Abstract
Marine particles in the ocean are exposed to diverse bacterial communities, and colonization and growth of attached bacteria are important processes in the degradation and transformation of the particles. In an earlier study, we showed that the initial colonization of model particles by individual bacterial strains isolated from marine aggregates was a function of attachment and detachment. In the present study, we have investigated how this colonization process was further affected by growth and interspecific interactions among the bacteria. Long-term incubation experiments showed that growth dominated over attachment and detachment after a few hours in controlling the bacterial population density on agar particles. In the absence of grazing mortality, this growth led to an equilibrium population density consistent with the theoretical limit due to oxygen diffusion. Interspecific interaction experiments showed that the presence of some bacterial strains (“residents”) on the agar particles either increased or decreased the colonization rate of other strains (“newcomers”). Comparison between an antibiotic-producing strain and its antibiotic-free mutant showed no inhibitory effect on the newcomers due to antibiotic production. On the contrary, hydrolytic activity of the antibiotic-producing strain appeared to benefit the newcomers and enhance their colonization rate. These results show that growth- and species-specific interactions have to be taken into account to adequately describe bacterial colonization of marine particles. Changes in colonization pattern due to such small-scale processes may have profound effects on the transformation and fluxes of particulate matter in the ocean.
Particles are important components in the turnover, decomposition, and sinking flux of both organic and inorganic matter and elements in aquatic systems (48). Macroscopic organic aggregates (i.e., marine snow) comprise particles from various sources and are a major component of the material caught in sediment traps. In addition to being a primary vehicle for transporting surface derived material to deep water and the ocean floor (1), particles also harbor dense microbial communities (20, 28), suggesting that they are important sites for biological processes, e.g., production, decomposition, and nutrient recycling in the water column (1, 48). Knowing the rates at which microbes colonize and grow on particles is therefore the key to understanding the transformation and degradation of particle aggregates and the related flux processes in aquatic systems.
Organic particles and the surrounding “phycospheres” (4, 8, 15) and “detritospheres” (9) represent micropatches of concentrated substrates that pelagic bacteria may exploit (3, 33). Large fractions (40 to 70%) of marine bacteria are motile (31), many of which have chemotactic behavior (11, 12) that allows them to cluster around patches of dissolved organic matter (DOM) or rapidly colonize particulate organic matter (24, 34). A recent model predicts that chemotactic bacteria grow 50% faster when clustering within nutrient patches around detrital particles (10). Thus, motility and foraging strategies can be important adaptations of heterotrophic bacteria and archaea to gradients of DOM and particulate organic matter in the ocean.
Previous modeling and experimental efforts with individual bacterial strains show that the encounter rate between particles and free bacteria is independent of processes on the particles and that the net result of colonization and detachment within few 10s of minutes leads to a predicted steady-state abundance of attached bacteria as a function of particle size (34). However, the predicted steady-state abundance is markedly different from reported abundances of bacteria on marine particle aggregates. This discrepancy suggests that other processes, such as growth and cell-cell interactions, may play an important role in modifying the population dynamics of bacteria on particles (18, 21). Recently it has been shown that bacteria isolated from marine particle aggregates produce communication signals (acylated homoserine lactones [AHLs]) that are involved in quorum sensing (26). AHLs are known to govern phenotypic traits of bacteria (e.g., biofilm formation, exoenzyme production, and antibiotic production) (25); thus, the production of AHLs by particle-colonizing bacteria strongly indicates the importance of intra- and interspecific interactions in regulating microbial dynamics on particle surfaces. In addition, hydrolytic enzyme activities of attached bacteria (37) may help liberate DOM from the particles, thereby attracting other chemotactic bacteria. On the other hand, some particle-associated bacteria display antagonistic activities towards other bacteria (39), which suggests that inhibition among attached bacteria can also affect the development of microbial communities on particles.
The present experiments were designed to study growth and interspecific interactions among particle-colonizing bacteria. Our main goal was to understand how these processes affect short-term and long-term microbial dynamics on particles, which is essential for understanding microbial transformation of particulate matter in the ocean.
MATERIALS AND METHODS
Bacterial cultures and basic experimental design.
All bacteria used in the experiments were originally isolated from natural marine particle aggregates. Isolates were grown in batch cultures enriched with Marine Broth (MB2216; Difco), and identified to the species level (Table 1). The DNA sequences of the bacteria will be presented elsewhere (H.-P. Grossart and T. Brinkhoff, unpublished data). Bacteria in exponential growth phase were used for experiments. Basic experimental design follows that of Kiørboe et al. (34). Model agar particles were suspended on thin glass threads in suspensions of bacteria, and changes in abundance of attached bacteria were monitored over time. Incubation containers were either 2- or 20-liter beakers. For long-term experiments the incubators were stirred slowly by magnetic stirrers to ensure (near-) time-independent colonization rate (34). All long-term experiments were conducted in a biological safety cabinet to minimize contamination. Although agar particles are different from natural particle aggregates, many of the underlying processes that govern bacterial colonization remain the same (34). Besides, the size and chemical constituent of agar particles can be easily manipulated, and their simple geometry and uniform surfaces greatly facilitate the experimental design. Thus, agar particles provide an excellent tool to develop a basic model for bacterial colonization of particles, upon which we could later expand and modify to account for other compounding factors.
TABLE 1.
Bacterial strains used in this studya
| Strain | Identification by GenBank alignment | % Homology to GenBank sequence |
|---|---|---|
| 11 | M58792 (Microscilla furvescens) | 90 |
| 13 | AF359546 (marine bacterium SCRIPPS 739) | 98 |
| 15 | AJ000647 (Marinobacter PCOB-2) | 98 |
| 22 | AJ318163 (uncultured alpha proteobacterium) | 94 |
| 30 | AJ294355 (uncultured Roseobacter 667-19) | 98 |
| 36 | AJ302707 (Marinobacter sp. strain ME108) | 91 |
| 38 | AJ296095 (Bacillus sp. strain OS-5) | 98 |
| 46 | AJ391199 (Bacillus sp. strain AS-38) | 98 |
| T5 | AJ2961582 (marine bacterium PP-154) | 99 |
| T5Mut | AJ296158 (marine bacterium PP-154) | 99 |
All strains were isolated from marine snow collected at the German Wadden Sea and grown in Marine Broth.
Short-term interspecific interactions.
Since all the strains used for the experiments were originally attached to natural particle aggregates, they presumably had equal capability to initiate colonization. However, the order of their arrival to a particle may be simply a matter of chance and yet may significantly affect the subsequent development of the microbial communities on the particles. Short-term incubations were conducted to investigate how the order of arrival among different strains affects the development of the whole bacterial communities on particles. Agar particles were first incubated in a suspension of strain 36, 38, or 46 for 80 min to establish a “resident” population. Then aliquots of strain 11 (“newcomer”) were added to the incubation tank. Strain 11 was prestained with SybrGold (Molecular Probes) so that the resident strain and the newcomer strain on the agar particles could be distinguished easily by epifluorescence microscopy (see below). Preliminary experiments showed that staining with SybrGold did not affect the colonization behavior of strain 11. For the control precolonized particles were replaced by clean particles. Changes in bacterial abundance on the particles were monitored for 160 min. Differences in the colonization by newcomer strain 11 between agar particles precolonized by different resident strains (strains 36, 38, and 46) or without any colonization were statistically evaluated in an analysis of covariance with separate slopes, using standard least squares as the fitting facility. All statistical analyses were performed using the software JMP 4.02.
Effects of resident cell density were studied by incubating agar particles with strain 13 or 22 for 0, 1.1, or 7.8 h to establish different initial resident cell densities. Aliquots of SybrGold-stained strain 11 were then moved to the incubation tank, and changes in bacterial abundance on the particles were monitored for >300 min. We expected that the colonization rate of strain 11 would decrease with higher resident densities of strains 13 and 22.
Interspecific interactions were studied in more detail with strains 11, 15, and 30. Agar particles were first incubated with one of the three strains (strain X) for 80 min. Half of the particles were then transferred to a suspension of strain X and half to a mixture of strains X and Y. A parallel incubation with new agar particles in a suspension of strain Y also started simultaneously. Changes in bacterial abundance on the particles were monitored for another 60 to 80 min, and strain X and strain Y in the mixture treatment were differentiated by fluorescent in situ hybridization (FISH) (see below). Ambient concentrations of strains X and Y were ∼106 cells ml−1 in both pure and mixed incubations, and the total ambient concentration in the mixture was thus about twice the concentrations in the pure suspensions. Because colonization rates and equilibrium abundances in short-term single-strain experiments scale with ambient concentration (34), the null hypothesis was that abundances of attached cells in the mixture would equal the sum of attached bacteria in the two single-strain incubations, indicating no interspecific interaction. This set of experiments allowed us to examine the mutual effects between resident and newcomer strains and whether the effects are reciprocal when the resident and the newcomer strains trade places.
Antagonistic interaction among the bacteria was studied using strain T5 and its mutant T5Mut. T5 produces the antibiotic substance tropodithietic acid, which inhibits the growth of strain 11 and 22 in agar diffusion assays, whereas T5Mut does not (T. Brinkhoff, G. Bach, T. Heidorn, L. Liang, A. Schlingloff, and M. Simon, submitted for publication). Sterile agar particles were incubated for 2 to 4 days in rolling tanks with a high concentration of T5 or T5Mut. Particles precolonized by T5 or T5Mut were then incubated in a suspension of strain 11 or 22 for 160 min. Some precolonized particles were transferred to sterile seawater to measure the detachment rate of T5 or T5Mut (34). Changes in total bacterial abundance on the particles after corrections for detachment yield the colonization rate of strain 11 or 22 in the presence of T5 or T5Mut. Differences between the T5 and T5Mut treatments would indicate the effects of antibiotics on the colonization behavior of strains 11 and 22.
Long-term growth and species-interaction.
To study the effect of growth, agar particles were incubated in a suspension of strain 11 or 13 for >3,000 min. A parallel incubation in a mixture of strains 11 and 13 was conducted to study the effects of long-term species-interaction. Again, ambient bacterial concentration in the mixture was the sum of concentrations in the pure-strain treatments. Agar particles were removed at fixed time points, and bacteria on the particles were enumerated by epifluorescence microscopy (see below). In the mixture treatment, strains 11 and 13 on the particles were distinguished by FISH.
Bacterial enumeration.
Agar particles were placed in a counting chamber made of an O-ring glued onto a microscopic slide. A drop of formalin and a drop of fluorescent stain (DAPI [1 mg 100 ml−1; Fluka] or SybrGold solution [1:100; Molecular Probes]) were added. For experiments with mixed bacterial strains, one of the added strains was prestained with SybrGold, whereas bacteria on agar particles were stained with DAPI so that the added prestained bacteria would fluoresce green and the other bacteria would fluoresce blue. The chamber was then closed with a coverslip, and the bacteria were enumerated at a magnification of ×1,000 with an epifluorescence microscope (Axioplan; Zeiss, Jena, Germany). Loss of bacteria during sample preparation was small (34). Aliquots (1-ml) of surrounding water were filtered onto 0.2-μm-pore-size Nuclepore filters for enumeration of free-living bacteria (DAPI direct count).
FDC.
Dividing cells were observed by epifluorescence microscopy and a video analysis system (Soft Imaging System). The frequency of dividing cells (FDC) was measured as a percentage of dividing cells.
FISH.
Agar particles were transferred into 2-ml Eppendorf tubes with 100 μl of fresh paraformaldehyde (4%) solution and stored at 4°C in the refrigerator until FISH analysis. Each agar particle was mounted onto Teflon microslides (Marienfeld KG, Bad Mergentheim, Germany). We used 16S rRNA oligonucleotide probes to determine the percentages of α-Proteobacteria (strains 13 and 30) and γ-Proteobacteria (strain 15) and of the Cytophaga-Fexibacterium-Bacteroides cluster (Cytophaga, strain 11). Specific sequences of oligonucleotide probes are given by Amann et al. (2) for α- and γ-Proteobacteria as well as Cytophaga.
RESULTS
Short-term interaction experiments.
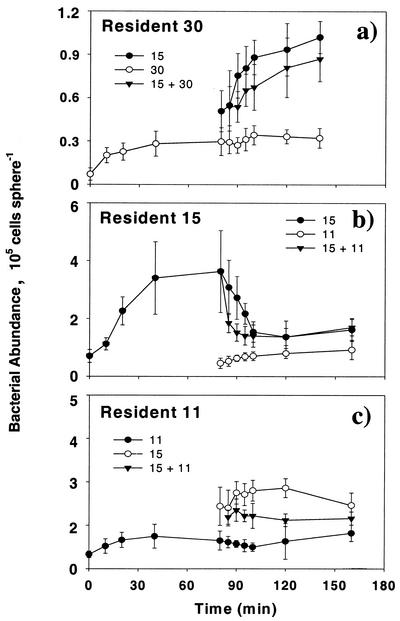
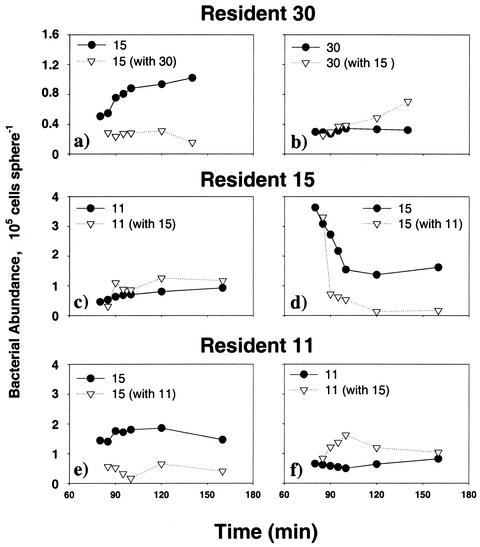
Short-term experiments were conducted with strains 30, 15, and 11. Colonization rate of the bacteria differed in presence or absence of another strain on the agar particles (Fig. 1). Despite the fact that ambient bacterial concentration was twice as high in the mixed-strain suspensions, in all cases interactions between the resident strain and the newcomer strain led to an intermediate equilibrium bacterial abundance between the equilibrium abundances of the respective individual strains. FISH analyses further revealed complex interactions that differed between species pairs. The colonization rate of strain 15 was reduced significantly when the particles were precolonized by strain 30 or 11 (Fig. 2a and e). Interestingly, the presence of strain 15 in the ambient water also enhanced the colonization rate of strain 30 on particles precolonized by strain 30 after 140 min (Fig. 2b). Similar enhancement was also observed with resident strain 11 in the first 120 min (Fig. 2f). Colonization rate of strain 11 was not affected by the presence of strain 15 on the particles (Fig. 2c); on the contrary, the presence of strain 11 in the ambient water reduced further colonization by strain 15 of particles precolonized by strain 15 (Fig. 2d).
FIG. 1.
Bacterial abundance on agar particles incubated in single or in mixed cultures. Particles were precolonized by strains 30 (a), 15 (b), or 11 (c). Bacteria were enumerated by epifluorescence microscopy (magnification, ×1,000) after DAPI staining. Error bars reflect standard deviations of three parallel measurements.
FIG. 2.
Same experiment as in Fig. 1. Abundances of a specific strain on agar particles in the presence or absence of another strain are compared in each panel. Resident and newcomer strains were differentiated by FISH. All panels at left are for newcomer strains; all panels at right are for resident strains.
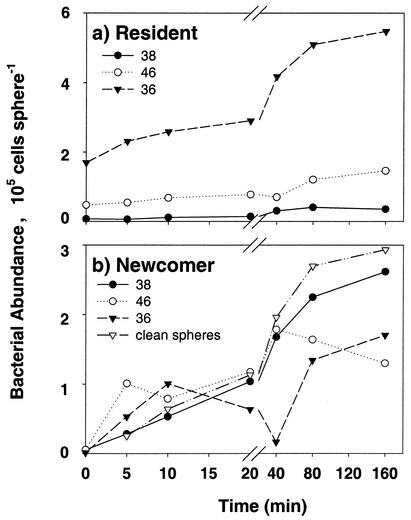
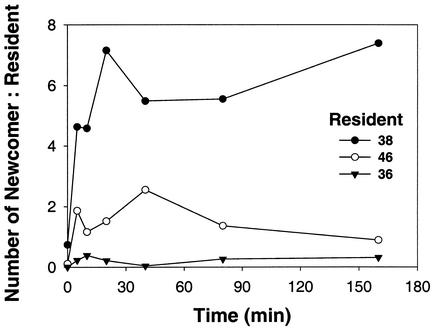
To test whether the presence of different resident strains may change the colonization pattern of a newcomer strain, we incubated strain 11 with clean agar particles or agar particles that had been precolonized by various strains (strains 36, 38, and 46; Fig. 3). The colonization pattern of strain 11 changed with time and with the resident strains (Fig. 3b). Statistical analysis showed that the abundance of strain 11 on precolonized and sterile agar particles increased significantly over time (P < 0.0001). The colonization of the newcomer strain 11 of the previously colonized agar particles was significantly less than that on particles without colonization (Fig. 3b; Table 2). Considering only the first 20 min of colonization, there were no significant differences between colonization rates of newcomer 11 on particles precolonized by strain 36 and sterile ones (Fig. 3b; Table 3). There were further clear differences between the strain 11 colonization rates of the previously colonized particles: those precolonized by strain 36 were colonized significantly more slowly by strain 11 than those precolonized by the strains 38 or 46 (Fig. 3b; Table 2). There was no significant difference in the colonization rates of the two latter strains. Again, the picture changes if one considers only the first 20 min of the experiments: there is no significant difference in colonization rates of newcomer strain 11 on particles precolonized by strains 36 or 38, while on strain 46 precolonized particles strain 11 colonizes more rapidly than on those inhabited by strains 36 and 38 (Fig. 3b; Table 3). The ability of a newcomer to invade a resident population can be better seen by plotting the temporal change in the ratio of newcomer cells to resident cells (Fig. 4). In our experiments, strain 11 quickly dominated over resident strain 38, leading to a newcomer/resident ratio of >1 after only 5 min. In comparison, strain 11 fared equally well as resident strain 46 toward the end of the experiments (ratio ≈ 1:1), whereas it was somewhat suppressed by resident strain 36 (ratio < 1) (Fig. 4).
FIG. 3.
Colonization by newcomer strain 11 of agar particles that had been precolonized by various resident strains (strains 38, 46, and 36). Clean agar particles were used as controls. (a) Abundance of the respective resident strains; (b) abundance of newcomer strain 11. For statistical analyses see Tables 2 and 3.
TABLE 2.
Abundance of strain 11 on precolonized particles, including all time points
| Resident strain |
P for comparison with strain:
|
||
|---|---|---|---|
| 36 | 38 | 46 | |
| 38 | <0.0001 | ||
| 46 | 0.0002 | NSa (0.2675) | |
| None | <0.0001 | <0.0001 | 0.0336 |
NS, not significant
TABLE 3.
Abundance of strain 11 on precolonized particles, including only time points <20 min
| Resident strain |
P for comparison with strain:
|
||
|---|---|---|---|
| 36 | 38 | 46 | |
| 38 | NSa (0.3175) | ||
| 46 | 0.0173 | <0.0001 | |
| None | NS (0.7271) | 0.0452 | <0.0001 |
NS, not significant.
FIG. 4.
Ratio between normalized abundance of newcomer strain (strain 11) and the respective resident strains. Note that initial abundances of the resident strains were different (for strain 38, 0.1 × 105 cells sphere−1; for strain 46, 0.5 × 105 cells sphere−1; for strain 36, 1.8 × 105 cells sphere−1).
Effects of resident cell density.
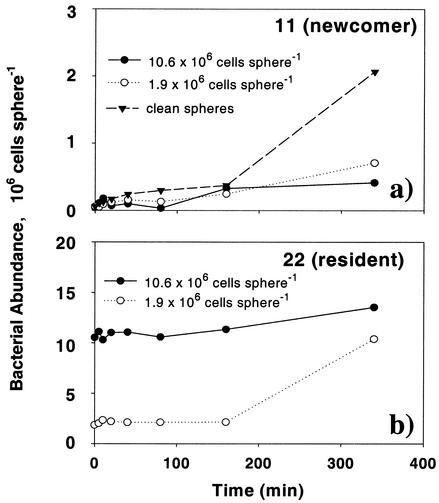
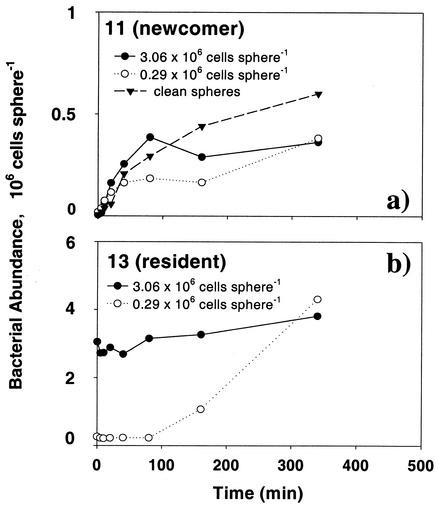
Preincubating the agar particles with strains 22 and 13 for 1.1 and 7.8 h yielded an order of magnitude difference in the initial resident populations (Fig. 5b and 6b). The presence of strains 22 and 13 (resident) on the agar particles did not affect the colonization by strain 11 (newcomer) within the first 160 min (Fig. 5a and 6a). Toward the end of the experiments, however, colonization by strain 11 was significantly lowered on spheres that were precolonized by strain 22 and 13 (Fig. 5a and 6a), during which time the resident populations had reached almost the same densities between the 1.1- and 7.8-h treatments (Fig. 5b and 6b). Thus, while there was a clear, negative effect due to the presence of resident strains 22 and 13 on the long-term abundance of strain 11, the effects of resident cell densities could not be discerned in the short-term colonization pattern.
FIG. 5.
Colonization by newcomer strain 11 of agar particles that had been precolonized by resident strain 22 (initial abundances 1.9 and 10.6 × 106 cells sphere−1). Clean particles were used as controls. (a) Abundance of newcomer strain 11 on precolonized particles and clean particles; (b) abundance of resident strain 22.
FIG. 6.
Colonization by newcomer strain 11 of agar particles that had been precolonized by resident strain 13 (initial abundances 0.29 and 3.06 × 106 cells sphere−1). Clean agar particles were used as controls. (a) Abundance of newcomer strain 11 on precolonized particles and clean particles; (b) abundance of resident strain 13.
Effects of antibiotic production.
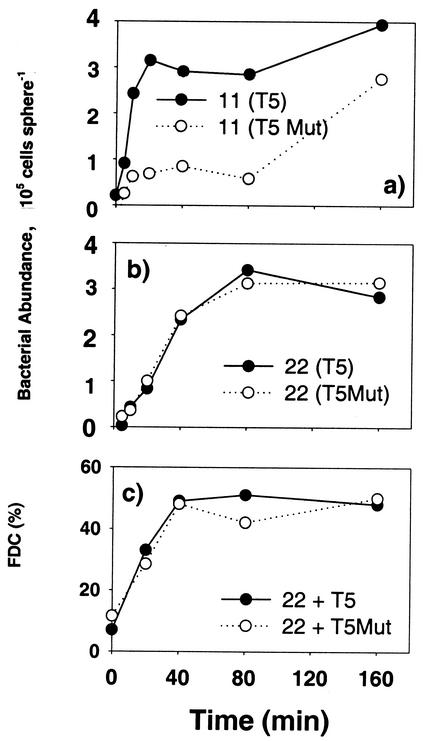
Strain 22 did not show any difference in colonization rate on particles precolonized with T5 or T5Mut (Fig. 7b and c). Measurements of the FDC also showed no negative effects of T5 on the growth of strain 22 within the experimental period (Fig. 7c). Contrary to our expectations, strain 11 colonized agar particles with T5 at a higher rate than those with T5Mut (Fig. 7a), suggesting attraction of strain 11 by T5. After 1 week of incubation the agar particles were completely solubilized by strain T5 but not by T5Mut, indicating high hydrolytic enzyme activity and presumably high release of DOM in the presence of T5.
FIG. 7.
Effects of the antibiotic-producing resident strain T5 and its antibiotic-free mutant (T5Mut) on colonization by newcomer strains 11 and 22. (a) Abundance of newcomer strain 11 in the presence of resident T5 or T5Mut; (b) abundance of newcomer strain 22 in the presence of T5 or T5Mut; (c) FDC of strain 22 plus T5 and strain 22 plus T5Mut.
Growth and long-term interactions.
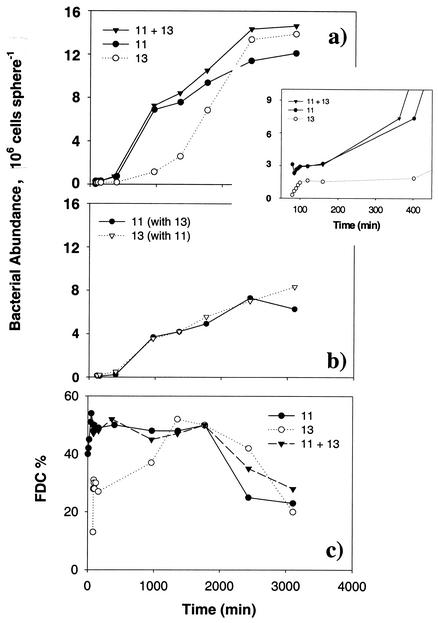
Colonization of and subsequent growth on clean agar particles by strains 11 and 13 was higher than that by strain 11 or 13 alone but not as high as the sum of the two in the single-strain incubation (Fig. 8a). The abundance of strain 11 was significantly higher than that of strain 13 during the first 2,000 min, after which time abundances of the two were similar (Fig. 8a). Abundance of bacteria on the agar particles in the single-strain treatments increased and almost reached that of the mixed-strain treatment near the end of the experiment. FISH analysis showed that in the mixed-strain treatment, the agar particles were colonized by almost equal numbers of strains 11 and 13 (Fig. 8b). The FDC was similar for strain 11 and the mixture of strains 11 and 13 throughout the whole experiment (Fig. 8c). However, the FDC of strain 13 was significantly lower than the others in the first 1,000 min (Fig. 8c).
FIG. 8.
Long-term incubation experiment. (a) Bacterial abundance on agar particles in single strain treatment (strain 11 or 13) or in mixed strain treatment (strains 11 and 13). The inset shows the colonization pattern during the first 400 min of incubation. (b) Respective abundance of strains 11 and 13 in the mixed-strain treatment as differentiated by FISH. (c) FDC of strains 11 and 13 in single- and mixed-strain treatments.
DISCUSSION
Field observations show that marine particle aggregates are often densely colonized by bacteria and other microbes, and activities of these microbes play pivotal roles in transforming and degrading these aggregates (1, 48). We have previously developed an encounter model to describe the initial phase of bacterial colonization of marine particles based on the bacterium's motility pattern, the hydrodynamic environment, and the detachment probability of attached bacteria (34). Our model predicts that the equilibrium abundance of bacteria between attachment and detachment on marine particle aggregates should scale with aggregate size to the power of ca. 1.45; however, field observations show a scaling to the power of <1 (34). Such a discrepancy between model prediction and field observations suggests that factors other than attachment and detachment need be considered in the colonization process. Since natural marine particle aggregates may be exposed to diverse bacterial communities for an extended time, interactions among different bacterial strains may change their colonization behaviors and growth. These interactions may take place at various levels and can be studied on our model particles in great detail.
Mutual interference.
When a newly formed marine particle is exposed to a mixed population of bacteria, every strain of bacteria should have equal opportunity to colonize that particle. In the absence of interference, the initial colonization rates of each strain should be independent of the presence of another strain such that the abundance of attached bacteria is proportional to the total ambient bacterial concentration (34). In our experiments with strain 11 alone and strain 13 alone, balance between attachment and detachment led to an initial equilibrium bacterial abundance on the agar particles within the first 200 min (Fig. 8a), consistent with our earlier observations (34). However, in the mixture treatment with strains 11 and 13, despite the higher ambient bacterial concentration, the initial colonization rate was lower than expected (i.e., the summation of the rates with the individual strains [Fig. 8a]), indicating interference between strains 11 and 13 in the colonization process. This interference may take place in ambient water or on the particle surface. Longer incubation revealed that strains 11 and 13 colonized the surface equally (Fig. 8b); FDC measurements also showed that both strains had similar growth rates after 1,000 min (Fig. 8c). Thus, it appeared that the two strains performed equally well once on the surface and that the interference likely took place in the ambient water. Since the initial colonization rate is dependent on the swimming speed and swimming pattern of the bacteria (34), this interference may be a result of altered swimming behavior of the bacteria in the mixture. Testing such a hypothesis requires further study on the bacteria's swimming behavior in single versus mixed populations.
Interactions between residents and newcomers.
Once a strain has established itself as resident on a particle, it may begin to alter the surface characteristics of the particle and thereby affect the subsequent colonization by other strains. For example, enzyme activities due to the resident bacteria may liberate DOM from the particle, which may attract other bacteria with chemosensory behavior, leading to higher initial colonization rate of newcomers (27, 32, 38). On the other hand, a well-established resident population may have a competitive edge over the newcomer in harvesting the limited resources on the particle surface, e.g., nutrients, oxygen, and habitable space (17, 37). Such was the case with strain 46 in our experiments: resident populations of strain 46 on the agar particles enhanced the initial colonization (first 20 min) of newcomer strain 11 relative to resident strain 38 and the control (Fig. 3b) but were more resistant to the longer-term accumulation of strain 11 on the agar particles (Fig. 3b and 4).
Longer-term experiments with resident strains 13 and 22 also showed that the growth of newcomer strain 11 on the agar particles was suppressed by the resident populations relative to the control (Fig. 5a and 6a). There also appeared to be a difference between strains 13 and 22 in their resistance to strain 11: at the end of the experiments (360 min), resident strain 13 reduced the accumulation of strain 11 by 30%, whereas resident strain 22 reduced that by 25% relative to the control (Fig. 5a and 6a). Such a difference may be partly due to the different growth rates of strain 13 and 22 on the particles. Thus, our results show that interactions between resident and newcomer is highly specific to the strains involved and the time scale under consideration.
Reciprocal effects.
Yet for any given pair of bacterial strains, which one will be the resident and which one will be the newcomer may be simply a matter of chance, i.e., which one reaches the particle first. Thus, our short-term experiments with strains 11, 15, and 30 were designed to test if the resident-newcomer interactions remain the same when the two strains switch positions (Fig. 1 and 2). In all cases, the abundance of attached bacteria in mixed strain treatment (X plus Y) was less than the summation of the individual strain treatments, indicating interference between the resident and the newcomer (Fig. 1). However, different strains were affected differently. Strain 15, whether as resident or newcomer, was always suppressed by the other strains (Fig. 2a, d, and e). Interestingly, the presence of strain 15 in the ambient water also stimulated the increase of resident strain 30 on the agar particles (Fig. 2b), but the reason for this is unknown. Strain 11 behaved differently as a resident than as a newcomer: as a newcomer, strain 11 was not affected by resident strain 15 (Fig. 2d). However, when the two strains switched places, resident strain 11 was initially stimulated by newcomer strain 15 (Fig. 2e). Our results therefore showed that although resident-newcomer interaction tends to lower the overall bacterial abundance on our model particles, individual strains may respond differently and, in some cases, benefit from the interaction. Because substrate utilization and breakdown is highly species specific among bacteria (29, 38, 39), species-specific response in resident-newcomer interactions will be an important factor in degradation and transformation of natural marine particles.
Antagonistic interactions.
A special case of species-specific interaction may involve certain strains that exhibit antagonistic effects on other strains. Marine particles are physically more stable and display less-resource-limiting habitats than the free water column. It may therefore be beneficial for attached bacteria to invest in defending against invaders, such as by releasing antibiotic substances. Indeed, many particle-associated bacteria exhibit antagonistic behavior against other bacteria in laboratory assays (37). In the present study, strain T5 exhibits antagonistic behavior against strains 11 and 22 in standard diffusion assays. Surprisingly, no negative effects were observed in our colonization experiments; colonization of agar particles by strain 11 was on the contrary enhanced by T5 during the first 160 min (Fig. 7). Unlike in standard diffusion assays where materials are immobilized on the two-dimensional surface of an agar plate, the agar particles in the present experiments were suspended in water and antagonistic substances produced by T5 may therefore diffuse away from the particles. Enzyme activity of T5 may further release DOM from the agar particles into the surrounding water and attract strain 11. It should be noted, however, that our colonization experiments with T5 lasted only 160 min, which may be shorter than the time required for any negative effects on growth to be observed. Nevertheless, our results caution against direct application of standard assay results to marine particle study where hydrodynamics around the particles may modify or diminish any antagonistic effects.
Long-term growth and species interactions.
On a longer time scale, attached bacteria (whether resident or newcomer) will grow on the particles. This growth will further alter the surface property of the particles due to rapid accumulation of bacterial biomass, release of cell metabolites, depletion of substrates, and in some cases secretion of biofilm matrix. In the present study, growth was evident after 200 min when bacterial abundance increased rapidly following the initial equilibrium (Fig. 5 and 8). However, in the long-term experiments with strains 11 and 13, bacterial abundances reached, after ∼2 days, a second equilibrium at ∼1.2 × 107 cells particles −1 (Fig. 8a). What could be limiting further increase in bacterial abundance on the particles? The agar particles in our experiments had an average diameter of 4 mm, giving a surface area of 5 × 107 μm2. A cell of strain 11 averaged 2.2 μm in diameter. At the maximum cell density, over 90% of the available surface of an agar particles will be covered by bacteria. Thus, continuous increase in bacterial abundance would be limited by available space. This analysis assumes that the bacteria form a monolayer on the surface. Microscopic observation revealed, however, that the bacteria formed multiple layers embedded in mucous at high densities. While forming multiple layers may somewhat lessen space limitation, bacterial growth could still be hampered by substrate limitation. Consider the case of oxygen. For a stationary 4-mm-diameter particle, oxygen is supplied by diffusion at a rate of 1.5 × 10−11 mol s−1 (assuming a diffusion coefficient for O2 of ca. 2 × 10−5 cm2 s−1 and an O2 saturation concentration of 3 × 10−7 mol cm−3). A 2-μm-diameter bacterium contains ca. 4 × 10−13 g of C cell−1. At a per capita growth rate of 3 × 10−5 s−1 (growth rate of ambient bacteria in our experiments) and an assumed growth efficiency of 0.5, the bacterium will metabolize 1.2 × 10−17 g of C cell−1 s−1, which is equivalent to a cell-specific oxygen demand of 1.25 × 10−18 mol of oxygen cell−1 s−1 (assuming a respiratory quotient of 0.8). The maximum bacterial population set by oxygen limitation is given by the equation oxygen supply/cell-specific oxygen demand = 1.2 × 107 cells particles −1. Thus, it is conceivable that long-term bacterial growth on the agar particles was limited by oxygen supply. It should be emphasized that this calculation applies only to stationary particles. For sinking particles, oxygen supply is enhanced by advection and therefore oxygen limitation would be less severe (43). Microsensor studies have shown that a stable anoxic condition around a particle aggregate can only be maintained by high and continuous carbon metabolism (45). Thus, for particle aggregates that are made of refractory organic material, nutrient limitation would be a more probable cause to restrict bacterial growth on the aggregates.
Particle aggregates as complex microcosms.
Traditionally marine particle aggregates are regarded as simple vehicles for sinking fluxes. Yet a newly emerging view suggests that marine particle aggregates are complex microcosms within which material and energy flows are controlled by interactions among different microbes and their environments (33, 37, 47). Our and other studies show that bacterial colonization of particle surfaces and subsequent biofilm formation are controlled by a variety of factors that include hydrodynamics, surface chemistry, physiology, and genotype of the cell. Succession of microbial populations on particles further depends on inter- and intraspecific interactions and the environmental condition (22, 42). For example, Streptococcus mutans and Lactobacillus casei form a mixed, stable community under glucose limitation at pH of 7.0; however, decrease of pH to 4.8 results in a dramatic reduction of S. mutans cell numbers (14). Likewise, the ability of the fungus Candida albicans to compete for glucose with oral bacteria also greatly varies with chemostat conditions (6).
Surface-active chemicals also influence microbial attachment, and cellular adsorption is mediated by cell surface hydrophobicity, bacterium-substratum charge interactions, surface roughness, and surface free energy (52, 53, 54). For example, pathogenic strains of Vibrio express chemotactic behavior toward specific mucous surfaces of fish depending on mucous chemistry, temperature, and other parameters (13). Other heterotrophic bacteria are also able to react very specifically to changes in environmental conditions. Mueller (40) showed that the specific rate coefficient for adsorption of two Pseudomonas species increased significantly when the organisms were starved. During that time the desorption probability was zero, demonstrating that not only attachment but also detachment probability may be strongly dependent on time and changes in the environment.
Our findings are in good agreement with recent literature on intra- and interspecific interactions among bacteria on biofilms in various aquatic habitats. Rickard et al. (46) determined phylogenetic relationships and coaggregation ability of freshwater biofilm bacteria. Their results demonstrate that both distantly and closely related strains coaggregated at intergeneric, intrageneric, and intraspecies levels. Coaggregation among biofilm bacteria has been shown to play an important role in microbial colonization processes and demonstrate different roles of various bacterial isolates during biofilm development (19). Antagonistic interactions among bacteriocin-producing enteric bacteria have been shown in dual species biofilms preventing the colonization of a potential competitor into preestablished biofilms (51). Stoodley et al. (50) even suggest that complex cell-cell interactions within prokaryotic communities are an ancient characteristic leading to highly structured bacterial communities similar to multicellular organisms.
The growth of attached bacteria is indeed dependent on the specific microenvironment created by the marine particles (7, 33, 48, 49). Attached bacteria also modify the surrounding microenvironment, thereby affecting the behavior and physiology of other microbes. Evidence of metabolic interactions between community members of biofilms have been found (41), and studies on bacterial growth frequently show inhibition of growth in the presence of another strain of bacterium due to production of repellents, antibiotics, or toxins (reviewed in reference 38). Other studies indicate increased bacterial growth in the presence of other bacteria due to production of probiotics (38). A more recent study shows that the presence of cyclic AMP and acylated homoserine lactones (AHLs) increases the growth of several heterotrophic bacteria (16). Production of AHLs has been demonstrated for marine snow bacteria (26), suggesting that AHL production by bacteria on marine snow is important for quorum sensing and regulation of physiological functions such as exoenzyme synthesis (25). Communication and regulation via AHL production is arguably more favorable for attached bacteria than for free-living bacteria, because the particles provide physically more stable platforms whereupon the chemicals can act. Formation of microcolonies or multiple cell layers on particle surfaces as well as the formation of biofilms with an extensive exopolymer matrix will further facilitate chemical communication among the bacteria (23). We have evidence that the AHL-producing strain 30 (26) showed higher growth in the presence of a second strain (Fig. 2b). Nevertheless, more tests will be required to confirm the role of AHLs in the interactions between strain 30 and the other bacteria. In addition to chemical interactions, formation of dense bacterial communities on particles may facilitate also plasmid exchange among the attached bacteria (36), leading to the evolution of a genetically and functionally diverse attached bacterial community.
While natural marine particles are important nursery ground for heterotrophic bacteria, they can also be a risky environment for the attached bacteria. Sedimentation of marine particles will transport the attached bacteria to the less favorable deep water. Frequent resuspension of particles in tidally driven shallow waters, e.g., the German Waddensea, from where our isolates originate, will expose the attached bacteria to fluctuating radiation and salinity with the possibility of impairing their physiological functions (5, 35). Natural marine particles are also colonized by bacterivorous protozoan (19) and are ingested by larger zooplankton and fish (27, 30, 32, 46), causing mortality to the attached bacterial populations. Laboratory observations show that decline of attached bacterial population on marine particle aggregates coincides with an increase in attached bacterivorous flagellates and ciliates (44). Indeed, bacterial abundance on natural marine particle aggregates is often lower than the theoretical growth limit, suggesting that grazing mortality is an important factor regulating bacterial population dynamics on the aggregates (47). Thus, understanding of the long-term microbial dynamics on marine particle aggregates will require coupling bacterial growth dynamics and predator-prey interactions (34a).
Acknowledgments
We thank Thorsten Brinkhoff for providing antibiotic-producing Ruegeria strain T5 and its antibiotic-free mutant T5Mut, Meinhard Simon for providing laboratory facilities at the ICBM of the University of Oldenburg, and Christian Schütt for his hospitality and logistic help at the Biologische Anstalt Helgoland of the Alfred Wegener Institute of Polar and Marine Research. We also thank Uffe H. Thygesen for help with software development.
We acknowledge financial support from the Danish Natural Science Council (to T.K.) (9801391 and 21-01-0549), the Carlsberg Foundation (to K.T.) (99053/20-950), the Danish Network for Fisheries and Aquaculture Research (to T.K.), the Humboldt foundation (to H.P.), and the ICBM of the University of Oldenburg.
REFERENCES
- 1.Alldredge, A. L., and M. W. Silver. 1988. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 20:41-82. [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:142-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 4.Azam, F., and J. W. Ammerman. 1984. Cycling of organic matter by bacterioplankton in the pelagic marine ecosystems: microenvironmental considerations, p. 345-360. In M. J. R. Fasham (ed.), Flows of energy and materials in marine ecosystems. Plenum Publishing Corp., New York, N.Y.
- 5.Barry, K. J., and N. R. Wainwright. 1997. Biosynthetic induction of a secondary metabolite by a marine bacterium under nutritional stress: potential role of the incomplete oxidation of an organic acid. Biol. Bull. 193:274-275. [DOI] [PubMed] [Google Scholar]
- 6.Basson, N. J. 2000. Competition for glucose between Candida albicans and oral bacteria grown in mixed culture in a chemostat. J. Med. Microbiol. 49:969-975. [DOI] [PubMed] [Google Scholar]
- 7.Baty, A. E., C. C. Eastburn, S. Techkarnjanaruk, A. E. Goodman, and G. G. Geesey. 2000. Spatial and temporal variations in chitinolytic gene expression and bacterial biomass production during chitin degradation. Appl. Environ. Microbiol. 66:3574-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, W., and R. Mitchell. 1972. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143:265-277. [Google Scholar]
- 9.Biddanda, B. A., and L. R. Pomeroy. 1988. Microbial aggregation and degradation of phytoplankton-derived detritus in seawater. I. Microbial succession. Mar. Ecol. Prog. Ser. 42:79-89. [Google Scholar]
- 10.Blackburn, N., F. Azam, and 32 O Hagström. 1997. Spatially explicit simulations of a microbial food web. Limnol. Oceanogr. 42:613-622. [Google Scholar]
- 11.Blackburn, N., T. Fenchel, and J. Mitchell. 1998. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science 282:2254-2256. [DOI] [PubMed] [Google Scholar]
- 12.Blackburn, N., and T. Fenchel. 1999. Influence of bacteria, diffusion and shear on micro-scale patches, and implications for bacterial chemotaxis. Mar. Ecol. Prog. Ser. 189:1-7. [Google Scholar]
- 13.Bordas, M. A., M. C. Balebona, J. M. Rodriguez-Maroto, J. J. Borrego, and M. A. Morinigo. 1998. Chemotaxis of pathogenic vibrio strains towards mucus surfaces of gilt-head sea bream (Sparus aurata L.). Appl. Environ. Microbiol. 64:1573-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowden, G. H., and I. R. Hamilton. 1989. Competition between Streptococcus mutans and Lactobacillus casei in mixed continuous culture. Oral Microbiol. Immunol. 4:57-64. [DOI] [PubMed] [Google Scholar]
- 15.Bowen, J. D., K. D. Stolzenbach, and S. W. Chisholm. 1993. Simulating bacterial clustering around phytoplankton cells in a turbulent ocean. Limnol. Oceanogr. 38:36-51. [Google Scholar]
- 16.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess, J. G., E. M. Jordan, M. Bregu, A. Mearns-Spragg, and K. G. Boyd. 1999. Microbial antagonism: a neglected avenue of natural products research. J. Biotechnol. 70:27-32. [DOI] [PubMed] [Google Scholar]
- 18.Busscher, H. J., R. Bos, and H. C. van der Mei. 1995. Initial microbial adhesion is a determinant for the strength of biofilm adhesion. FEMS Microbiol. Lett. 128:229-234. [DOI] [PubMed] [Google Scholar]
- 19.Buswell, C. M., Y. M. Herlihy, P. D. Marsh, C. W. Keevil, and S. A. Leach. 1997. Coaggregation amongst aquatic biofilm bacteria. J. of Appl. Microbiol. 83:477-484. [Google Scholar]
- 20.Caron, D. A., P. G. Davis, L. P. Madin, and J. N. McSieburth. 1982. Heterotrophic bacteria and bacterivorous protozoans in oceanic macroaggregates. Science 218:795-797. [DOI] [PubMed] [Google Scholar]
- 21.Characklis, W. G., G. A. McFeters, and K. C. Marshall. 1990. Physiological ecology in biofilm systems, p. 341-394. In W. G. Characklis and K. C. Marshall (ed.), Biofilms. John Wiley & Sons, New York, N.Y.
- 22.Dang, H., and C. R. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decho, A. W. 1990. Microbial exopolymer secretion in ocean environments: their role(s) in food webs and marine processes. Oceanogr. Mar. Biol. Annu. Rev. 28:73-153. [Google Scholar]
- 24.Fenchel, T. 2001. Eppur si muove: many water column bacteria are motile. Aquat. Microb. Ecol. 24:197-201. [Google Scholar]
- 25.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signaling. Nature Rev. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 26.Gram, L., H.-P. Grossart, A. Schlingloff, and T. Kiørboe. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green, E. P., and M. J. Dagg. 1997. Mesozooplanktom association with medium to large marine snow aggregates in the northern Gulf of Mexico. J. Plankton Res. 19:435-447. [Google Scholar]
- 28.Grossart, H.-P., and M. Simon. 1998. Bacterial colonization and microbial decomposition of limnetic organic aggregates (lake snow). Aquat. Microb. Ecol. 15:127-140. [Google Scholar]
- 29.Grossart, H.-P., and H. Ploug. 2001. Microbial degradation of organic carbon and nitrogen on diatom aggregates. Limnol. Oceanogr. 46:267-277. [Google Scholar]
- 30.Grossart, H.-P., T. Berman, M. Simon, and K. Pohlmann. 1998. Occurrence and microbial dynamics of macroscopic organic aggregates (lake snow) in Lake Kinneret, Israel, in fall. Aquat. Microb. Ecol. 14:59-67. [Google Scholar]
- 31.Grossart, H.-P., L. Riemann, and F. Azam. 2001. Bacterial motility in the sea and its ecological implications. Aquat. Microb. Ecol. 25:247-258. [Google Scholar]
- 32.Kiørboe, T. 2000. Colonization of marine snow aggregates by invertebrate zooplankton: abundance, scaling and possible role. Limnol. Oceanogr. 45:479-484. [Google Scholar]
- 33.Kiørboe, T., H. Ploug, and U. H. Thygesen. 2001. Fluid motion and solute distribution around sinking aggregates. 1. Small-scale fluxes and heterogeneity of nutrients in the pelagic environment. Mar. Ecol. Prog. Ser. 211:1-13. [Google Scholar]
- 34.Kiørboe, T., H.-P. Grossart, H. Ploug, and K. Tang. 2002. Mechanisms and rates of bacterial colonization of sinking aggregates. Appl. Environ. Microbiol. 68:3996-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Kiørboe, T., K. Tang, H.-P. Grossart, and H. Ploug. 2003. Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl. Environ. Microbiol. 69:3036-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitano, M., T. Nishio, S. Kawai, and H. Morishita. 1986. Effects of salt environment on the drug resistance of marine bacterium, M-32. Annu. Rep. Osaka City Inst. Public Health Environ. Sci. 49:87-92. [Google Scholar]
- 36.Licht, T. R., B. B. Christensen, K. A. Krogfelt, and S. Molin. 1999. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145:2615-2622. [DOI] [PubMed] [Google Scholar]
- 37.Long, R., and F. Azam. 2001. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 67:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Brock, biology of microorganisms. 9th ed. Prentice-Hall, Inc., Upper Saddle River, N.J.
- 39.Martinez, J., D. C. Smith, G. F. Steward, and F. Azam. 1996. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat. Microb. Ecol. 10:223-230. [Google Scholar]
- 40.Mueller, R. F. 1996. Bacterial transport and colonization in low nutrients environments. Water Res. 30:2681-2690. [Google Scholar]
- 41.Müller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okabe, S., C. M. Santegoeds, Y. Watanabe, and D. de Beer. 2002. Successional development of sulfate-reducing bacterial populations and their activities in an activated sludge immobilized agar gel film. Biotechnol. Bioeng. 68:119-130. [DOI] [PubMed] [Google Scholar]
- 43.Ploug, H., and H. P. Grossart. 1999. Bacterial production and respiration in suspended aggregates: a matter of incubation method. Aquat. Microb. Ecol. 20:21-29. [Google Scholar]
- 44.Ploug, H., and H. P. Grossart. 2000. Bacterial growth and grazing on diatom aggregates: respiratory carbon turnover as a function of aggregatesize and sinking velocity. Limnol. Oceanogr. 45:1467-1475. [Google Scholar]
- 45.Ploug, H., M. Kühl, B. Buchholz-Cleven, and B. B. Jørgensen. 1997. Anoxic aggregates-an ephemeral phenomenon in the pelagic environment? Aquat. Microb. Ecol. 13:285-294. [Google Scholar]
- 46.Rickard, A. H., S. A. Leach, L. S. Hall, C. M. Buswell, N. J. High, and P. S. Handley. 2002. Phylogenteic relationships and coaggregation ability of freshwater biofilm bacteria. Appl. Environ. Microbiol. 68:3644-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shanks, A. L., and E. W. Edmondson. 1990. Vertical flux of metazoans (holoplankton, meiofauna, and larval invertebrates) due to their association with marine snow. Limnol. Oceanogr. 35:455-463. [Google Scholar]
- 48.Simon, M., H. P. Grossart, B. Schweitzer, and H. Ploug. 2002. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28:175-211. [Google Scholar]
- 49.Sternberg, C., B. B. Christensen, T. Johansen, A. T. Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 51.Tait, K., and I. W. Sutherland 2002. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J. Appl. Microbiol. 92:345-352. [DOI] [PubMed] [Google Scholar]
- 52.Vanhaeke, E., J. P. Remon, M. Moors, F. Raes, D. de Rudder, and A. van Petighem. 1990. Kinetics of Pseudomonas aeruginosa adhesion to 304- and 316-L-stainless steel: role of cell surface hydrophobicity. Appl. Environ. Microbiol. 56:788-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Loosdrecht, M. C. M., J. Lyklema, W. Norde, and A. J. B. Zehnder. 1990. Influence of interfaces on microbial activity. Microbiol. Rev. 54:75-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitekettle, W. K. 1991. Effects of surface-active chemicals on microbial adhesion. J. Ind. Microbiol. 7:105-116. [Google Scholar]