7-E1, φ14-E10, φ22-A2 |
| 9-B4 | L. plantarum | − | − | − | φ9-B4, φ14-E10, φ22-A2, φ22-D10 |
| 14-A4 | L. plantarum | − | − | − | φ14-A4, φ14-E10, φ22-A2 |
| 14-C8 | L. plantarum or L. pentosus | − | − | + | φ14-C8, φ7-C4, φ22-D10, φ60-D8 |
| 14-E10 | L. plantarum | − | − | + | φ14-E10 |
| 14-F3 | L. paraplantarum | − | − | + | φ14-F3, φ14-E10, φ14-H4, φ22-A2 |
| 14-H4 | L. paraplantarum | − | − | + | φ14-H4 |
| 22-A2 | L. plantarum | − | − | + | φ22-A2 |
| 22-D10 | L. plantarum or L. pentosus | − | − | + | φ22-D10, φ7-C4, φ14-C8, φ22-E2, φ60-D8 |
| 22-E2 | L. plantarum | − | − | + | φ22-E2, φ14-C8, φ22-D10 |
| 60-E4 | L. plantarum | − | − | + | φ60-E4, φ22-D10, φ60-D8 |
| 60-E8 | L. plantarum | − | − | + | φ60-E8, φ22-D10 |
The first number of each host designation indicates the fermentation day when the host was isolated. All hosts were catalase negative.
Phage typing is defined as the pattern of sensitivity of a bacterium to lysis by a series of phages.
FIG. 4.
Restriction profiles of the ITS PCR product obtained from 10 phage hosts. Lane 1, L. mesenteroides 1-A4; lane 2, L. mesenteroides 1-F8; lane 3, L. mesenteroides 3-A4; lane 4, L. mesenteroides 3-B1; lane 5, L. pseudomesenteroides 3-B11; lane 6, L. brevis 7-E1; lane 7, L. plantarum 14-C8; lane 8, L. plantarum 22-D10; lane 9, L. fallax 3-G1; lane 10, Weissella strain 3-H2; lane M, a 100 bp DNA ladder.
Biochemical tests showed that all hosts were catalase negative (Table 4). All Leuconostoc and Weissella strains produced gas from glucose. In addition, Leuconostoc and Weissella strains also produced a characteristic slime of dextran from sucrose. None of the Lactobacillus strains except the L. brevis strain (7-E1) produced gas. Neither the L. plantarum nor the L. brevis strain produced slime colonies on sucrose agar. In contrast to most lactobacilli, most leuconostocs did not have malolactic activity.
It was found that all of the hosts isolated from days 1 and 3 belong to the genus Leuconostoc or Weissella, and hosts isolated after day 3 all belong to Lactobacillus. Among day 1 and 3 hosts, a number of strains, including three L. fallax strains, were sensitive only to a single phage. Most host strains isolated on or after day 7 were L. plantarum and were sensitive to two or more phages. Some hosts, such as 22-D10, were sensitive to up to five distinct phages. There was, with one exception, no overlapping susceptibility either between Leuconostoc species or between Lactobacillus species. The exception was L. brevis strain 7-E1, which was susceptible to one L. brevis phage (φ7-E1) and two L. plantarum phages (φ14-E10 and φ22-A2). Surprisingly, two phages (φ14-E10 and φ22-A2) were capable of infecting both L. brevis (7-E1) and L. plantarum (14-E10 or 22-A2).
In 2001, a total of 111 phage isolates were obtained from the three tanks by using the 28 distinct host strains isolated in 2000 as the principal hosts (Table 5). Twenty-four (86%) of these 28 hosts were found to be susceptible to the phages present in two or three tanks at selected time points in 2001. Phages φ1-F8, φ7-E1, φ14-C8, and φ22-D10 were frequently isolated in 2000 (Table 3). Their hosts were used at multiple time points in 2001 to investigate the presence and persistence of these phages in three different fermentation tanks. Table 5 shows that all of these hosts were susceptible to phages from any of the three tanks at one or more time points. Phages attacking 7-E1, 14-C8, and 22-D10 were found in all three tanks from day 14 or 22 to day 60. In addition to distinct hosts, several other host isolates, which were considered the same as the distinct ones based on phage typing, were also included for testing phages to see if they gave the same results. For instance, 7-E1 and 14-A2 were identical or related strains, based on phage typing (Table 2). Both isolates were used for testing day 14 brines from three tanks. They gave the same results (Table 5), suggesting that the two isolates were the same. With the same method, 14-C8 and 22-F3 gave identical results, and so did 22-D10 and 60-D8. The exceptions were 1-F8 and 3-B5. Strain 1-F8 was found to be susceptible to day 3 samples from any of the three tanks, whereas 3-B5 was susceptible to only two of the three samples. Host 7-E1 was found to be susceptible to only one of the three day 7 samples. However, it showed susceptibility to two day 9 samples and then to all three day 14 samples and remained susceptible thereafter, indicating that the phage titer in these tanks increased from day 7 to 22 and remained above the detection limit (2 PFU/ml) thereafter. A total of 24 phage isolates were obtained from the three tanks on day 60 by using eight bacterial isolates, including three redundant host isolates (14-C8 and 22-F3 were considered the same LAB strain as 60-H11, and 22-D10 was the same as 60-D8).
TABLE 5.
Phage isolation from three commercial sauerkraut fermentation tanks in 2001
| Day | Host | Phage isolation from tanka:
|
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| 1 | 1-A3 | + | + | + |
| 1-A4 | + | + | − | |
| 1-B8 | + | + | + | |
| 1-C4 | + | + | + | |
| 1-C5 | + | + | + | |
| 1-E10 | + | + | + | |
| 1-F7 | + | + | + | |
| 1-F8 | + | + | − | |
| 1-F10 | + | + | + | |
| 3 | 3-A4 | + | + | + |
| 3-B1 | + | + | + | |
| 3-B11 | + | + | + | |
| 3-E11 | + | + | + | |
| 3-G1 | + | + | + | |
| 3-G10 | + | + | + | |
| 3-H2 | + | − | − | |
| 1-F8 | + | + | + | |
| 3-B5 | + | + | − | |
| 7 | 7-C4 | + | + | + |
| 7-E1 | + | − | − | |
| 9 | 9-B4 | − | + | + |
| 7-E1 | + | − | + | |
| 14 | 14-A4 | − | − | − |
| 7-E1 | + | + | + | |
| 14-C8 | + | + | + | |
| 14-E10 | − | − | − | |
| 14-F3 | − | − | − | |
| 14-H4 | + | + | + | |
| 14-A2 | + | + | + | |
| 22 | 22-A2 | − | − | − |
| 7-E1 | + | + | + | |
| 14-C8 | + | + | + | |
| 22-F3 | + | + | + | |
| 22-D10 | + | + | + | |
| 22-E2 | + | + | + | |
| 22-G10 | + | + | + | |
| 60 | 60-E4 | + | + | + |
| 7-E1 | + | + | + | |
| 14-C8 | + | + | + | |
| 22-F3 | + | + | + | |
| 22-D10 | + | + | + | |
| 60-D8 | + | + | + | |
| 60-E8 | + | + | + | |
| 60-H11 | + | + | + | |
+, one or more phages were isolated; −, no phases were isolated.
Phage characterization.
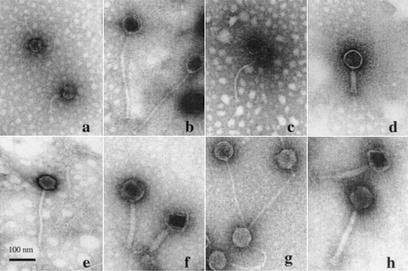
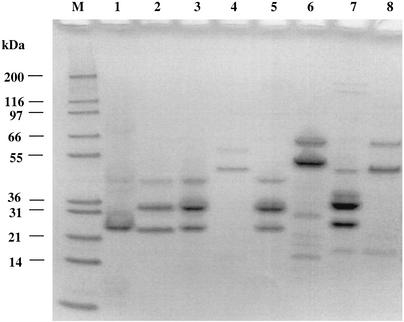
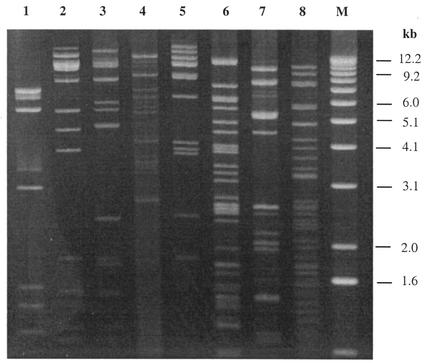
Eight of the 26 distinct phages isolated in 2000 (φ1-A4, φ1-F8, φ3-A4, φ3-B1, φ3-B11, φ7-E1, φ14-C8, and φ22-D10) were selected for further characterization. These phages appeared two or more times during phage isolation process (Tables 2 and 3), representing frequently occurring or predominant phages. The electron micrographs (Fig. 5) show that the eight phages all had tails and isometric heads. Morphological features were used for the characterization of these phages (1), which differed in host range (Table 3) and are summarized in Table 6. The major structural proteins of the eight phages were analyzed by SDS-PAGE. Several different structural protein profiles were observed (Fig. 6) for phages φ1-A4, φ3-B1, φ7-E1, φ14-C8, and φ22-D10. The remaining phages showed similar protein profiles. HindIII restriction digestion analysis of the eight phage genomes revealed eight unique restriction banding patterns (Fig. 7).
FIG. 5.
Electron micrographs of eight phages isolated from commercial sauerkraut fermentations. CsCl-purified phage preparations were negatively stained with 2% uranyl acetate (pH 4.0). (a) φ1-A4; (b) φ1-F8; (c) φ3-A4; (d) φ3-B1; (e) φ3-B11; (f) φ7-E1; (g) φ14-C8; (h) φ22-D10. Magnification, ×50,000.
TABLE 6.
Morphological features of eight selected phages isolated from a commercial sauerkraut fermentationa
| Phage | Family | Head diam (nm) | Tail
|
|
|---|---|---|---|---|
| Length (nm) | Width (nm) | |||
| φ1-A4 | Siphoviridae | 53 ± 2 | 131 ± 7 | 10 ± 1 |
| φ1-F8 | Siphoviridae | 71 ± 3 | 351 ± 14 | 12 ± 1 |
| φ3-A4 | Siphoviridae | 60 ± 2 | 288 ± 10 | 9 ± 1 |
| φ3-B11 | Siphoviridae | 60 ± 3 | 273 ± 11 | 13 ± 2 |
| φ14-C8 | Siphoviridae | 70 ± 2 | 292 ± 10 | 13 ± 2 |
| φ3-B1 | Myoviridae | 78 ± 4 | 155 ± 6 | 23 ± 2 |
| φ7-E1 | Myoviridae | 87 ± 4 | 149 ± 6 | 26 ± 3 |
| φ22-D10 | Myoviridae | 85 ± 5 | 271 ± 9 | 25 ± 3 |
Each value is the mean and the standard deviation of five independent measurements.
FIG. 6.
SDS-PAGE analysis of structural proteins from eight phages isolated from a commercial sauerkraut fermentation. Lane M, molecular mass markers; lane 1, φ1-A4; lane 2, φ1-F8; lane 3, φ3-A4; lane 4, φ3-B1; lane 5, φ3-B11; lane 6, φ7-E1; lane 7, φ14-C8; lane 8, φ22-D10.
FIG. 7.
HindIII restriction digestion analysis of DNAs from eight phages isolated from a commercial sauerkraut fermentation. Lane 1, φ1-A4; lane 2, φ1-F8; lane 3, φ3-A4; lane 4, φ3-B1; lane 5, φ3-B11; lane 6, φ7-E1; lane 7, φ14-C8; lane 8, φ22-D10; lane M, 1-kb DNA ladder.
DISCUSSION
Knowledge of the diversity, abundance, and properties of phages in vegetable fermentations is essential for developing phage control strategies for achieving high and consistent quality of fermented products. In the United States, commercial sauerkraut production is usually carried out in bulk tanks without addition of starter cultures (15). Because of the increasing interest in reducing waste brine disposal, low-salt fermentation is being developed. This will require greater control of the nonlactic population and may involve the use of starter cultures. Since the fermentation system is not sterile, the starter cultures may be susceptible to the infection by phages naturally present in these environments. Therefore, phage-control strategies may be needed to ensure the viability of starter cultures until the end of the fermentation. To our knowledge, this is the first reported study of the bacteriophage ecology of vegetable fermentations.
The results from microbial and chemical analyses indicated that the fermentations in the four commercial sauerkraut fermentation tanks were normal and consistent with those described by Fleming et al. (14). The phage-host relationships shown in Table 2 suggest that there are two phage-host systems in the fermentation corresponding to the shift occurring between day 3 and day 7 after the start of fermentation, from heterofermentative to homofermentative LAB populations. In 2000, the 23 phage isolates and corresponding host isolates obtained on days 1 and 3 were in one phage-host system (system I), whereas the phage and host isolates isolated on or after day 7 formed from the other phage-host system (system II). Within the same phage-host system, multiple phage isolates attacked the same host strain, and several hosts were sensitive to the same phage. A most striking observation, however, was that phages isolated from days 1 and 3 did not infect hosts isolated on or after day 7, whereas phages isolated after day 3 did not attack hosts isolated on day 1 or day 3. The restricted infectivity of phages to hosts from specific days clearly indicated that microbial populations shifted between day 3 and day 7, which was confirmed by a parallel study on bacterial ecology in these fermentations (detailed data will be published elsewhere). The appearance of a new group of phages was closely correlated to bacterial succession, which was supported by the host identification data. Among the 28 distinct host strains, 15 were obtained on days 1 and 3 (Table 4). All of these 15 strains belong to Leuconostoc or Weissella. The hosts isolated on or after day 7 were all lactobacilli (Table 4).
Sauerkraut fermentation is a dynamic biochemical system. The chemical composition and microbial ecology of the system are continuously changing. The numbers and types of phages appearing may reflect the activity of both phages and their hosts because phage infection is usually both density dependent and species specific. The fact that all of the 23 Leuconostoc and Weissella phage isolates were obtained from days 1 and 3 (Table 2) and no Leuconostoc and Weissella phages were isolated after day 3 suggests that Leuconostoc and Weissella strains were active only during the first 3 days but died off thereafter. It has been suggested that viral infection can contribute significantly (22%; range, 4.5 to 45%) to overall bacterial mortality in many marine and freshwater environments (5). Results from this study showed that an average of 12% (23 of the 192) of the randomly picked LAB isolates (mainly Leuconostoc) from days 1 and 3 were susceptible to phage infection. Such a significant phage infection could contribute greatly to the overall mortality in the Leuconostoc population, thereby influencing the dynamics of LAB populations in the fermentation. In contrast, no Lactobacillus phages were isolated until day 7, indicating that the Lactobacillus population was very small at the early stage of fermentation. The number of isolated Lactobacillus-specific phages increased from two on day 7 to nine on day 14 and then decreased to four on day 60 (Table 2), indicating the changes in the populations of lactobacilli. The correlation between the appearance of new groups of phages and the bacterial succession suggests that phages may play a role in the succession, rising with the corresponding host population and eventually causing its elimination.
It was noteworthy that 9 of the 26 distinct phages were independently isolated more than once on the same day or different days in 2000 (Table 3). Phage φ7-E1 was isolated six times (four times on day 14), whereas φ14-C8 appeared among phage isolates four times (twice each on days 14 and 60), suggesting that these phages, as well as their hosts, were predominant on day 14 and/or day 60. Four phage isolates were obtained from day 60, when the brine pH was very low (3.5), indicating that these phages and their hosts were very persistent in the acidic environment.
Host range is an important aspect of phage diversity that has ecological repercussions. Many phages infected two or more bacterial strains (Table 3). Phages φ14-C8, φ14-E10, and φ22-A2 were capable of infecting five or six strains. The broad-host-range phenomenon has also been observed for a number of phages infecting LAB starter strains used in the dairy industry (4, 8, 12). It was reported that Streptococcus thermophilus phage DT1 is capable of infecting 10 strains (12). Some proteins on the cell surface, such as LamB on Escherichia coli, serve as specific cell surface receptors not only for λ phage but also for a series of other phages (9, 30). Some complex phages can use more than one receptor and therefore have alternative routes of uptake into cells (9). Broad-host-range phages may promote genetic diversity and genetic exchange in microbial communities (18). Table 3 also shows that a number of phages were capable of cross-infecting each other's hosts. Surprisingly, two L. plantarum phages (φ14-E10 and φ22-A2) were also capable of infecting the L. brevis strain (7-E1). To our knowledge, this interspecies cross-infection has not been described in the literature. Comparative phage genomics has suggested that phages may have evolved through exchange of functional modules, individual genes, or gene segments via various genetic recombination events (6, 20). It is possible that horizontal gene transfer may have occurred between some phages of L. plantarum and L. brevis origins, as these hosts usually share the same ecological niche. Alternatively, a common ancestor phage infecting one species may have acquired the capacity, not necessarily via horizontal gene transfer, to infect the other bacterial species (17, 28). It may also be possible that L. brevis and L. plantarum exhibited similar receptors on cell surfaces. In contrast to the phages having a broad host range, several phages, including three Weissella phages, exhibited a very restricted host range and infected only a single strain. The large variety of phage types (Table 3) indicated the complex phage ecology in commercial sauerkraut fermentation. More work is needed to determine if these phages are lytic or lysogenic.
Based on the restriction banding patterns of ITS PCR products, the 10 selected phage hosts (Fig. 4) belong to six different species in three genera, including Leuconostoc, Weissella, and Lactobacillus. The resulting host identifications were confirmed by 16S ribosomal DNA sequence analysis. L. fallax is a recently recognized LAB species in sauerkraut fermentation (3, 31). The three L. fallax strains and one Weissella strain isolated in this study were sensitive to only one specific phage. In contrast, host 22-D10 was susceptible to five distinct L. plantarum phages isolated on days 7, 14, 22, and 60, while L. brevis phage φ7-E1 attacked only bacterial strain 7-E1. The host 7-E1 (L. brevis) was susceptible to two L. plantarum phages (φ14-E10 and φ22-A2), suggesting that 7-E1 may have either two different types of receptors or one type of receptors that could be recognized by both L. brevis and L. plantarum phages.
The 111 phage isolates obtained from three fermentation tanks in 2001 were the same as, or very similar to, those isolated in 2000, because they attacked the same set of LAB hosts, suggesting that these phages are commonly present in sauerkraut fermentations. The large number and variety of phage isolates confirmed a complex phage ecology in sauerkraut fermentation, with some phages reoccurring. This result was consistent with that from the previous year. The temporal relationship between phages and their hosts suggests that phages could influence the number, types, and sequences of microbial populations in the fermentation. Hosts 7-E1 and 14-C8 (Table 5) were sensitive to phages in all three tanks from day 14 to day 60 in 2001, suggesting that these hosts and their phages were very common and persistent in sauerkraut fermentation. Twenty-four phage isolates were obtained on day 60 (pH ≤3.5) from the three fermentation tanks in 2001, indicating that these phages were very stable in such a low-pH environment. Although several phages were frequently isolated in 2000 and 2001, no evidence showed that a single phage dominated the fermentation system. Each type of microorganism is involved in the microbial succession and contributes to the final characteristic properties of the fermented products.
The eight characterized phages belonged to either the Myoviridae or Siphoviridae family (Fig. 5), each having distinct morphological features (Table 6). The principal hosts of these phages included four L. mesenteroides strains (1-A4, 1-F8, 3-A4, and 3-B1), one L. pseudomesenteroides strain (3-B11), two L. plantarum strains (14-C8 and 22-D10), and one L. brevis strain (7-E1) (Table 4). The SDS-PAGE analysis showed that several of them had a similar structural protein profile (Fig. 6). However, the restriction analysis (Fig. 7) indicated that the eight phages were genetically distinct. The data from these eight selected phages provided a glimpse of the genetic diversity of phage population in commercial sauerkraut fermentations. The genetic diversity of LAB phages were also observed in dairy fermentations. An ecological survey in a cheese factory (8) showed a wide range of restriction patterns of phage isolates attacking S. thermophilus strains, reflecting the diversity of phages normally found in their environment.
More research is warranted to continue to unravel the ecological role of phages and to evaluate their impact on the vegetable fermentation process. Additionally, it may be necessary to study the fermentations at other geographic locations to examine phage diversity. In conclusion, results from this 2-year study demonstrate for the first time the complex phage ecology in commercial sauerkraut fermentations. The appearance of a new group of phages was correlated closely to bacterial succession. The data suggest that phages may influence the succession of LAB, due to the diversity of phages discovered, and the correlation of phage and LAB populations. The results indicate that phage infection is common in sauerkraut fermentations, with some phages predominating and reoccurring. This suggests that a phage control strategy may be essential in low-salt sauerkraut fermentations which rely on starter cultures.
Acknowledgments
This investigation was supported in part by a research grant from Pickle Packers International, Inc., St. Charles, Ill..
We thank Bush Brothers & Company (Shiocton, Wis.) for their cooperation during this 2-year study. We also thank Roger Thompson and Laura Reina for technical assistance, Todd R. Klaenhammer and Eric Altermann for helpful advice, and Dora Toler for excellent secretarial assistance.
Mention of a trademark or proprietary product does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture or North Carolina Agricultural Research Service, nor does it imply approval to the exclusion of other products that may be suitable.
Footnotes
Paper no. FSR02-39 of the Journal Series of the Department of Food Science, North Carolina State University.
REFERENCES
- 1.Ackermann, H.-W. 2001. Frequency of morphological phage descriptions in the year 2000. Brief Rev. Arch. Virol. 146:843-857. [DOI] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1993. Sucrose agar, p. 854. In C. P. Lawrence (ed.), Handbook of microbiological media. CRC Press, London, England.
- 3.Barrangou, R., S.-S. Yoon, F. Breidt, H. P. Fleming, and T. R. Klaenhammer. 2002. Identification and characterization of Leuconostoc fallax strains isolated from an industrial sauerkraut fermentation. Appl. Environ. Microbiol. 68:2877-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhimani, R. S., and Y. M. Freitas. Isolation and characterization of the bacteriophages of lactic streptococci. J. Dairy Sci. 74:1461-1471.
- 5.Binder, B. 1999. Reconsidering the relationship between virally induced bacterial mortality and frequency of infected cells. Aquat. Microb. Ecol. 18:207-215. [Google Scholar]
- 6.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-491. [DOI] [PubMed] [Google Scholar]
- 7.Breidt, F., and H. P. Fleming. 1996. Identification of lactic acid bacteria by ribotyping. J. Rapid Methods Automation Microbiol. 4:219-233. [Google Scholar]
- 8.Bruttin, A., F. Desiere, N. d'Amico, J.-P. Guérin, J. Sidoti, B. Huni, S. Lucchini, and H. Brüssow. 1997. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 63:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charbit, A., and M. Hofnung. 1985. Isolation of different bacteriophages using the LamB protein for adsorption on Escherichia coli K-12. J. Virol. 53:667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daeschel, M. A., R. F. McFeeters, H. P. Fleming, T. R. Klaenhammer, and R. B. Sanozky. 1984. Mutation and selection of Lactobacillus plantarum strains that do not produce carbon dioxide from malate. Appl. Environ. Microbiol. 47:419-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daeschel, M. A., R. E. Andersson, and H. P. Fleming. 1987. Microbial ecology of fermenting plant materials. FEMS Microbiol. Rev. 46:357-367. [Google Scholar]
- 12.Duplessis, M., and S. Moieau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 13.Fleming, H. P. 1982. Fermented vegetables, p. 227-258. In A. H. Rose (ed.), Economic microbiology, vol. 7. Fermented foods. Academic Press, Inc., New York, N.Y.
- 14.Fleming, H. P., R. F. McFeeters, J. L. Etchells, and T. A. Bell. 1984. Pickled vegetables, p. 663-681. In M. L. Speck (ed.), Compendium of methods for the microbiological examination of foods, 2nd ed. American Public Health Association, Washington, D.C.
- 15.Fleming, H. P., R. F. McFeeters, and E. G. Humphries. 1988. A fermentor for study of sauerkraut fermentation. Biotechnol. Bioeng. 31:189-197. [DOI] [PubMed] [Google Scholar]
- 16.Fleming, H. P., R. F. McFeeters, and M. A. Daeschel. 1992. Fermented and acidified vegetables, p. 929-952. In C. Vanderzant and D. F. Splittstoesser (ed.), Compendium of methods for the microbiological examination of foods, 3rd ed. American Public Health Association, Washington, D.C.
- 17.Haggard-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, E. C., H. S. Schrader, B. Rieland, T. L. Thompson, K. W. Lee, K. W. Nickerson, and T. A. Kokjohn. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, Z., F. Breidt, H. P. Fleming, E. Altermann, and T. R. Klaenhammer. Isolation and characterization of a Lactobacillus plantarum bacteriophage, φJL-1, from a cucumber fermentation. Int. J. Food Microbiol., in press. [DOI] [PubMed]
- 20.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald, L. C., H. P. Fleming, and H. M. Hassan. 1990. Acid tolerance of Leuconostoc mesenteroides and Lactobacillus plantarum. Appl. Environ. Microbiol. 56:2120-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mundt, J. O., W. F. Graham, and I. E. McCarty. 1967. Spherical lactic acid-producing bacteria of southern-grown raw and processed vegetables. Appl. Microbiol. 15:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mundt, J. O., and J. L. Hammer. 1968. Lactobacilli on plants. Appl. Microbiol. 16:1326-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mundt, J. O. 1970. Lactic acid bacteria associated with raw plant food material. J. Milk Food Technol. 33:550-553. [Google Scholar]
- 25.Pederson, C. S., and M. N. Albury. 1969. The sauerkraut fermentation. New York State Agric. Exp. Sta. Tech. Bull. 824:1-84. [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Stamer, J. R., B. O. Stoyla, and B. A. Dunckel. 1971. Growth rates and fermentation patterns of lactic acid bacteria associated with the sauerkraut fermentation. J. Milk Food Technol. 34:521-525. [Google Scholar]
- 28.Stanley, E., G. F. Fitzgerald, C. Le Marrec, B. Fayard, and D. van Sinderen. 1997. Sequence analysis and characterization of φO1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology 143:3417-3429. [DOI] [PubMed] [Google Scholar]
- 29.Vaughn, R. H. 1985. The microbiology of vegetable fermentations. p. 49-109. In B. J. B. Wood (ed.), Microbiology of fermented foods, vol. 1. Elsevier, New York, N.Y.
- 30.Werts, C., V. Michel, M. Hofnung, and A. Charbit. 1994. Adsorption of bacteriophage lambda on the LamB protein of E. coli K-12: point mutations in gene J of lambda responsible for extended host range. J. Bacteriol. 176:941-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon, S. S., R. Barrangou-Poueys, F. Breidt, Jr., T. R. Klaenhammer, and H. P. Fleming. 2002. Isolation and characterization of bacteriophages from fermenting sauerkraut. Appl. Environ. Microbiol. 68:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]