Abstract
The objective of this study was to develop a rapid, reproducible, and robust method for detecting Salmonella enterica serotype Enteritidis in poultry samples. First, for the extraction and purification of DNA from the preenrichment culture, four methods (boiling, alkaline lysis, Nucleospin, and Dynabeads DNA Direct System I) were compared. The most effective method was then combined with a real-time PCR method based on the double-stranded DNA binding dye SYBR Green I used with the ABI Prism 7700 system. The specificity of the reaction was determined by the melting temperature (Tm) of the amplicon obtained. The experiments were conducted both on samples of chicken experimentally contaminated with serotype Enteritidis and on commercially available poultry samples, which were also used for comparisons with the standard cultural method (i.e., ISO 6579/2001). The results of comparisons among the four DNA extraction methods showed significant differences except for the results from the boiling and Nucleospin methods (the two methods that produced the lowest threshold cycles). Boiling was selected as the preferred extraction method because it is the simplest and most rapid. This method was then combined with SYBR Green I real-time PCR, using primers SEFA-1 and SEFA-2. The specificity of the reaction was confirmed by the Tm, which was consistently specific for the amplicon obtained; the mean peak Tm obtained with curves specific for serotype Enteritidis was 82.56 ± 0.22°C. The standard curve constructed using the mean threshold cycle and various concentrations of serotype Enteritidis (ranging from 103 to 108 CFU/ml) showed good linearity (R2 = 0.9767) and a sensitivity limit of less than 103 CFU/ml. The results of this study demonstrate that the SYBR Green I real-time PCR constitutes an effective and easy-to-perform method for detecting serotype Enteritidis in poultry samples.
In the last 20 years, Salmonella enterica serotype Enteritidis has become the most common cause of food poisoning in the United States (19) and in most European countries (4, 20), including Italy (26, 27); the most commonly implicated foods are eggs and poultry (11, 29, 38). Although various control measures have been adopted throughout the food production chain (2, 6, 32), the microbiological testing of poultry products during production and processing still plays a significant role in preventing food-borne infection. However, the standard cultural method for detecting salmonella (i.e., ISO 6579/2001) requires up to 5 days to produce results. To reduce the time required for testing, different methods have been developed (7, 18, 35), including tests based on novel reagents, yet these tests are generally used to supplement rather than replace existing methods. The exception is the methodology based on PCR, which has progressively been replacing biochemical and agglutination tests (13, 33). However, PCR methods include a final confirmation phase, such as electrophoresis, which requires a certain level of skill and which is both laborious and time-consuming, thus limiting the number of samples that can be analyzed (34). In recent years, a method based on PCR with an automatic confirmation phase has been developed. This method, known as real-time fluorogenic 5′-nuclease PCR, has been used to detect a number of pathogenic microorganisms, including Salmonella (5, 13), in foods (3, 34). The method uses the 5′-nuclease activity of Taq DNA polymerase to hydrolyze an internal fluorogenic probe for monitoring the amplification of DNA targets. However, its application requires the availability of primers and probes that must be selected according to very rigid conditions, which cannot always be easily applied. The use of the double-stranded DNA (dsDNA) binding dye SYBR Green I for the detection of PCR products has overcome this limitation by allowing real-time PCR to be applied without the need for probes linked to fluorescent molecules (1, 14, 24); protocols that are already in use for classic PCR can thus be used with only slight modifications (9).
In the absence of probes, the specificity of the reaction is determined by the melting temperature (Tm) of the amplicon obtained, defined as the temperature at which 50% of the DNA amplicon is in a double-stranded configuration. The Tm depends on various factors, including the concentration of the dsDNA, the amplicon length, and the nucleotide sequence (22). However, the effectiveness of this method depends on the capacity of the extraction and purification procedure to remove from the food those substances that inhibit the activity of polymerase DNA (15, 23, 25).
The objective of the present study was to develop a rapid, reproducible, and robust method by first comparing four methods of DNA extraction and purification for their effectiveness and then combining the most effective method with a real-time PCR method based on the dsDNA binding dye SYBR Green I as used with the ABI Prism 7700 system to detect Salmonella serotype Enteritidis in poultry. To develop the method, the experiments were conducted on samples of chicken experimentally contaminated with serotype Enteritidis. To determine the method's effectiveness, numerous samples of commercially available poultry were analyzed, and the results were compared to those obtained by the standard cultural method.
MATERIALS AND METHODS
Serotype Enteritidis culture.
The strain used was S. enterica serotype Enteritidis ATCC 13076, grown in Triptone soy broth (Oxoid, Basingstoke, Hampshire, United Kingdom) at 37°C for 24 h. The broth culture was then washed three times by means of centrifugation (8,000 × g for 8 min) in a solution of 0.8% NaCl. The microbial suspensions were standardized by turbidimetry (40% transmittance at a wavelength of 540 nm; Bausch & Lomb Spectronic 20 turbidimeter). A parallel count of CFU of the serotype Enteritidis strain was performed on Triptone soy agar (Oxoid) and yielded about 8 log CFU/ml.
Food samples.
A sample of chicken experimentally spiked with serotype Enteritidis was used for the development of the SYBR Green I real-time PCR method and melting curve analysis. The experiments were repeated three times.
Sixty samples of poultry (both chicken and turkey), including muscle, skin, and internal organs, randomly sampled at a commercial poultry processing plant, were analyzed to determine the effectiveness of the SYBR Green I real-time PCR method, and results were compared to those of the standard cultural method (ISO 6579/2001).
Preparation of the samples. (i) Experimentally spiked sample.
Twenty-five grams of poultry homogenized with 225 ml of buffered peptone water (BPW; Oxoid) in a Stomacher Lab Blender 400 (U.A.C. House, London, United Kingdom) was incubated at 37°C for 24 h to allow for bacterial growth. The preenrichment broth was then divided into three aliquots. (a) The first aliquot was used to confirm the absence of Salmonella by means of the standard cultural method (ISO 6579/2001). (b) The second aliquot was spiked with an appropriate quantity of serotype Enteritidis suspension to obtain a final concentration of 107 CFU/ml. The spiked preenrichment broth was subdivided into two aliquots: one was subjected to immunomagnetic concentration by means of paramagnetic particles coated with anti-Salmonella antibodies (Dynabeads anti-Salmonella; Dynal, Oslo, Norway), according to the manufacturer's instructions (20 μl of Dynabeads plus 1 ml of the preenriched samples). The other aliquot was not subjected to immunomagnetic concentration. (c) The third aliquot was used as a negative control.
The two spiked aliquots (the one that was subjected to immunomagnetic concentration and the one that was not) and the aliquot used as a negative control were subjected to four methods for extracting and purifying the DNA: boiling, alkaline lysis, Nucleospin, and Dynabeads DNA Direct System I.
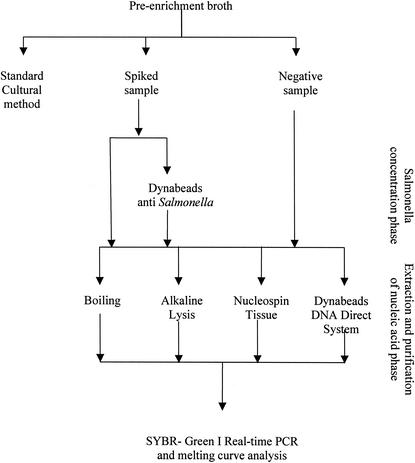
All 12 aliquots (3 per method) were then used for the SYBR Green I real-time PCR and melting curve analysis. The experimental scheme is shown in Fig. 1.
FIG. 1.
Experimental scheme used in this study.
(ii) Samples from the poultry processing plant.
Twenty-five grams of each sample was homogenized in 225 ml of BPW in a Stomacher Lab Blender 400 and incubated at 37°C for 24 h. After incubation, the preenrichment broth was subdivided into two aliquots: one aliquot was analyzed with the standard cultural method (ISO 6579/2001), and the other aliquot was subjected to boiling (the selected DNA extraction and purification method, as described below) and to SYBR Green I real-time PCR and melting curve analysis.
Extraction and purification of DNA. (i) Boiling.
One milliliter of the preenriched sample was transferred to a microcentrifuge tube with a capacity of 1.5 ml. The cell suspension was centrifuged for 10 min at 14,000 × g. The supernatant was discarded carefully. The pellet was resuspended in 300 μl of DNase-RNase-free distilled water (Sigma, Sigma-Aldrich s.r.l., Gallarate, Milan, Italy) by vortexing. The tube was centrifuged at 14,000 × g for 5 min, and the supernatant was discarded carefully. The pellet was resuspended in 200 μl of DNase-RNase-free distilled water (Sigma) by vortexing. The microcentrifuge tube was incubated for 15 min at 100°C and immediately chilled on ice. The tube was centrifuged for 5 min at 14,000 × g at 4°C. The supernatant was carefully transferred to a new microcentrifuge tube and incubated again for 10 min at 100°C and chilled immediately on ice. An aliquot of 5 μl of the supernatant was used as the template DNA in the PCR. The procedure was performed according to the method outlined by B. Malorny (http://www.pcr.dk/DNA-purification.htm).
(ii) Alkaline extraction.
One milliliter of the preenriched sample was transferred to a microcentrifuge tube with a capacity of 1.5 ml. The cell suspension was centrifuged for 10 min at 14,000 × g. The supernatant was discarded carefully. The pellet was resuspended in 50 μl of 0.05 N NaOH. The microcentrifuge tube was centrifuged for 5 min at 14,000 × g at 4°C. The supernatant was carefully transferred to a new microcentrifuge tube and supplemented with 8 μl of 1 M Tris-HCl buffer (pH 7.00). The microcentrifuge tube was centrifuged for 2 min at 14,000 × g at 4°C. DNase-RNase-free distilled water was then added to achieve a final volume of 200 μl (Sigma). A 5-μl aliquot of the supernatant was used as the template DNA in the PCR.
(iii) Nucleospin tissue (Macherey-Nagel GmbH and Co. KG, Düren, Germany).
One milliliter of the preenriched sample was transferred to a microcentrifuge tube with a capacity of 1.5 ml and centrifuged for 5 min at 5,200 × g. The DNA was extracted by following the manufacturer's instructions in a final volume of 200 μl of elution buffer (10 mM Tris-HCl; 1 mM EDTA, pH 8.00; Macherey-Nagel). A 5-μl aliquot of the supernatant was used as the template DNA in the PCR.
(iv) Dynabeads DNA Direct System I (Dynal).
One milliliter of the preenriched sample was treated according to the manufacturer's instructions. DNase-RNase-free distilled water was then added to achieve a final volume of 200 μl (Sigma). A 5-μl aliquot of the supernatant was used as the template DNA in the PCR.
SYBR Green I real-time PCR and melting curve analysis.
In the SYBR Green I real-time PCR, the amplification of the DNA target is measured in terms of the increment in the quantity of fluorescence determined at the end of each amplification cycle. In brief, SYBR Green I binds with the minor groove of dsDNA, greatly enhancing the fluorescence. The fractional cycle in which the increase in the fluorescence generated by the accumulation exceeds 10 standard deviations of the mean baseline fluorescence, with a selected range of cycles, is referred to as the threshold cycle (CT) (17, 36).
The results were visualized using the software Sequence Detector 1.7 provided with the ABI Prism 7700 system (Applied Biosystems, Foster City, Calif.), and the standard curve was confirmed by using Microsoft Excel 2000.
The specificity of the reaction is given by the Tms of the amplification products immediately after the last reaction cycle. The Tm, which is specific for each amplicon, is determined during the phase of slow heating, from 60 to 95°C in 19 min 59 s, during which time there occurs a rapid decrease in the fluorescence due to the denaturation of the amplicons, which results in the formation of single filaments of DNA and the successive detachment of the SYBR Green I. The melting curve was visualized with the software Dissociation Curve 1.0 provided with the ABI Prism 7700 system (Applied Biosystems).
(i) Choice of primers and optimization of primer concentrations.
The primer pair SEFA-1 (5′-GCAGCGGTTACTATTGCAGC-3′) and SEFA-2 (5′-CTGTGACAGGGACATTTAGCG-3′) (30, 37) was used (M-Medical-Genenco, Milan, Italy). These primers, which generate a 310-bp product, were selected in the region sefA, which is the region that codifies a major subunit protein of a novel fimbrial structure on the surface of serotype Enteritidis (31) and which is found in few other Salmonella serotypes (8).
To determine the optimal concentration of primers, preliminary tests were performed using equimolecular 300, 50, 25, and 5 nM concentrations of the primers SEFA-1 and SEFA-2.
(ii) Determining the sensitivity limit.
The sensitivity limit was determined by constructing the standard curve obtained from the CT values of the chicken sample in the preenrichment broth after incubation at 37°C for 24 h, supplemented with reducing concentrations of serotype Enteritidis (108, 107, 106, 105, 104, and 103 CFU/ml). One nonspiked sample was used as a negative control.
(iii) Amplification.
All amplification reactions were performed in a total volume of 50 μl with an ABI Prism 7700 sequence detector (Applied Biosystems) with 96-well microwell plates (MicroAmp; Applied Biosystems). In each well, we placed 5 μl of purified DNA, 25 μl of SYBR Green I PCR Master Mix (Applied Biosystems), a 50 nM concentration of the primer SEFA-1, a 50 nM concentration of the primer SEFA-2, and, to reach a total volume of 50 μl per well, DNase-RNase-free distilled water (Sigma). The reaction mixture was run online at 50°C for 2 min and 95°C for 10 min, followed by 35 cycles at 95°C for 1 min, 55°C for 1 min, and 72°C for 60 s, with an extension phase of 1 cycle at 95°C for 1 min, 60°C for 1 min, and 95°C for 1 min (ramp time, 19.59 min).
Standard cultural method.
In accordance with the standard cultural method (ISO 6579/2001), after incubation at 37°C for 24 h, 2 ml of preenrichment broth (BPW) was transferred into 20 ml of selenite-cystine broth (Oxoid), and 0.1 ml of BPW was transferred into 10 ml of Rappaport-Vassiliadis soya peptone medium (Oxoid); these broth cultures were then incubated, respectively, at 37 ± 1 and 41.5 ± 1°C for 24 h.
After incubation, each broth culture was streaked onto two selective media (Brillant Green agar and desoxycolate citrate agar; both from Oxoid) and incubated at 37°C for 24 to 48 h.
Suspected Salmonella colonies were biochemically identified using the API 20E system (bioMérieux sa, Marcy-l'Etoile, France); serotyping was performed using commercially available specific antisera (Statens Serum Institut, Copenhagen, Denmark).
Statistical analysis.
One-way analysis of variance with Bonferroni post hoc comparisons was used for the evaluation of the results. The null hypothesis was rejected if the significant difference between values was determined to have a probability (P value) of less than 0.05.
RESULTS
The concentration of primers chosen for the experiments was 50 nM. This concentration was chosen because it provided the lowest CT (15.062) and because the CT values obtained at lower primer concentrations (at 5 and 25 nM) were significantly higher (P < 0.05; data not shown).
Table 1 shows the CT values obtained by the four DNA extraction and purification methods either in combination with or without the immunomagnetic concentration of Salmonella. The only statistically significant difference was observed for Dynabeads DNA Direct System I, for which the CT value obtained with immunomagnetic concentration was higher than that obtained without immunomagnetic concentration. Based on these results, the decision was made not to include immunomagnetic concentration as part of the methodology.
TABLE 1.
Comparison of the statistical differencesa in CT values for four methods of DNA extraction and purification with or without immunomagnetic concentration of Salmonella Dynabeads
| DNA extraction and purification method | Mean CT ± SDc
|
P value | |
|---|---|---|---|
| Not treated with Salmonella Dynabeads | Treated with Salmonella Dynabeads | ||
| Boiling | 19.36 ± 0.24 | 19.43 ± 0.33 | NSb |
| Alkaline lysis | 19.89 ± 0.24 | 19.66 ± 0.31 | NS |
| Nucleospin | 19.28 ± 0.11 | 19.54 ± 0.53 | NS |
| Dynabeads DNA Direct System I | 23.32 ± 0.17 | 32.00 ± 3.31 | <0.05 |
Differences were analyzed by a post hoc Bonferroni comparison.
NS, not significant.
Means of three determinations ± standard deviations.
The one-way analysis of variance of the mean differences in CT values among the four DNA extraction and purification methods without immunomagnetic concentration of Salmonella (Dynabeads) showed a statistically significant difference (P < 0.0001). The post hoc Bonferroni comparison showed significant differences for all comparisons (P < 0.05) except for the methods of boiling and Nucleospin (the two methods that produced the lowest CTs).
The standard curve constructed using the mean CT obtained with boiling for various concentrations of serotype Enteritidis (ranging from 103 to 108 CFU/ml) showed a good linearity of response (R2 = 0.9767). The sensitivity limit of the reaction was less than 103 CFU/ml.
Of the 60 samples taken from the poultry processing plant, 11 were found to be positive when they were analyzed with the standard cultural method. For these 11 samples, Table 2 shows the serotypes identified by the standard method and the CT and Tm values from the SYBR Green I real-time PCR analysis. Three of 11 the samples were identified as serotype Enteritidis by the standard method, and only these three samples were positive for serotype Enteritidis when they were subjected to SYBR Green I real-time PCR.
TABLE 2.
Serotypes identified by the standard cultural method (ISO 6579/2001) and CT and Tm values obtained by SYBR Green I real-time PCR
| Salmonella-positive sample | Salmonella enterica serotype according to ISO 6579/2001 method | SYBR Green I real-time PCR
|
|
|---|---|---|---|
| Mean CT ± SD | Tm (°C)a | ||
| 1 | Newrochelle | >35 | NS |
| 2 | Infantis | >35 | NS |
| 3 | Newrochelle | >35 | NS |
| 4 | Enteritidis | 16.80 ± 0.127 | 82.7 |
| 5 | London | >35 (36.74 ± 3.701) | NS |
| 6 | London | >35 (38.24 ± 3.048) | NS |
| 7 | Infantis | >35 | NS |
| 8 | Enteritidis | 14.24 ± 0.070 | 82.5 |
| 9 | Enteritidis | 15.43 ± 0.572 | 82.6 |
| 10 | Newrochelle | >35 | NS |
| 11 | Typhimurium | >35 (35.73 ± 0.814) | NS |
NS, not shown.
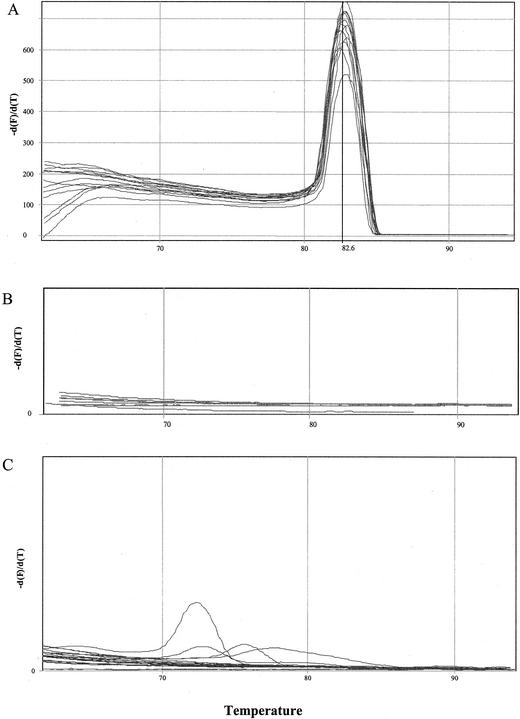
The mean peak Tm obtained with 83 curves specific for serotype Enteritidis was 82.56 ± 0.22°C (range, 81.80 to 82.90°C). Some characteristic curves are shown in Fig. 2A. The mean peak Tm of the amplicons of the three poultry samples naturally contaminated with serotype Enteritidis confirmed the specificity of the reaction (Table 2).
FIG. 2.
Melting curve of serotype Enteritidis after 35 cycles (A); serotype London, serotype Typhimurium, and negative controls after 35 cycles (B); and serotype London, serotype Typhimurium, and negative controls after 40 cycles (C).
The negative controls and the samples contaminated with serotypes other than serotype Enteritidis did not show peaks in the Tm when they were subjected to 35 cycles of amplification (Fig. 2B), whereas they did show some peaks (for example, with serotype London and serotype Typhimurium) when they were subjected to 40 cycles, although the Tms were lower than those for serotype Enteritidis (Fig. 2C).
DISCUSSION
In contrast to other studies (16, 21), we found that the immunomagnetic concentration conducted on the preenrichment culture showed no advantages in terms of purifying DNA and that a single phase is sufficient for eliminating the inhibiting substances present in the sample.
The four methods tested for the extraction and purification of DNA, with the exception of Dynabeads DNA Direct System I, produced similar results. From these methods, boiling was chosen because of its simplicity and rapidity of execution.
The specificity of the reaction was confirmed by the determination of the Tm, which was consistently specific for the amplicon obtained. Determining the Tm has the marked advantage of eliminating the phase of electrophoresis, which is time-consuming, carries the risk of laboratory contamination with nucleic acid due to post-PCR manipulation (10), and requires the use of ethidium bromide, which, being a potent mutagenic agent, is not suitable for routine use (28). However, variations of more than 1°C can occur in the minimum and maximum Tms, as has also been reported from other studies (9, 36). These variations are in part due to a nonhomogeneous distribution of the temperature in the thermocycler. It is thus recommended that two positive controls be used for each experiment: one placed at the center of the block and the other placed at a more external position (as described by M. Wagner, D. Schoder, and M. Kuhn, http://www.pcr.dk/Ny_mappe/o11015 Thermocycler Guidelines III - final version.doc).
The effectiveness of the proposed method was, moreover, confirmed by the analysis of 60 poultry samples, for which neither false-positive nor false-negative results were obtained.
The standard curve showed a strict inverse correlation between the CT and the concentration of Salmonella in the samples, and the limit of sensitivity of the method was less than 103 CFU/ml. Based on the CT values found in the samples containing serotype Enteritidis under our experimental conditions, the concentration of the microorganism in the preenrichment culture after 18 to 20 h of incubation at 37°C was always greater than 108 CFU/ml. This result suggests that the incubation period of the preenrichment medium can be reduced, thus shortening the time required for this method while maintaining its effectiveness and rendering it particularly useful for determining the effectiveness of the hazard analysis and critical point systems used in the food industry to reduce the numbers of pathogenic germs (12).
Acknowledgments
We are grateful to Mark Kanieff for linguistic revision.
This work was supported by the Istituto Superiore di Sanità project Strategie per una Accurata e Rapida Risposta ai Pericoli Microbici Veicolati da Alimenti and by the Fifth Framework of the European Commission project QLK1-CT-1999-00226 (Validation of Diagnostic PCR for Detection of Foodborne Pathogens).
REFERENCES
- 1.Aarts, H. J., R. G. Joosten, M. H. Henkens, H. Stegeman, and A. H. van Hoek. 2001. Rapid duplex PCR assay for the detection of pathogenic Yersinia enterocolitica strains. J. Microbiol. Methods 47:209-217. [DOI] [PubMed] [Google Scholar]
- 2.Alterkruse, S. F., L. K. Tollefson, and K. Bögel. 1993. Control strategies for Salmonella enteritidis in five countries. Food Control 4:10-16. [Google Scholar]
- 3.Bassler, H. A., S. J. A. Flood, K. J. Livak, J. Marmaro, R. Knorr, and C. A. Batt. 1995. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl. Environ. Microbiol. 61:3724-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler, A. J., B. M. Hargis, and R. M. Tsolis. 2000. Tracing the origins of Salmonella outbreaks. Science 287:50-52. [Online.] http://www.sciencemag.org/cgi/content/full/287/5450/50. [DOI] [PubMed] [Google Scholar]
- 5.Chen, S., A. Yee, M. Griffiths, C. Larkin, C. T. Yamashiro, R. Behari, C. Paszko-Kolva, K. Rahn, and S. A. De Grandis. 1997. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int. J. Food Microbiol. 35:239-250. [DOI] [PubMed] [Google Scholar]
- 6.Davies, R., M. Breslin, J. E. Corry, W. Hudson, and V. M. Allen. 2001. Observations on the distribution and control of Salmonella species in two integrated broiler companies. Vet. Rec. 149:227-232. [DOI] [PubMed] [Google Scholar]
- 7.De Medici, D., G. Pezzotti, C. Marfoglia, D. Caciolo, G. Foschi, and L. Orefice. 1998. Comparison between ICS-Vidas, MSRV and standard cultural method for Salmonella recovery in poultry meat. Int. J. Food Microbiol. 45:205-210. [DOI] [PubMed] [Google Scholar]
- 8.Doran, J. L., S. K. Collinson, S. C. Clouthier, T. A. Cebula, W. H. Koch, J. Burian, P. A. Banser, E. C. Todd, and W. W. Kay. 1996. Diagnostic potential of sefA DNA probes to Salmonella enteritidis and certain other O-serogroup D1 Salmonella serovars. Mol. Cell. Probes 10:233-246. [DOI] [PubMed] [Google Scholar]
- 9.Egygor, A., K. T. Carli, and C. B. Unal. 2002. Implementation of real-time PCR to tetrathionate broth enrichment step of Salmonella detection in poultry. Lett. Appl. Microbiol. 34:37-41. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 11.Hargis, B. M., D. J. Caldwell, R. L. Brewer, D. E. Corrier, and J. R. Deloach. 1995. Evaluation of the chicken crop as a source of Salmonella contamination for broiler carcasses. Poult. Sci. 74:1548-1552. [DOI] [PubMed] [Google Scholar]
- 12.Hofstra, H., J. M. van der Vossen, and J. van der Plas. 1994. Microbes in food processing technology. FEMS Microbiol. Rev. 15:175-183. [DOI] [PubMed] [Google Scholar]
- 13.Hoorfar, J., P. Ahrens, and P. Rådström. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser, K., M. Rabodonirina, and S. Picot. 2001. Real time quantitative PCR and RT-PCR for analysis of Pneumocystis carinii hominis. J. Microbiol. Methods 45:113-118. [DOI] [PubMed] [Google Scholar]
- 15.Lantz, P. G., B. Hahn Hagerdal, and P. Rådström. 1994. Sample preparation methods in PCR-based detection of food pathogens. Trends Food Sci. Technol. 5:384-389. [Google Scholar]
- 16.Li, X., N. Boudjellab, and X. Zhao. 2000. Combined PCR and slot blot assay for detection of Salmonella and Listeria monocytogenes. Int. J. Food Microbiol. 56:167-177. [DOI] [PubMed] [Google Scholar]
- 17.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meer, R. R., and D. L. Park. 1995. Immunochemical detection methods for Salmonella spp., Escherichia coli O157:H7, and Listeria monocytogenes in foods. Rev. Environ. Contam. Toxicol. 142:1-12. [DOI] [PubMed] [Google Scholar]
- 19.Olsen, S. J., L. C. MacKinnon, J. S. Goulding, N. H. Bean, and L. Slutsker. 2000. Surveillance for foodborne-disease outbreaks—United States, 1993-1997. Morb. Mortal. Wkly. Rep. 49:1-62. [PubMed] [Google Scholar]
- 20.Rabsch, W., H. Tschäpe, and A. J. Bäumler. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3:237-247. [DOI] [PubMed] [Google Scholar]
- 21.Rijpens, N., L. Herman, F. Vereecken, G. Jannes, J. De Smedt, and L. De Zutter. 1999. Rapid detection of stressed Salmonella spp. in dairy and egg products using immunomagnetic separation and PCR. Int. J. Food Microbiol. 46:37-44. [DOI] [PubMed] [Google Scholar]
- 22.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1997. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245:154-160. [DOI] [PubMed] [Google Scholar]
- 23.Romero, C., and I. Lopez-Goñi. 1999. Improved method for purification of bacterial DNA from bovine milk for detection of Brucella spp. by PCR. Appl. Environ. Microbiol. 65:3735-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruzsovics, A., B. Molnar, Z. Unger, Z. Tulassay, and L. Pronai. 2001. Determination of Helicobacter pylori cagA, vacA genotypes with real-time PCR melting curve analysis. J. Physiol. Paris 95: 369-377. [DOI] [PubMed] [Google Scholar]
- 25.Scheu, P. M., K. Berghof, and U. Stahl. 1998. Detection of pathogenic and spoilage microorganisms in food with the polymerase chain reaction. Food Microbiol. 15:13-31. [Google Scholar]
- 26.Scuderi, G., M. Fantasia, E. Filetici, and M. P. Anastasio. 1996. Foodborne outbreaks caused by salmonella in Italy, 1991-4. Epidemiol. Infect. 116:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scuderi, G., and S. Gabriella. 2000. A review of the salmonellosis surveillance systems in Italy: evolution during the course of time within the international framework. Eur. J. Epidemiol. 16:861-868. [DOI] [PubMed] [Google Scholar]
- 28.Singer, V. L., T. E. Lawlor, and S. Yue. 1999. Comparison of SYBR Green I nucleic acid gel stain mutagenicity and ethidium bromide mutagenicity in the Salmonella/mammalian microsome reverse mutation assay (Ames test). Mutat. Res. 439:37-47. [DOI] [PubMed] [Google Scholar]
- 29.Suzuky, S. 1994. Pathogenicity of Salmonella enteritidis in poultry. Int. J. Food Microbiol. 21:89-105. [DOI] [PubMed] [Google Scholar]
- 30.Szabo, E. A., and B. M. Mackey. 1999. Detection of Salmonella enteritidis by reverse transcription-polymerase chain reaction (PCR). Int. J. Food Microbiol. 51:113-122. [DOI] [PubMed] [Google Scholar]
- 31.Turcotte, C., and M. J. Woodward. 1993. Cloning, DNA nucleotide sequence and distribution of the gene encoding the SEF14 fimbrial antigen of Salmonella enteritidis. J. Gen. Microbiol. 139:1477-1485. [DOI] [PubMed] [Google Scholar]
- 32.Van de Giessen, A. W., A. J. H. A. Ament, and S. H. W. Notermans. 1994. Intervention strategies for Salmonella enteritidis in poultry flocks: a basic approach. Int. J. Food Microbiol. 21:145-154. [DOI] [PubMed] [Google Scholar]
- 33.Vaneechoutte, M., and J. Van Eldere. 1997. The possibilities and limitations of nucleic acid amplification technology in diagnostic microbiology. J. Med. Microbiol. 46:188-194. [DOI] [PubMed] [Google Scholar]
- 34.Vishnubhatla, A., D. Y. C. Fung, R. D. Oberst, M. P. Hays, T. G. Nagaraja, and S. J. Flood. 2000. Rapid 5′ nuclease (TaqMan) assay for detection of virulent strains of Yersinia enterocolitica. Appl. Environ. Microbiol. 66:4131-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wawerla, M., A. Stolle, B. Schalch, and H. Eisgruber. 1999. Impedance microbiology: applications in food hygiene. J. Food Prot. 62:1488-1496. [DOI] [PubMed] [Google Scholar]
- 36.Wilhelm, J., M. Hahn, and A. Pingoud. 2000. Influence of DNA target melting behavior on real-time PCR quantification. Clin. Chem. 46:1738-1743. [PubMed] [Google Scholar]
- 37.Woodward, M. J., and S. E. Kirwan. 1996. Detection of Salmonella enteritidis in eggs by the polymerase chain reaction. Vet. Rec. 138:411-413. [DOI] [PubMed] [Google Scholar]
- 38.World Health Organization. 1990. Report on WHO consultation on salmonellosis control in agriculture, Orvieto Italy, 9-12 April 1990. VPH/SCA/90. World Health Organization, Geneva, Switzerland.