Abstract
Biosurfactants are a unique class of compounds that have been shown to have a variety of potential applications in the remediation of organic- and metal-contaminated sites, in the enhanced transport of bacteria, in enhanced oil recovery, as cosmetic additives, and in biological control. However, little is known about the distribution of biosurfactant-producing bacteria in the environment. The goal of this study was to determine how common culturable surfactant-producing bacteria are in undisturbed and contaminated sites. A series of 20 contaminated (i.e., with metals and/or hydrocarbons) and undisturbed soils were collected and plated on R2A agar. The 1,305 colonies obtained were screened for biosurfactant production in mineral salts medium containing 2% glucose. Forty-five of the isolates were positive for biosurfactant production, representing most of the soils tested. The 45 isolates were grouped by using repetitive extragenic palindromic (REP)-PCR analysis, which yielded 16 unique isolates. Phylogenetic relationships were determined by comparing the 16S rRNA gene sequence of each unique isolate with known sequences, revealing one new biosurfactant-producing microbe, a Flavobacterium sp. Sequencing results indicated only 10 unique isolates (in comparison to the REP analysis, which indicated 16 unique isolates). Surface tension results demonstrated that isolates that were similar according to sequence analysis but unique according to REP analysis in fact produced different surfactant mixtures under identical growth conditions. These results suggest that the 16S rRNA gene database commonly used for determining phylogenetic relationships may miss diversity in microbial products (e.g., biosurfactants and antibiotics) that are made by closely related isolates. In summary, biosurfactant-producing microorganisms were found in most soils even by using a relatively limited screening assay. Distribution was dependent on soil conditions, with gram-positive biosurfactant-producing isolates tending to be from heavy metal-contaminated or uncontaminated soils and gram-negative isolates tending to be from hydrocarbon-contaminated or cocontaminated soils.
Biosurfactants are unique amphipathic molecules with properties that have been explored for a variety of industrial and bioremediation applications (4, 6, 19, 28, 37, 47). From a clinical perspective, it is well known that some biosurfactants have antibiotic activity (5, 6, 60) and that at least one biosurfactant, rhamnolipid produced by Pseudomonas aeruginosa, has a role in the pathogenesis of this opportunistic pathogen (50). Recently, several groups have presented intriguing data suggesting that biosurfactants are important for microbial growth and survival in the environment. For example, surfactin production is necessary for fruiting body formation by Bacillus subtilis (9). Rhamnolipid is necessary for normal biofilm formation by P. aeruginosa (16, 45).
Despite this work, our understanding of biosurfactants as a class of molecules remains limited. This is partially because the present body of knowledge has been developed around a relatively small number of well-characterized biosurfactants. Contributing to this is the lack of a concerted effort to perform a comprehensive screening for biosurfactants and the microorganisms that produce them. Such an effort is hampered by the fact that common genes or regulatory pathways do not exist among the different types of biosurfactant producers. Thus, molecular approaches alone are not useful in screening for biosurfactant producers; instead, screening must be done by using an activity measurement such as surface tension analysis. Surface tension is a parameter that is commonly used to describe the effectiveness of a surfactant. Our work with biosurfactants has indicated that (i) under identical growth conditions, the same amount and type of biosurfactant are produced and this is reflected in an identical surface tension measurement; (ii) a single isolate often generates chemical variations of the same surfactant, resulting in the production of a surfactant mixture with an associated characteristic surface tension (i.e., closely related isolates may produce mixtures with slightly different homologues or with different ratios of the same homologues, and again, these will each have characteristic surface tension values); and (iii) if two different organisms produce surfactants under identical growth conditions but with differing surface tension, then the assumption can be made that the surfactants produced are different. For different genera or different species, this may mean a totally different chemical structure. For closely related isolates, this means that while the surfactant type is the same, there are subtle chemical differences in the molecules produced. It is important to point out that even small differences in the structure of a surfactant can have profound effects on its function and its potential industrial applications.
A further difficulty with screening for biosurfactant producers is that biosurfactant production depends both on the type of carbon source present and on the types and amounts of other nutrients in the screening medium (1, 17, 18, 36, 46, 56). Thus, the screening medium used will influence whether or not surfactant is produced and further will influence the makeup of the mixture produced and the amount produced.
The present work is an initial attempt to systematically screen for biosurfactant-producing microorganisms and to evaluate their phylogenetic diversity. Twenty soil samples were collected from both contaminated sites (i.e., with organics and/or metals) and undisturbed sites in the arid southwestern United States. The soil samples were screened for biosurfactant producers by using a combination of cultural and molecular methods. Putative biosurfactant-producing isolates were cultured from the various soils and grouped using repetitive extragenic palindromic (REP)-PCR fingerprinting. A phylogenetic analysis of each unique isolate indicated by REP-PCR was then performed by PCR amplification of the 16S rRNA gene (16S-PCR) followed by sequencing. A phylogenetic tree was constructed by combining the results of the present study with a survey of biosurfactant-producing eubacteria and archaea found in the literature to examine the diversity of biosurfactant producers.
MATERIALS AND METHODS
Soils.
Twenty soil samples were collected and stored at 4°C (Table 1). Soils were classified as uncontaminated, contaminated with organics (petroleum), contaminated with metals, or cocontaminated with organics and metals.
TABLE 1.
Soil characteristics and screening results
| Soil type/location (sample no.) | Contamination | Texture | TOCa or OMb (%) | CFU/g of soilc | No. of isolates screenedd | Biosurfactant- producing isolatese |
|---|---|---|---|---|---|---|
| Undisturbed | ||||||
| Tucson, Ariz. (1) | None | Sandy | 0.43a | (9.9 ± 3.8) × 105 | 88 | 2 |
| Tucson, Ariz. (2) | None | Sandy loam | 4.59a | (7.1 ± 57.7) × 106 | 72 | 1 |
| Tucson, Ariz. (3) | None | Loamy sand | 1.27a | (3.4 ± 38.0) × 107 | 77 | 1 |
| Tucson, Ariz. (4) | None | Sandy loam | 0.12a | (5.3 ± 34.2) × 106 | 82 | 4 |
| Elgin, Ariz. (5) | None | Loam | 1.88a | (2.9 ± 11.4) × 106 | 72 | 1 |
| Tucson, Ariz. (6) | None | Sandy loam | 0.2a | (1.2 ± 17.4) × 106 | 75 | 4 |
| Hydrocarbon contaminated | ||||||
| Tucson, Ariz. (7) | Motor oil | NDh | ND | (1.6 ± 23.0) × 107 | 91 | 4 |
| Blythe, Calif. (8) | Waste oil | ND | ND | (8.1 ± 64.3) × 107 | 81 | 0 |
| Layton, Utah (9) | JP4,f PAHg | Fine to coarse sand w/ interbedded gravel | ND | (1.5 ± 22.0) × 106 | 42 | 0 |
| Layton, Utah (10) | JP4, chlorinated solvents, PAH | Fine to coarse sand w/ interbedded gravel | ND | (1.6 ± 42.8) × 107 | 36 | 0 |
| Tucson, Ariz. (11) | Gasoline | Silty clay | ND | (4.9 ± 81.3) × 106 | 43 | 0 |
| Willcox, Ariz. (12) | Mineral oil | Sandy clay | ND | (4.4 ± 39.6) × 106 | 64 | 3 |
| Metal contaminated | ||||||
| Green Valley, Ariz. (13) | Pb, Cd | Sandy loam | 0.55a | (4.1 ± 40.5) × 105 | 71 | 7 |
| Wickenburg, Ariz. (14) | Cr, Hg | Sandy loam | 0.30b | (3.7 ± 51.6) × 106 | 54 | 0 |
| Tucson, Ariz. (15) | Pb | Sandy loam | ND | (2.0 ± 30.1) × 107 | 47 | 0 |
| Tucson, Ariz. (16) | Pb, Cd, Cr | Sandy loam | ND | (3.6 ± 33.1) × 107 | 53 | 0 |
| Bellemount, Ariz. (17) | As, Ba, Be, Cd, Cr, Pb | Sandy loam | 3.78a | (2.0 ± 21.9) × 106 | 54 | 1 |
| Cocontaminated | ||||||
| Tucson, Ariz. (18) | Pb, waste oil | Sandy loam | ND | (8.9 ± 2.5) × 106 | 83 | 2 |
| San Manuel, Ariz. (19) | Flue dust, hydrocarbon waste | ND | ND | (3.4 ± 26.1) × 106 | 63 | 1 |
| San Manuel, Ariz. (20) | Flue dust, diesel fuel | Alluvial fill | ND | (7.1 ± 75.7) × 106 | 57 | 14 |
TOC, total organic carbon.
OM, organic matter.
Plate counts were not significantly different in the day 3, 7, 14, and 21 soil slurries.
Total number of colonies screened from day 3, 7, 14, and 21 soil slurries.
Total number of biosurfactant-producing isolates obtained from the day 3, 7, 14, and 21 soil slurries.
JP4, jet fuel.
PAH, polyaromatic hydrocarbon.
ND, not determined.
Culturable screening for biosurfactant-producing isolates.
Soils were screened for biosurfactant-producing isolates by using the following procedure. A 5-g sample of each soil was placed into a 250-ml flask containing 50 ml of tap water and incubated at 23°C on a shaker at 200 rpm for 21 days. On days 3, 7, 14, and 21, a sample from each soil slurry was serially diluted, plated on R2A agar (Becton Dickinson Company, Cockeysville, Md.), and incubated for 1 week. After incubation, plates were enumerated, and morphologically different bacteria were selected for biosurfactant screening (approximately 15 to 30 isolates per sampling time). Isolated colonies were inoculated into 5-ml mineral salts medium (MSM) containing 2% glucose as the sole carbon and energy source. The MSM was a mixture of solution A and solution B. Solution A contained (per liter) 2.5 g of NaNO3, 0.4 g of MgSO47H2O, 1.0 g of NaCl, 1.0 g of KCl, 0.05 g of CaCl22H2O, and 10 ml of concentrated phosphoric acid (85%). This solution was adjusted to pH 7.2 with KOH pellets. Solution B contained (per liter) 0.5 g of FeSO47H2O, 1.5 g of ZnSO47H2O, 1.5 g of MnSO4H2O, 0.3 g of K3BO3, 0.15 g of CuSO45H2O, and 0.1 g of Na2MoO42H2O. One milliliter of solution B was added to 1,000 ml of solution A to form the MSM. The broth cultures were incubated with shaking (200 rpm) for 7 to 9 days at 23°C. The cell suspensions were then tested for the presence of surfactant by using the qualitative drop-collapse method (7).
Qualitative measurement of surface tension.
Qualitative drop-collapse tests were performed in the polystyrene lid of a 96-microwell 12.7- by 8.5-cm plate (VWR, Cerritos, Calif., or Biolog, Hayward, Calif.) (7). The lids have 96 circular wells (internal diameter, 8 mm). A thin coat of 10W-40 oil (Pennzoil, Oil City, Pa.; 1.8 μl/well) was applied to each well. The coated wells were equilibrated for 24 h at 23°C, and then a 5-μl aliquot of supernatant was delivered into the center of each well. If the drop remained beaded, the result was scored as negative. If the drop spread and collapsed, the result was scored as positive for the presence of biosurfactant. Cultures were tested in triplicate. The MSM alone had a negative drop-collapse test.
Quantitative measurement of surface tension.
All isolates that tested positive in the drop-collapse test and were identified as unique by REP-PCR analysis were then tested quantitatively for biosurfactant production with the du Nouy ring method (7). The isolates were grown in 25 ml of MSM amended with 2% glucose. Cell suspensions were centrifuged (10,000 × g; Beckman [Palo Alto, Calif.] model J2-21 centrifuge), and the cell-free supernatant was placed into a clean glass 50-ml beaker. A Surface Tensiomat (model 21; Fisher Scientific, Pittsburgh, Pa.) was used to measure the surface tension. Between each pair of measurements, the platinum wire ring used to measure surface tension was rinsed three times with water, three times with acetone, and then allowed to dry.
Analysis of biosurfactant producers.
A total of 1,305 isolates were screened, and of these, 45 isolates demonstrated the ability to produce biosurfactants. The 45 biosurfactant-producing isolates were Gram stained and fingerprinted by using REP analysis as previously described by Versalovic et al. (58) with modifications. For all PCRs, cell lysates were prepared as follows. One milliliter of bacterial culture was centrifuged at 14,000 × g for 2 min, decanted, and resuspended in 1 ml of distilled water. One hundred microliters of resuspended cells was heat lysed at 98°C for 10 min in a GeneAmp 2400 PCR system thermal cycler (Perkin-Elmer, Foster City, Calif.). Raw extracts were used in subsequent PCRs without further purification. For REP analysis, a 25-μl PCR mixture contained a 0.5 μM concentration of each primer (REP1R-1 5′ III ICG ICG ICA TCI GGC 3′; REP2-1 5′ ICG ICT TAT CIG GCC TAC 3′), a 1.25 mM concentration of each deoxynucleoside triphosphate (dNTP), 1× buffer consisting of 10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2 (pH 8.9), 5.0% dimethyl sulfoxide (DMSO), 2.5 U of Taq DNA polymerase (Roche, Indianapolis, Ind.), and 2.5 μl of cell lysate. The PCR program used for amplification was 95°C for 5 min, followed by 35 cycles consisting of 95°C for 0.5 min, 45°C for 0.5 min, and 72°C for 4 min and a single final extension step consisting of 72°C for 16 min. Fingerprints were visualized on a 3.0% agarose gel (NuSieve 3:1 agarose; FMC BioProducts, Rockland, Maine).
Phylogenetic analysis of each isolate that showed a unique REP fingerprint was performed by using 16S-PCR. The forward primer was 27f (5′AGA GTT TGA TCC TGG CTC AG 3′) and the reverse primer was 1492r (5′ TAC GGT TAC CTT GTT ACG ACT T 3′) (34, 61). Each 50-μl PCR mixture contained a 0.5 μM concentration of each primer, a 0.2 mM concentration of each dNTP, 1× buffer (consisting of 10 mM Tris-HCl, 50 mM KCl, 2.0 mM MgCl2 [pH 8.3]), 5.0% DMSO, 1.0 U of Taq DNA polymerase (Roche), and 5.0 μl of cell lysate. The PCR program used for amplification was 95°C for 5 min followed by 30 cycles consisting of 94°C for 1 min, 63°C for 1 min, and 72°C for 1 min 15 s, and a single final extension step consisting of 72°C for 5 min. The entire 1,500-bp product was visualized on a 1.0% agarose gel (SeaKem LE agarose; FMC BioProducts). The PCR product was subsequently purified by using a Qiaquick PCR Purification Kit (Qiagen, Valencia, Calif.) and sequenced at the Genomic Analysis and Technology Core Facility (Division of Biotechnology, University of Arizona Research Laboratories, Tucson). Sequence analysis resolved two approximately 500-bp regions of each 16S rRNA gene corresponding to the 5′ and 3′ ends. Thus, internal primers were designed to sequence the remaining internal 500 bp of the gene. Primers were designed by using OLIGO primer analysis software, version 6.1 (Molecular Biology Insights, Inc., Cascade, Colo.). Separate internal primers were designed for gram-positive isolates, the Flavobacterium isolate, the Pseudomonas sp. isolates, and the P. aeruginosa isolates. The gram-positive internal primer set was GP404 forward (5′ TAG GGA AGA ACA AGT ACC 3′) and GP1048 reverse (5′ ACT TAA CCC AAC ATC TCA C 3′). The Flavobacterium internal primer set was MTN11 forward (5′ CGG GAA TAA ACC TCT TTA 3′) and MTN11 reverse (5′ CTG GCA ACT AAA CAT AGG 3′). The Pseudomonas sp. internal primers were STB17 forward (5′ GGA GGA AGG GTT GTA GAT 3′) and STB17 reverse (5′ CGT GCT GGT AAC TAA GGA 3′) and the P. aeruginosa internal primers were BHP7-6 forward (5′ GAA GGG CAG TAA GTT AAT AC 3′) and BHP7-6 reverse (5′ GAG GTG CTG GTA ACT AAG 3′).
Sequencher software version 3.0 was used to align and to obtain a consensus sequence for each isolate (Gene Codes Corporation, Ann Arbor, Mich.). A representative consensus sequence for each of the 16 unique isolates was compared to known sequences using the BLAST program (2). Finally, the 16S rRNA gene nucleotide sequence of each unique isolate was submitted to the National Center for Biotechnology Information (NCBI) database and assigned an accession number.
Construction of the phylogenetic tree.
Sequences representing a broad range of biosurfactant-producing microorganisms from this study and from the literature were retrieved from the NCBI 16S rRNA gene database and downloaded in FASTA format. Sequences were aligned using ClustalX version 1.81 (54). The regions of the 16S rRNA gene sequences before the 125-bp position and after the 1,550-bp position were excluded from the alignment analysis. Lack of sequence data in these regions caused the alignment software to create large gap regions in the sequences, which would have created biases in subsequent phylogenetics analysis. Finally, the tree was constructed with PAUP* beta version 4.0 software by using the neighbor-joining method with the Hasegawa-Kishino-Yano (HKY85) substitution model (53).
Detection of the rhlB gene.
Rhamnolipid is a biosurfactant known to be produced only by P. aeruginosa. The rhamnosyl transferase I (rhlB) gene, part of the rhamnolipid biosynthesis pathway, is unique to P. aeruginosa. Detection of this gene in DNA extracts was used to confirm the putative homology of isolates with P. aeruginosa, according to the 16S rRNA gene sequence, and to distinguish them from other Pseudomonas species. Template DNA extracts were prepared as above. A PCR was performed to determine the presence of rhlB in the extracts by using the primers kpd1 and kpd2. The forward primer, kpd1, is homologous to the 1,030- to 1,048-bp region of the rhlB gene and has the following sequence: 5′ GCC CAC GAC CAG TTC GAC 3′. The reverse primer, kpd2, is homologous to the 1,256- to 1,238-bp region of the rhlB gene, and has the following sequence: 5′ CAT CCC CCT CCC TAT GAC 3′. The PCRs were 50 μl in volume and contained an optimized PCR buffer (10 mM Tris-HCl, 50 mM KCl, 1 mM MgCl2, pH 9.2), a 0.5 μM concentration of each primer, a 0.2 mM concentration of each dNTP, 5% DMSO, 2.5 U of Taq DNA polymerase (Roche), and 5 μl of template DNA. The reactions were exposed to 94°C for 2 min, then to 30 cycles of 94°C for 15 s, 54°C for 15 s, and 72°C for 15 s, and finally to a final extension of 72°C for 2 min in a GeneAmp 2400 PCR system thermal cycler (Perkin-Elmer). The PCR products were visualized by electrophoresis through a 2.0% SeaKem LE agarose gel (FMC BioProducts). Resolution of the expected 226-bp PCR product was considered indicative of the presence of the rhlB gene in the template DNA and confirmed that the DNA came from P. aeruginosa.
Thin-layer chromatography.
Rhamnolipid production was confirmed by thin-layer chromatography as described previously (62). Briefly, the solvent system used was chloroform-methanol-water (65:25:4). After development of the thin-layer chromatography plate, it was sprayed with anthrone reagent to visualize rhamnolipid. In this system, monorhamnolipid has an Rf value of 0.72 and dirhamnolipid has an Rf value of 0.46.
RESULTS
The twenty soil samples screened for biosurfactant producers were collected primarily from arid soils in southern Arizona. Most had a sandy loam texture, and total organic carbon content ranged from 0.12 to 4.59% (Table 1). The soils had culturable populations ranging from (4.1 ± 0.41) × 105 to (8.1 ± 0.64) × 107 CFU/g of dry soil (Table 1). The initial screening on R2A yielded a total of 1,305 isolates, which were grown in MSM-glucose broth for a week and then tested qualitatively for biosurfactant production with the drop-collapse test. This resulted in 45 putative biosurfactant-producing isolates or a total of 3.4% of the isolates tested under these screening conditions. Of the 20 soils tested, 13 contained biosurfactant producers, including all six undisturbed soils and all three cocontaminated soils. However, only two of the six hydrocarbon-contaminated and two of the five metal-contaminated soils contained biosurfactant-producers.
Of the 45 putative biosurfactant producers, 47% were gram negative and 53% were gram positive (Table 2). Upon examining the types of soils and their contaminants, the following pattern was revealed: gram-positive isolates dominated in the undisturbed and metal-contaminated soils (95%), and gram-negative isolates dominated in the hydrocarbon-contaminated and cocontaminated soils (83%). This distribution may represent the ability of the microorganisms to survive in these soils or may be a response to the type of contaminant present. Further investigation is needed to determine whether this pattern holds for soils from nonarid regions.
TABLE 2.
Distribution of positive biosurfactant-producing isolates
| Soils (no. of soils tested) | No. of biosurfactant-producing isolatesa | Gram negative | Gram positive |
|---|---|---|---|
| Undisturbed (6) | 13 | 1 | 12 |
| Hydrocarbon contaminated (6) | 7 | 6 | 1 |
| Metal contaminated (5) | 8 | 0 | 8 |
| Cocontaminated (3) | 17 | 14 | 3 |
This represents the total number of biosurfactant-producing isolates obtained from each group of soils.
REP-PCR grouped the 45 putative biosurfactant producers into 16 unique fingerprint patterns. These 16 isolates were further characterized in two ways. First, the surface tension of culture supernatants for each isolate grown under identical conditions was measured, with values ranging from 27.3 ± 0.3 to 49.4 ± 0.5 (Table 3). Second, the 16 isolates were identified based on their 16S rRNA gene sequences and classified into three genera, Pseudomonas, Bacillus, and Flavobacterium, with homologies of 97% or greater in each case suggesting good matches (Table 3).
TABLE 3.
Biosurfactant-producing isolates with unique REP fingerprints
| Isolate (accession no.) | Nearest relative | Percent sequence identity, accession no. for nearest relative | Surface tension (mN/m) | Soil sample no.a |
|---|---|---|---|---|
| GA1-5 (AY162126) | B. subtilis | 99, AB018484 | 49.4 ± 0.5 | 6 |
| HAZ2 (AY162127) | B. subtilis | 99, AB018484 | 39.2 ± 0.4 | 13 |
| HAZ14 (AY162128) | B. subtilis | 99, AB018484 | 33.3 ± 0.3 | 13 |
| WP1-21 (AY162129) | B. subtilis | 99, AB018484 | 33.9 ± 0.2 | 18 |
| STB29 (AY162130) | B. subtilis | 100, AY030331, AY030330, AJ276351, Z99104 | 31.3 ± 0.4 | 7 |
| BHP6-1 (AY162131) | B. subtilis | 99, AY030331, AY030330, AJ276351, Z991044 | 31.4 ± 0.2 | 19 |
| MA12 (AY162132) | B. subtilis | 99, AB018486 | 31.3 ± 0.2 | 3 |
| BZ15 (AY162133) | B. subtilis | 99, AF318900, AB018486 | 33.1 ± 0.4 | 4, 13, 18 |
| GA1-17 (AY162134) | B. licheniformis | 99, AY030335 | 39.6 ± 0.6 | 1, 4, 5, 6, 17 |
| HAZ6-17 (AY162135) | B. licheniformis | 99, AY030335 | 42.6 ± 0.5 | 13 |
| GA8 (AY162136) | B. licheniformis | 99, AB039328, X68416 | 42.6 ± 0.2 | 6 |
| MTN11 (AY162137) | Flavobacterium sp. | 97, AF388029 | 40.6 ± 0.0 | 2 |
| MO2b (AY162138) | P. aeruginosa | 99, AF331663, AF227866, AF094718, AF094713, AE004844 | 32.5 ± 0.2 | 12 |
| BHP7-6b/AY162139 | P. aeruginosa | 99, AF331663, AF227866, AF094718, AF094713, AE004844 | 31.8 ± 0.2 | 20 |
| STB17c/AY162140 | Pseudomonas sp. | 99, AY029759 | 28.7 ± 0.4 | 7 |
| BHP7-11c/AY162141 | Pseudomonas sp. | 99, AF105389 | 27.3 ± 0.3 | 20 |
The soil sample numbers listed for each isolate indicate from which soil or soils the isolate was obtained.
Isolates were positive for the rhlB gene and for rhamnolipid production.
Isolates were negative for the rhlB gene and for rhamnolipid production.
Four Pseudomonas isolates were obtained during the soil screening. Two were phylogenetically positioned as P. aeruginosa, while two were positioned only to the genus level. All four were subjected to PCR analysis for the rhlB gene. The P. aeruginosa isolates tested, MO2 and BHP7-6, were positive for the rhlB gene, while the two Pseudomonas spp. were not. Rhamnolipid production by MO2 and BHP7-6 was confirmed by thin-layer chromatography analysis.
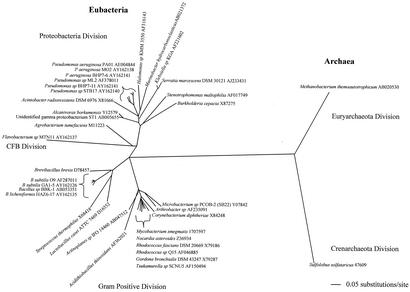
The data from this study were combined with known biosurfactant producers from the literature and used to construct a phylogenetic tree. As shown in Fig. 1, biosurfactant-producing organisms were found in at least three of the major divisions of the Eubacteria, including Proteobacteria, Firmicutes (gram positive), and Cytophaga-Flexibacter-Bacteroides, as well as the two major divisions of the Archae, i.e., Crenarchaeota and Euryarchaeota. Isolates from this study were distributed between the Proteobacteria, Firmicutes, and Cytophaga-Flexibacter-Bacteroides divisions.
FIG. 1.
Phylogenetic tree was based on the 16S rRNA gene sequence from microorganisms representing biosurfactant producers isolated in this study as well as known biosurfactant producers from the literature. The unrooted tree was created by using the neighbor-joining method. CFB, Cytophaga-Flexibacter-Bacteroides.
Finally, both REP-PCR and sequencing analysis were performed on two closely related biosurfactant-producing isolates from our laboratory to determine the level of discrimination of these two analyses for unique biosurfactant products. The isolates were P. aeruginosa ATCC 9027 and P. aeruginosa IGB83, both of which produce rhamnolipid biosurfactants. These isolates are known to produce chemically distinct rhamnolipids, with ATCC 9027 producing only monorhamnolipid and IGB83 producing a mixture of mono- and dirhamnolipid. Results showed that the REP fingerprint patterns were different for these two strains but that the 16S rRNA gene sequences were identical (data not shown).
DISCUSSION
As has been observed for other natural products such as antibiotics, phylogenetic analysis revealed that great diversity exists among biosurfactant-producing microorganisms, suggesting that biosurfactant production is an important survival tool (Fig. 1). Since biosynthesis genes for different surfactant types are entirely unrelated (6, 27, 52), biosurfactant production appears to have evolved in an independent yet parallel fashion. For example, rhamnolipid (P. aeruginosa) biosynthesis genes are completely different from surfactin (B. subtilis) biosynthesis genes, yet these species both produce highly effective surfactant molecules. While the significance of these molecules to their producing species is not completely known (similar to antibiotics), a majority of the biosynthesis genes are conserved at the species level, thus indicating their importance.
It is currently known that the type of biosurfactant made is dictated by the producing microorganism. One major class of biosurfactants is the glycolipids, which includes rhamnolipids, trehalose lipids, and sophorose lipids. Rhamnolipids are produced only by P. aeruginosa (30, 33); trehalose lipids are produced only by a number of closely related genera, including Rhodococcus, Nocardia, Corynebacterium, Tsukamurella, Gordonia, Mycobacterium, and Arthrobacter, all belonging to the Firmicutes division (3, 12, 31, 44); and sophorose lipids are produced by several species of Candida (14, 15, 22). A newly identified glycolipid group called mannosylerthritol lipids are produced by Candida and Ustilago maydis (25, 26, 51). A second major class of biosurfactant is the lipoproteins, which includes surfactin, iturin, fengycin, and lichenysin, which are produced only by Bacillus sp. (24, 42, 57). Other genera that produce lipoproteins include Actinoplanes, Arthrobacter, Pseudomonas, and Serratia (23, 32, 35, 38, 39, 59). Polymeric biosurfactant producers have been isolated from Eubacteria, Eucaryote, and Archaea and include the following genera: Acinetobacter, Bacillus, Candida, Halomonas, Methanobacterium, Phormidium, Pseudomonas, Saccharomyces, and Sulfolobus (8, 10, 11, 13, 20, 43, 48, 49, 55).
In this study, three biosurfactant-producing genera, Pseudomonas, Bacillus, and Flavobacterium, were obtained from the screened soils. Of these, the isolation of Flavobacterium MTN11 as a surfactant producer is novel, and the biosurfactant is currently being purified for structure elucidation. The four Pseudomonas isolates obtained were screened for rhamnolipid production by using primers for the rhlB gene. Only the two P. aeruginosa isolates were positive for the rhlB gene, and rhamnolipid production was confirmed by thin-layer chromatography analysis. Other Pseudomonas species have been reported to produce a variety of lipoproteins including viscosin, tensin, syringomycin, and syringopeptin, as well as polymeric biosurfactants such as Biosur-Pm and PM factor (10, 21, 23, 32, 39, 41, 43). The remaining 11 isolates obtained in this study were Bacillus spp. This genus has been reported to produce a variety of lipoprotein surfactants. B. subtilis usually produces a mixture composed of surfactin, iturin, and fengycin, each of which has several isoforms (29, 42). Bacillus licheniformis produces lichenysin, which also has a number of isoforms (24).
It is possible that other biosurfactant-producing populations were present but not enriched by the screening conditions used for the soils tested in this study. The screening method employed used glucose as the sole carbon and energy source. Glucose was selected for the following reasons: (i) it is known to support the production of a variety of biosurfactants, and (ii) there is interest in stimulating in situ production of biosurfactants for remediation applications, and glucose is a nontoxic carbon source appropriate for this purpose. However, it must be emphasized that biosurfactant production is dependent on the carbon source and media selected. Thus, if another medium and carbon source were chosen, it is probable that a different population of biosurfactant-producers would have been enriched. Despite the limitations of the screening method used, a diverse group of biosurfactant-producing organisms was obtained, including a novel biosurfactant producer, Flavobacterium sp. strain MTN11. These results suggest that a more exhaustive screening may yield other new biosurfactant-producing microorganisms.
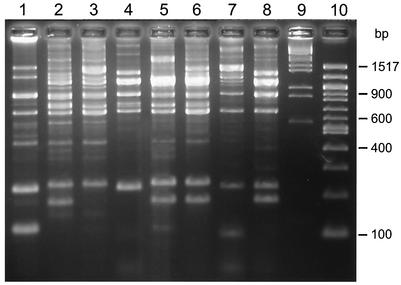
One fascinating aspect of this work that requires further exploration is that several of the 16 isolates that were differentiated by REP-PCR had identical NCBI database matches. For example, Bacillus subtilis GA1-5, HAZ2, HAZ14, and WP1-21 had 99% sequence identity with the NCBI database isolate with accession number AB018484. This does not mean that these four sequences are identical; indeed, they vary from each other by up to 8 bp. For instance, when comparing HAZ2 and GA1-5, there is a difference of only 1 bp in the 1,466 bp that were sequenced. Thus, if all 45 isolates were merely sequenced instead of first grouping them by REP-PCR, then only 10 unique isolates would have been identified rather than 16. So the use of REP-PCR in this case was critical in identifying the uniqueness of these two isolates (as shown in Fig. 2, the REP-PCR analyses for HAZ2 and GA1-5 have different fingerprinting patterns).
FIG. 2.
REP analysis of all the isolates that matched with B. subtilis. Lane 1, strain MA12; lane 2, strain HAZ2; lane 3, strain HAZ14; lane 4, strain WP1-21; lane 5, GA1-5; lane 6, strain BHP6-1; lane 7, strain BZ15; lane 8, strain STB29; lane 9, molecular weight marker III; lane 10, 100-bp marker.
To continue with this example, the surface tension reduction activities of the biosurfactants produced by HAZ2 and GA1-5 were different. The HAZ2 culture supernatant reduced surface tension from 72 mN/m (water) to 39.2 ± 0.4 mN/m, while the GA1-5 culture supernatant reduced surface tension to only 49.4 ± 0.5 mN/m. The difference in these surface tension activities is significant; surface tension reduction to 49 is considered minimally active, while a surface tension reduction to 39 is considered moderately active. Thus, while it is likely that HAZ2 and GA1-5 both produce a lipopeptide surfactant, given their close relationship to B. subtilis, the surfactant or surfactant mixture produced by each of these microorganisms is distinct. As previously explained, it is important that even slight differences in biosurfactant structure or in the mixture of the biosurfactants produced can have great consequences on activity in the environment or in clinical settings or in industrial applications.
To further support this example, two P. aeruginosa strains extensively used in our laboratory were examined. The first is P. aeruginosa ATCC 9027, which produces only monorhamnolipid, and the second is P. aeruginosa IGB83, which produces a mixture of mono- and dirhamnolipid. The physicochemical properties of monorhamnolipid are quite different from those of the mixture of mono- and dirhamnolipid. Specifically, monorhamnolipid is less soluble, sorbs to surfaces more strongly, solubilizes hydrocarbons to a greater extent, and is able to bind cationic metals up to 10 times more strongly (40, 63, 64). Thus, potential industrial applications are very different (6). These two strains were separated based on REP fingerprints but not by their 16S rRNA gene sequences, which were identical. Thus, the REP analysis in this case was critical to distinguishing closely related strains that produce distinctly different surfactant mixtures.
In summary, biosurfactants produced by very similar isolates may have subtle differences that are useful in different applications. It is therefore important that any screening process be discriminatory enough to ensure that even closely related isolates are examined to reveal details about the structural diversity of biosurfactants and likely other natural products as well. The screening process used in this study was a combination of cultural and molecular techniques. Even though the initial screening step employed was limited in scope by the culture medium used, results showed that biosurfactant-producing bacteria are widely distributed in both undisturbed and contaminated soils. In addition, a novel biosurfactant-producing Flavobacterium isolate was obtained.
Acknowledgments
We thank Wendy Moore for her expertise in the phylogenetic tree analysis used to create Fig. 1.
This work was supported in part by grant number 2 P42 ESO4940-11 from the National Institute of Environmental Health Sciences, NIH, and in part by grant CHE-0133237 from the National Science Foundation.
REFERENCES
- 1.Adamczak, M., and W. Bednarski. 2000. Influence of medium composition and aeration on the synthesis of biosurfactants produced by Candida antarctica. Biotechnol. Lett. 22:313-316. [Google Scholar]
- 2.Altschul, S. F., W. Wich, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Asselineau, J., and G. Lanéelle. 1998. Mycobacterial lipids: a historical perspective. Front. Biosci. 3:3164-3174. [DOI] [PubMed] [Google Scholar]
- 4.Banat, I. M., R. S. Makkar, and S. S. Cameotra. 2000. Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 53:495-508. [DOI] [PubMed] [Google Scholar]
- 5.Bechard, J., K. C. Eastwell, P. L. Sholberg, G. Mazza, and B. Skura. 1998. Isolation and partial chemical characterization of an antimicrobial peptide produced by a strain of Bacillus subtilis. J. Agric. Food Chem. 46:5355-5361. [Google Scholar]
- 6.Bodour, A. A., and R. M. Maier. 2002. Biosurfactants: types, screening methods, and applications, p. 750-770. In G. Bitton (ed.), Encyclopedia of environmental microbiology, 1st ed. John Wiley and Sons, Inc., Hoboken, N.J.
- 7.Bodour, A. A., and R. M. Miller-Maier. 1998. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J. Microbiol. Methods 32:273-280. [Google Scholar]
- 8.Bouchotroch, S., E. Quesada, I. Izquierdo, M. Rodriguez, and V. Béjar. 2000. Bacterial exopolysaccharides produced by newly discovered bacteria belonging to the genus Halomonas, isolated from hypersaline habitats in Morocco. J. Ind. Microbiol. Biotechnol. 24:374-378. [Google Scholar]
- 9.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burd, G., and O. P. Ward. 1996. Physicochemical properties of PM-factor, a surface-active agent produced by Pseudomonas marginalis. Can. J. Microbiol. 42:243-251. [DOI] [PubMed] [Google Scholar]
- 11.Cameron, D. R., D. G. Cooper, and R. J. Neufeld. 1988. The mannoprotein of Saccharomyces cerevisiae is an effective bioemulsifier. Appl. Environ. Microbiol. 54:1420-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, K.-S., S.-H. Kim, and T.-H. Lee. 1999. Purification and characterization of biosurfactant from Tsukamurella sp. 26A. J. Microbiol. Biotechnol. 9:32-38. [Google Scholar]
- 13.Cirigliano, M. C., and G. M. Carman. 1985. Purification and characterization of liposan, a bioemulsifier from Candida lipolytica. Appl. Environ. Microbiol. 50:846-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper, D. G., and D. A. Paddock. 1984. Production of a biosurfactant from Torulopsis bombicola. Appl. Environ. Microbiol. 47:173-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutler, A. J., and R. J. Light. 1979. Regulation of hydroxydocosanoic and sophoroside production in Candida bogoriensis by the level of glucose and yeast extract in the growth medium. J. Biol. Chem. 254:1944-1950. [PubMed] [Google Scholar]
- 16.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davila, A.-M., R. Marchal, and J.-P. Vandecasteele. 1997. Sophorose lipid fermentation with differentiated substrate supply for growth and production phases. Appl. Microbiol. Biotechnol. 47:496-501. [Google Scholar]
- 18.Davis, D. A., H. C. Lynch, and J. Varley. 1999. The production of surfactin in batch culture by Bacillus subtilis ATCC 21332 is strongly influenced by the conditions of nitrogen metabolism. Enzyme Microb. Technol. 25:322-329. [Google Scholar]
- 19.Desai, J. D., and I. M. Banat. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fattom, A., and M. Shilo. 1985. Production of emulycan by Phormidium J-1: its activity and function. FEMS Microbiol. Ecol. 31:3-9. [Google Scholar]
- 21.Fogliano, V., M. Gallo, F. Vinale, A. Ritieni, G. Randazzo, M. Greco, R. Lops, and A. Graniti. 1999. Immunological detection of syringeopeptins produced by Pseudomonas syringae pv. lachrymans. Physiol. Mol. Plant Pathol. 55:255-261. [Google Scholar]
- 22.Gorin, P. A. J., J. F. T. Spencer, and A. P. Tulloch. 1961. Hydroxy fatty acid glycosides of sophorose from Torulopsis magnoliae. Can. J. Chem. 39:816-855. [Google Scholar]
- 23.Henriksen, A., U. Anthoni, T. H. Nielsen, J. Sørensen, C. Christophersen, and M. Gajhede. 2000. Cyclic lipoundecapeptide tension from Pseudomonas fluorescens strain 96.578. Acta Crystallogr. C 56:113-115. [DOI] [PubMed] [Google Scholar]
- 24.Jenny, K., O. Käppeli, and A. Fiechter. 1991. Biosurfactants from Bacillus licheniformis: structural analysis and characterization. Appl. Microbiol. Biotechnol. 36:5-13. [DOI] [PubMed] [Google Scholar]
- 25.Kim, H.-S., B.-D. Yoon, D.-H. Choung, H.-M. Oh, T. Katsuragi, and Y. Tani. 1999. Characterization of a biosurfactant, mannosylerythritol lipid produced from Candida sp. SY16. Appl. Microbiol. Biotechnol. 52:713-721. [DOI] [PubMed] [Google Scholar]
- 26.Kitamoto, D., S. Akiba, C. Hioki, and T. Tabuchi. 1990. Extracellular accumulation of mannosylerythritol lipids by a strain of Candida antarctica. Agric. Biol. Chem. 54:31-36. [Google Scholar]
- 27.Konz, D., S. Doekel, and M. A. Marahiel. 1999. Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J. Bacteriol. 181:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosaric, N. 2001. Biosurfactants and their application for soil bioremediation. Food Technol. Biotechnol. 39:295-304. [Google Scholar]
- 29.Kowall, M., J. Vater, B. Kluge, T. Stein, P. Franke, and D. Ziessow. 1998. Separation and characterization of surfactin isoforms produced by Bacillus subtilis OKB 105. J. Colloid Interface Sci. 204:1-8. [DOI] [PubMed] [Google Scholar]
- 30.Lang, S., and D. Wullbrandt. 1999. Rhamnose lipids—biosynthesis, microbial production and application potential. Appl. Microbiol. Biotechnol. 51:22-32. [DOI] [PubMed] [Google Scholar]
- 31.Lang, S., and J. C. Philp. 1998. Surface-active lipids in rhodococci. Antonie Leeuwenhoek 74:59-70. [DOI] [PubMed] [Google Scholar]
- 32.Laycock, M. V., P. D. Hildebrand, P. Thibault, J. A. Walter, and J. L. C. Wright. 1991. Viscosin, a potent peptidolipid biosurfactant and phytopathogenic mediator produced by a pectolytic strain of Pseudomonas fluorescens. J. Agric. Food Chem. 39:483-489. [Google Scholar]
- 33.Maier, R. M., and G. Soberón-Chávez. 2000. Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential environmental applications. Appl. Microbiol. Biotechnol. 54:625-633. [DOI] [PubMed] [Google Scholar]
- 34.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eucaryotic 16S-like rRNA-coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 35.Morikawa, M., H. Daido, T. Takao, S. Murata, Y. Shimonishi, and T. Imanaka. 1993. A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. J. Bacteriol. 175:6459-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulligan, C. N., G. Mahmourides, and B. F. Gibbs. 1989. The influence of phosphate metabolism on biosurfactant production by Pseudomonas aeruginosa. J. Biotechnol. 12:199-210. [Google Scholar]
- 37.Mulligan, C. N., R. N. Yong, and B. F. Gibbs. 1999. On the use of biosurfactants for the removal of heavy metals from oil-contaminated soil. Environ. Prog. 18:50-54. [Google Scholar]
- 38.Nakagawa, Y., and T. Matsuyama. 1993. Chromatographic determination of optical configuration of 3-hydroxy fatty acids composing microbial surfactants. FEMS Microbiol. Lett. 108:99-102. [Google Scholar]
- 39.Nielsen, T. H., C. Christophersen, U. Anthoni, and J. Sørensen. 1999. Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 86:80-90. [DOI] [PubMed] [Google Scholar]
- 40.Ochoa-Loza, F. J. 1998. Physico-chemical factors affecting rhamnolipid (biosurfactant) application for removal of metal contaminants from soil. Ph.D. dissertation. University of Arizona, Tucson.
- 41.Persson, A., E. Österberg, and M. Dostalek. 1988. Biosurfactant production by Pseudomonas fluorescens 378: growth and product characteristics. Appl. Microbiol. Biotechnol. 29:1-4. [Google Scholar]
- 42.Peypoux, F., J. M. Bonmatin, and J. Wallach. 1999. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51:553-563. [DOI] [PubMed] [Google Scholar]
- 43.Phale, P. S., H. S. Savithri, N. A. Rao, and C. S. Vaidyanathan. 1995. Production of biosurfactant “Biosur-Pm” by Pseudomonas maltophila CSV89: characterization and role in hydrocarbon uptake. Arch. Microbiol. 163:424-431. [Google Scholar]
- 44.Rapp, P., H. Bock, V. Wray, and F. Wagner. 1979. Formation, isolation and characterization of trehalose dimycolates from Rhodococcus erythropolis grown on n-alkanes. J. Gen. Microbiol. 115:491-503. [Google Scholar]
- 45.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphospate kinase is essential for biofilm development, quorum sensing, and virulence. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robert, M., M. E. Mercadé, M. P. Bosch, J. L. Parra, M. J. Espuny, M. A. Manresa, and J. Guinea. 1989. Effect of the carbon source on biosurfactant production by Pseudomonas aeruginosa 44T1. Biotechnol. Lett. 11:871-874. [Google Scholar]
- 47.Ron, E. Z., and E. Rosenberg. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg, E., and E. Z. Ron. 1999. High- and low-molecular-mass microbial surfactants. Appl. Microbiol. Biotechnol. 52:154-162. [DOI] [PubMed] [Google Scholar]
- 49.Shepherd, R., J. Rockey, I. W. Sutherland, and S. Roller. 1995. Novel bioemulsifiers from microorganisms for use in foods. J. Biotechnol. 40:207-217. [DOI] [PubMed] [Google Scholar]
- 50.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 51.Spoeckner, S., V. Wray, M. Nimtz, and S. Lang. 1999. Glycolipids of the smut fungus Ustilago maydis from cultivation on renewable resources. Appl. Microbiol. Biotechnol. 51:33-39. [Google Scholar]
- 52.Sullivan, E. R. 1998. Molecular genetics of biosurfactant production. Curr. Opin. Biotechnol. 9:263-269. [DOI] [PubMed] [Google Scholar]
- 53.Swofford, D. L. 2002. PAUP* phylogenetic analysis using parsimony and other methods. 4.0b10 PPC. Sinauer Associates, Sunderland, Mass.
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trebbau de Acevedo, G., and M. J. McInerney. 1996. Emulsifying activity in thermophilic and extremely thermophilic microorganisms. J. Ind. Microbiol. 16:1-7. [Google Scholar]
- 56.Uchida, Y., S. Misawa, T. Nakahara, and T. Tabuchi. 1989. Factors affecting the production of succinnoyl trehalose lipids by Rhodococcus erythropolis SD-74 grown on n-alkanes. Agric. Biol. Chem. 53:765-769. [Google Scholar]
- 57.Vanittanakom, N., and W. Loeffler. 1986. Fengycin—a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot. (Tokyo) 39:888-901. [DOI] [PubMed] [Google Scholar]
- 58.Versalovic, J., M. Schneider, F. J. DeBruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 59.Vértesy, L., E. Ehlers, H. Kogler, M. Kurz, J. Meiwes, G. Seibert, M. Vogel, and P. Hammann. 2000. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. II. Isolation and structural characterization. J. Antibiot. (Tokyo) 53:816-827. [DOI] [PubMed] [Google Scholar]
- 60.Vollenbroich, D., G. Pauli, M. Özel, and J. Vater. 1997. Antimycoplasma properties and application in cell culture of surfactin, a lipopeptide antibiotic from Bacillus subtilis. Appl. Environ. Microbiol. 63:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Y., and R. M. Miller. 1994. Effect of a Pseudomonas rhamnolipid biosurfactant on cell hydrophobicity and biodegradation of octadecane. Appl. Environ. Microbiol. 60:2101-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., and R. M. Miller. 1995. Effect of rhamnolipid (biosurfactant) structure on solubilization and biodegradation of n-alkanes. Appl. Environ. Microbiol. 61:2247-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., W. J. Maier, and R. M. Miller. 1997. Effects of rhamnolipids on the dissolution, bioavailability, and biodegradation of phenanthrene. Environ. Sci. Technol. 31:2211-2217. [Google Scholar]