Abstract
A large, intact sulfide chimney, designated Finn, was recovered from the Mothra Vent Field on the Juan de Fuca Ridge in 1998. Finn was venting 302°C fluids on the seafloor and contained complex mineralogical zones surrounding a large open central conduit. Examination of microorganisms within these zones, followed by community analysis with oligonucleotide probes, showed that there were variations in the abundance and diversity of eubacteria and archaea from the exterior to the interior of the chimney. The microbial abundance based upon epifluorescence microscopy and quantitative fatty acid analyses varied from >108 cells/g of sulfide 2 to 10 cm within the chimney wall to <105 cells/g in interior zones. Direct microscopic observation indicated that microorganisms were attached to mineral surfaces throughout the structure. Whole-cell hybridization results revealed that there was a transition from a mixed community of eubacteria and archaea near the cool exterior of the chimney to primarily archaea near the warm interior. Archaeal diversity was examined in three zones of Finn by cloning and sequencing of the 16S rRNA gene. The majority of sequences from the exterior of the chimney were related to marine group I of the Crenarchaeota and uncultured Euryarchaeota from benthic marine environments. In contrast, clone libraries from interior regions of the chimney contained sequences closely related to methanogens, Thermococcales, and Archaeoglobales, in addition to uncultured crenarchaeal phylotypes obtained from deep subsurface sites. These observations of microbial communities within an active hydrothermal chimney provide insight into the microbial ecology within such structures and may facilitate follow-up exploration into expanding the known upper temperature limits of life.
Volcanic and tectonic activity associated with midocean ridge spreading centers supports widespread hydrothermal activity that occurs over lateral distances of hundreds of kilometers and extends deep into the subseafloor (15). Thermal and chemical gradients created by mixing of hydrothermal fluids with seawater and fluid interactions with heated rock support microbial life uniquely adapted to specific extreme environments (30, 48). Sulfide edifices present in marine hydrothermal systems are the seafloor products of hydrothermal circulation, where they serve as conduits for hot, metal-enriched fluids. These structures have steep temperature and chemistry gradients similar to those suspected to exist in the subseafloor, but the gradients are condensed into much smaller spatial scales (15, 18, 30). Previously, researchers have investigated the microbial biomass and diversity within the walls of vent chimneys and associated structures by using culture-based, microscopic, and molecular methods (24, 26, 44, 50, 51). Thermophilic and hyperthermophilic microorganisms are commonly cultured from pieces of chimneys from the deep-sea environment (24, 30, 44). In addition, intact cells have been observed in small pieces of sulfide chimneys, porous beehive structures, and flanges associated with large chimney structures (24, 26, 50, 51), and detailed genetic libraries describing the largely unknown microbial communities that inhabit these environments have been constructed (50, 51). However, few studies have attempted to address the relationship of these microorganisms to the environmental settings present within the various structures.
The Edifice Rex sulfide recovery project in 1997 and 1998 resulted in retrieval of the largest and most pristine hydrothermal vent chimneys yet obtained from the deep-sea environment (14). A black smoker sulfide chimney, designated Finn, was among the structures recovered. Finn was venting 302°C hydrothermal fluid prior to removal from the seafloor. Its exterior was bounded by 2 to 10°C seawater, which created a ∼300°C gradient across the 5- to 42-cm-thick chimney wall. Within this gradient there were environments with intermediate temperatures and chemistry that could support diverse microbial communities (6, 38). Thus, the recovery of Finn provided a unique opportunity to explore the relationships between environmental conditions and the microbiology of deep-sea hydrothermal systems.
In this paper we describe the microbial communities within discrete mineralogical zones from Finn. These zones represent diverse physical-chemical environments due to their spatial location within the chimney wall, mineral composition, pore structure, and degree of mixing between hydrothermal fluids and seawater (54, 55). Petrological and microbiological techniques were employed with replicate samples obtained from defined mineralogical zones to examine the relationships among microbial abundance, diversity, and the microenvironmental conditions present within the walls of the active sulfide chimney. In most previous studies the workers have examined the microbial and abiological components of hydrothermal vent systems separately. In contrast, we used an integrated approach that considered the interrelated geomicrobial processes that operate at multiple scales. Our results show that the abundance and diversity of microbial communities varied significantly in different regions of Finn and that communities composed primarily of Crenarchaeota inhabited high-temperature interior regions of the chimney structure. The data are discussed below in terms of microenvironments present within the chimney structure, which may select for and sustain different physiological and metabolic groups of microorganisms.
MATERIALS AND METHODS
Sample collection and subsampling.
The Mothra Field (48°55N, 129°06W), at a water depth of 2,270 m, is one of five active hydrothermal fields along the Endeavour Segment of the Juan de Fuca Ridge, located approximately 300 km west of Vancouver Island, Canada. Mothra is the largest vent field along the Endeavour Segment and consists of five complexes of steep-sided, porous sulfide pinnacles that rise up to 20 m above the surrounding seafloor (31). Mothra contains multiple chimneys venting 30 to 302°C fluids (31). Four large sulfide structures were recovered from the Faulty Towers (Fig. 1) and Crab Basin complexes within the Mothra Field as part of the Edifice Rex sulfide recovery project (14). The four structures included an inactive structure (Phang), two structures venting warm diffuse fluids (Roane, Gwenen), and a black smoker chimney (Finn) venting 302°C fluids (Fig. 1) (14).
FIG. 1.
West-looking photomosaic of the Faulty Towers complex in the Mothra Vent Field composed of electronic still images taken with the ROV JASON (adapted from the study of Delaney et al. [14]). The textured nature of some of the chimneys reflects the dense growth of macrofaunal communities that colonize the exterior surfaces of active vent structures. Three structures from the Faulty Towers complex (Finn, Roane, and Phang) were recovered as part of the Edifice Rex Sulfide Recovery Project (14). Scale bar = 13 m.
The recovered portion of Finn was 1.6 m long and weighed 1,300 kg in air upon recovery. The chimney wall thickness varied from 5 to 42 cm along the length of the structure. The central conduit of the chimney was approximately 15 to 25 cm wide. Four discrete horizontal transects across the chimney wall of Finn were taken for analysis, and they ranged in thickness from 12 to 20 cm. The transects sampled had the same mineralogical gradients but varied in terms of location, abundance of fractures, and the mineralogy of the exterior surface. Subsamples from these transects were analyzed immediately, flash-frozen in liquid nitrogen, or fixed in 4% paraformaldehyde (12) for future analysis.
Precautions were taken during handling and sampling of the chimneys to preserve their integrity for microbiological analyses and to eliminate potential sources of contamination. Sulfide chimneys were shipboard 2 to 4 h after removal from the seafloor. Large (length, 10 to 25 cm) samples were removed from Finn by using sterilized tools and brought to the laboratory. Anaerobic conditions were maintained by placement of these samples immediately into an N2-filled chamber. Subsamples for microbiological analyses were obtained from mineralogical zones interior to the transects after the outer 0.5 cm of rock material was removed (45). Fixed samples were rinsed in 70% ethanol prior to microscopic analysis to remove transient cells.
Cell extraction and enumeration.
Cell counts were obtained by extracting microorganisms from 5 to 15 g (wet weight) of fixed sulfide material and concentrating the cells by the method of Harmsen et al. (24). This procedure was amended by addition of 30 s of sonication with a 20-kHz microtip ultrasonic probe (Misonix Inc.) following each extraction step to promote cell dissociation from the mineral surfaces. The optimal number of extraction steps (three steps) was defined as the number which yielded the greatest number of cells with the smallest amount of interfering coextracted material. Additional cycles did not significantly improve cell recovery but contributed to greater contamination from small mineral particles. Following extraction, cells were counted by filtration onto 0.2-μm-pore-size black polycarbonate filters and staining with DAPI (4′,6-diamidino-2-phenylindole) (Sigma) (40). The filters were subsequently examined by using an epifluorescence microscope with a filter for DAPI. The criteria used for counting particles as cells were blue fluorescence after staining with DAPI, cell diameters between 0.5 and 5.0 μm, and round, rod-shaped, or filamentous morphology. In certain cases, dual staining with DAPI and acridine orange was used (17). Each subsample from the four transects was extracted at least twice independently. At least 30 fields/filter, typically containing 10 to 30 cells/field, were counted for three filters per extraction. For samples with small amounts of biomass (<105 cells/g) either 50 fields or 200 cells were counted. Unextracted, ground sulfide material was also examined by staining with DAPI to characterize the architecture of attached microbial communities within the rock matrix.
Lipid analysis.
Polar lipid fatty acid (PLFA) and ether lipid biomarkers provided an independent measurement of biomass within the structure. Microbial lipids were extracted from approximately 5.0 g of frozen sulfide and separated by using the modified method of Bligh and Dyer (10, 57) by Microbial Insights Inc. (Rockford, Tenn.), and they were analyzed by gas chromatography-mass spectrometry (limit of detection, 50 pmol). PLFA were analyzed for subsamples obtained throughout the mineralogical gradients mentioned above. Ether lipids were analyzed only for samples from the exterior regions of the sulfide structure.
Oligonucleotide probe design.
Probes specific for the archaeal orders Thermococcales and Methanococcales were designed by comparison of alignments of the complete sequences of the 16S rRNA gene of target organisms with sequences from the closest nontarget relatives (obtained from the National Center for Biotechnology Information [NCBI]) by using the BioEdit sequence alignment editor, version 5.0.9 (22). Potential probe sequences were submitted to the Probe Match function of the Ribosomal Database Project II (35) and to BLAST (NCBI) (3) to examine their utility as probes for the organisms of interest. Probes that were unique to the target organisms and were free of structural concerns (21) were synthesized and labeled at the 5′ end with the indocarbocyanine dye Cy3 (Qiagen-Operon). Probes targeting thermophilic and hyperthermophilic Methanococcales (MC688; 5′-GGATCAACRCATTTCACCGC-3′) and Thermococcales (TC589; 5′-CGCCAGGGACTTACGGGC-3′) species were the results of this process. MC688 and TC589 were empirically tested by hybridization at various stringencies with target and nontarget laboratory cultures (12, 50). MC688 was combined with an additional Methanococcus-targeted probe, MC1109 (41), in an attempt to ensure comprehensive coverage of the Methanococcales.
FISH.
Fluorescence in situ hybridization (FISH) was performed with domain- and group-specific oligonucleotide probes (EUB338, ARC915, EUK502, EURY498, CREN499, MC688, MC1109, TC589) (Table 1) labeled with the fluorescent dye Cy3 or 6-FAM (5′-fluorescein phosphoramidite) (Qiagen-Operon) by the method of Bond and Banfield (12), with minor modifications. A control probe complementary to EUB338 (non-EUB338) was also employed to evaluate nonspecific probe binding. FISH specificity at various formamide concentrations was evaluated by hybridization to multiple target and nontarget reference cells (12). The empirically determined optimal formamide concentration for each probe was used in subsequent reactions and is shown in Table 1. When multiple probes were used in a single experiment, the more stringent conditions (higher formamide concentration) were employed. Hybridizations were performed either directly with fixed, ground rock material or with extracted cells filtered onto 0.2-μm-pore-size black polycarbonate filters (37). Poly(A) (0.1 mg/ml; Sigma) and bovine serum albumin (0.2 mg/ml; Sigma) were added to the hybridization solutions due to the presence of small mineral particles in the cell extracts, which tended to bind the probes (24). Following hybridization, samples were rinsed in distilled water, air dried, counterstained with DAPI, and mounted with Vectashield (Vector Laboratories). Only cells that were simultaneously stained with DAPI and a probe were counted in FISH experiments. Counting with subdomain probes (EURY498, CREN499, TC589, MC688, MC1109) typically yielded cell densities below statistically optimal values (<10 cells/field). Therefore, at least 50 fields/filter were counted for FISH experiments.
TABLE 1.
Oligonucleotide probes used in this study and optimized experimental conditions
| Probe | OPD namea | Specificity | Sequence (5′-3′)b | Formamide concn (%)c | Reference |
|---|---|---|---|---|---|
| ARC915 | S-D-Arch-0915-a-A-20 | Archaea | GTGCTCCCCCGCCAATTCCT | 30 | 46 |
| EUB338 | S-D-Bact-0338-a-A-18 | Eubacteria | GCTGCCTCCCGTAGGAGT | 30 | 5 |
| EUK502 | S-D-Euk-0502-a-A-16 | Eukarya | ACCAGACTTGCCCTCC | 15 | 4 |
| EURY498 | S-K-Eury-0498-a-A-14 | Euryarchaeota | CTTGCCCRGCCCTT | 10 | 13 |
| CREN499 | S-K-Cren-0499-a-A-18 | Crenarchaeota | CCAGRCTTGCCCCCCGCT | 20 | 13 |
| MC688 | S-G-Mcocc-0688-a-A-20 | Methanococcales | GGATCAACRCATTTCACCGC | 30 | This study |
| MC1109 | S-G-Mcocc-1109-a-A-20 | Methanococcales | GCAACATRGGGCACGGGTCT | 30 | 41 |
| TC589 | S-G-Tcocc-0589-a-A-18 | Thermococcales | CGCCAGGGACTTACGGGC | 25 | This study |
Oligonucleotide Probe Database (OPD) nomenclature (2).
R = G or A.
Optimal deionized formamide concentration in hybridization solutions.
Microscopy.
Fluorescently stained samples were examined with a Zeiss UEM epifluorescence microscope (Carl Zeiss Inc.) with filters for DAPI (excitation wavelength, 365 nm; emission wavelength, 397 nm), fluorescein isothiocyanate (excitation wavelength, 450 to 490 nm; emission wavelength, 520 nm), and Cy3 (excitation wavelength, 545 nm; emission wavelength, 565 nm). Frozen samples were analyzed with a JEOL model 6100 scanning electron microscope (SEM) with a cryogenically cooled stage and an energy-dispersive X-ray spectrometer (EDS) at the Image and Chemical Analysis Laboratory at Montana State University (Bozeman).
DNA extraction and purification.
Frozen rock samples were ground into 5- to 10-mm-diameter particles with a sterilized mortar and pestle, and total nucleic acids were extracted and purified by the method of Edwards et al. (20) and stored at −20°C until they were used.
PCR.
16S rRNA genes were amplified from purified DNA by using the universal archaeal primers Arch21F and Arch958R (16). Each PCR mixture (20 μl) contained 2.5 mM MgCl2, 0.8 mM deoxynucleoside triphosphates, each primer at a concentration of 0.25 μM, 1× PCR buffer (Promega), and 1 U of Taq polymerase (Promega). The following protocol was used for primers Arch21F and Arch958R: an initial denaturation step of 94°C for 5 min and then 30 to 35 cycles of 94°C for 20 s, 55°C for 45 s, and 72°C for 2 min, followed by a final extension step of 72°C for 10 min. To minimize PCR bias due to a high number of cycles, parallel PCRs were stopped after various numbers of cycles (20, 25, 30, 35, and 40 cycles), and the products were quantified on a 1% (wt/vol) agarose gel stained with SYBR green (Molecular Probes). The lowest number of cycles that produced sufficient amplification product was chosen (typically 25 to 30 cycles) and used for subsequent reactions. Eight to 10 parallel reaction mixtures were pooled, purified on a 1% agarose gel, extracted with a QIAquick gel extraction kit (Qiagen), and used in subsequent cloning steps.
Cloning of archaeal 16S rRNA genes.
Amplified double-stranded DNA from environmental samples was cloned into a plasmid vector in Escherichia coli by using a TOPO-TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. A total of 150 to 200 clones were selected per sample and stored on agar plates containing ampicillin (100 μg/ml). Clones were grown in 100 μl of Luria-Bertani broth for 1 h with shaking (220 rpm) at 37°C and screened for the presence of the correct-size insert by PCR by using the vector-specific primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′). Each reaction mixture (50 μl) contained 1 μl of clone culture, 1.5 mM MgCl2, 0.8 mM deoxynucleoside triphosphates, each primer at a concentration of 0.4 μM, 1× PCR buffer (Promega), and 1 U of Taq polymerase (Promega). The PCR conditions were an initial denaturation step of 94°C for 2 min and then 30 cycles of 94°C for 30 s, 54°C for 45 s, and 72°C for 2 min, followed by a final annealing step at 72°C for 10 min. PCR products were visualized on a 1% (wt/vol) agarose gel stained with SYBR green.
Restriction fragment length polymorphism analysis.
PCR-amplified archaeal inserts were digested with the restriction enzymes RsaI and HaeIII (Promega) according to the manufacturer's instructions. The DNA fragments were separated on a 2.5% agarose gel electrophoresed in Tris-borate-EDTA buffer and stained with SYBR green. The different banding patterns were noted, and a representative clone for each pattern was chosen for sequencing.
Sequencing and phylogenetic analysis.
Sequences were obtained from cleaned, PCR-amplified DNA by using the sequence analysis facility at the University of Washington Marine Molecular Biotechnology Laboratory. Two hundred nanograms of M13-amplified DNA was sequenced with primers Arch21F, Arch515R (5′-TTACCGCGGCKGCTGRCAC-3′), and Arch958R by using a Thermosequenase II dye terminator sequencing kit (Amersham Pharmacia Biotech Inc.) and were analyzed by using a MegaBACE 1000 sequencer (Molecular Dynamics). Approximately 600 nucleotide bases of sequence from representative clones was used in the phylogenetic analyses, and only homologous positions were included in the comparisons. Chromatographs were analyzed with Sequencher, version 4.0.5 (Gene Codes Corp.), and were checked for chimeric sequences by using the CHIMERA_CHECK program of Ribosomal Database Project II (RDP II) (35). Nonchimeric sequences were generally aligned by using the RDP Sequence Aligner program (35). Processed sequences were checked against the NCBI database by using the Advanced BLAST search program (NCBI) (3) and were manually aligned with the closest matches obtained from this program by using BioEdit, version 5.0.9 (22). Sequence similarity was calculated by using BioEdit, and representative sequences of each phylotype (similarity, >97%) were used to construct distance (NEIGHBOR and FITCH) trees by using the Phylip software package (version 3.5; obtained from J. Felsenstein, University of Washington, Seattle), with negative branch lengths prohibited in the analyses. Bootstrap analysis (with SEQBOOT) was used to provide confidence estimates of tree topologies.
Mineralogical analyses.
Subsequent to retrieval and subsampling of Finn for microbiological analyses, the structure was cut axially and vertically. The mineralogy and physical structures interior to the walls of Finn were mapped on slabbed surfaces at subcentimeter scales. These results were integrated with analyses of petrographic thin sections, which were taken from multiple transects across the chimney wall. Mineral abundance, petrogenesis, and porosity were characterized by transmitted and reflected light microscopy. In concert, results of these analyses provided a basis for interpreting the petrology, flow regimens, and growth history of the sulfide structure. A more detailed geological description of Finn is the focus of another study, and the results will be presented elsewhere (39; D. S. Kelley, J. R. Delaney, M. O. Schrenk, and J. A. Baross, Eos Trans. AGU Fall Meet. Suppl. 80[86], abstr. B42B-10, 1999; Kelley and Delaney, unpublished data).
Nucleotide sequence accession numbers.
The sequences reported in this study have been deposited in the GenBank database under accession numbers AY165965 to AY166119 (Z1 and Z2) and AY182063 to AY182135 (Z3).
RESULTS
Sample description.
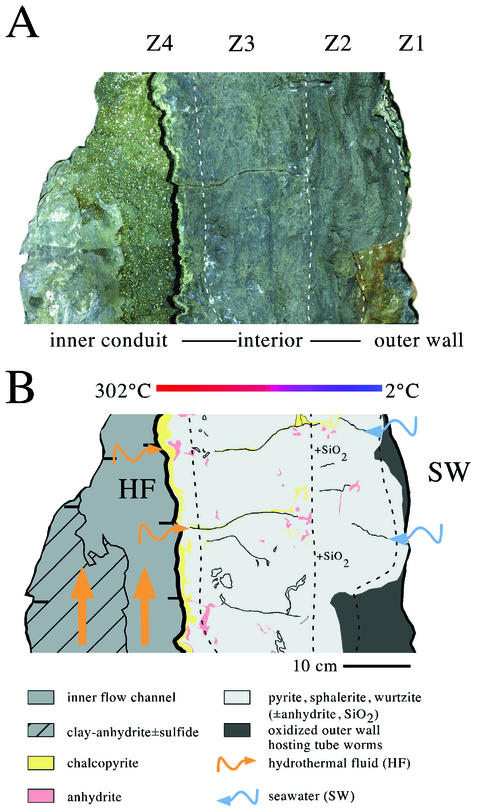
Once shipboard, several of the chimneys were still warm (Gwenen, >60°C; Roane, >90°C; and Finn, steaming), and reducing conditions were evident within pore spaces of Finn because of the odor of H2S emitted when the rock was split. The internal structure of Finn consisted of a chalcopyrite (CuFeS2)-lined central conduit surrounded by complex porous zones predominantly composed of pyrite (FeS2) and wurtzite-sphalerite (ZnS), silicified sulfide (pyrite, ZnS, and SiO2), and oxidized, tube worm-influenced outer wall material (Fig. 2 and Table 2). The mineralogical regions described above were designated zone 4 (Z4) (interior) through zone 1 (Z1) (exterior) and are referred to by these designations below. Rare, mineralized fractures originated from both the exterior and interior of the structure. Subsampling was based upon spatial relationships within the chimney wall and the mineralogy of the various zones.
FIG. 2.
High-resolution photomosaic of a cross-section through a cut face of Finn (A) and corresponding schematic diagram (B) showing key mineralogical and structural features. Fractures originating from the exterior and interior portions of the chimney indicate that there was active fluid entrainment into and out of the chimney wall. The temperature of exiting hydrothermal fluid was measured in situ and was determined to be 302°C. Measurement of the temperature of diffuse flow along the exterior surface of the chimney typically yielded values between 2 and 20°C.
TABLE 2.
FISH-direct count analysis of microbial populations within Finna
| Type of analysis | Z1
|
Z2
|
Z3
|
Z4
|
||||
|---|---|---|---|---|---|---|---|---|
| Cell concn (cells/g [dry wt] of sulfide)b | % of probed cellsc | Cell concn (cells/g [dry wt] of sulfide)b | % of probed cellsc | Cell concn (cells/g [dry wt] of sulfide)b | % of probed cellsc | Cell concn (cells/g [dry wt] of sulfide)b | % of probed cellsc | |
| DAPI | (1.9 ± 0.2) × 106 | (1.7 ± 0.2) × 108 | (1.9 ± 0.3) × 107 | (2.1 ± 1.2) × 105 | ||||
| ARCH915 | (6.3 ± 1.1) × 105 | 33 | (1.1 ± 0.3) × 108 | 65 | (1.2 ± 0.1) × 107 | 63 | (4.8 ± 2.7) × 104 | 23 |
| EUB338 | (1.0 ± 0.3) × 106 | 53 | (4.4 ± 1.3) × 107 | 26 | (3.8 ± 1.1) × 105 | 2 | ND | 0 |
| EUK502 | (3.8 ± 1.2) × 104 | 2 | NDd | 0 | ND | 0 | ND | 0 |
| CREN499 | (1.7 ± 0.3) × 105 | 9 | (5.8 ± 0.8) × 107 | 34 | (8.3 ± 1.2) × 106 | 44 | (2.8 ± 0.4) × 104 | 11 |
| EURY498 | (3.0 ± 0.9) × 105 | 16 | (2.5 ± 0.6) × 107 | 15 | (2.8 ± 0.8) × 106 | 15 | (8.3 ± 2.0) × 103 | 3 |
| TC589 | (1.6 ± 1.6) × 104 | <1 | (3.0 ± 1.0) × 106 | 2 | (3.6 ± 2.4) × 105 | 2 | (1.9 ± 1.9) × 103 | <1 |
| MC688 + MC1109 | (2.3 ± 1.2) × 104 | <1 | (6.0 ± 1.2) × 106 | 4 | (9.6 ± 3.6) × 105 | 5 | (1.7 ± 1.7) × 103 | <1 |
| Lipid | 3.4 × 106e | 1.7 × 108e | ND | ND | ||||
The predominant mineralogy of the zones was as follows: Z1, pyrite and ZnS (wurtzite and sphalerite) with or without marcasite and with or without iron oxyhydroxides; Z2, pyrite, ZnS (wurtzite and sphalerite), and amorphous silica with or without barite and with or without clay; Z3, pyrite and ZnS (wurtzite and sphalerite) with or without chalcopyrite and with or without anhydrite; and Z4, chalcopyrite with or without anhydrite and with or without ZnS (wurtzite and sphalerite). The average porosities of the mineralized zones based on petrological thin sections examined at a magnification of × 50 (500-μm field of view) were as follows: Z1, 30 to 40%; Z2, 15 to 20%; Z3, 20 to 25%; and Z4, 5 to 10%.
For all data except the lipid analyses, the data are means ± 95% confidence intervals for four transects. The limit of detection was 1.6 × 103 cells/g.
Percentage of probed cells in the DAPI-stained total population.
ND, not detected.
Lipid-derived prokaryotic cell density determined by using the conversion factor of Balkwill et al. (7). The limit of detection was 1.25 × 106 cells/g.
Microscopy.
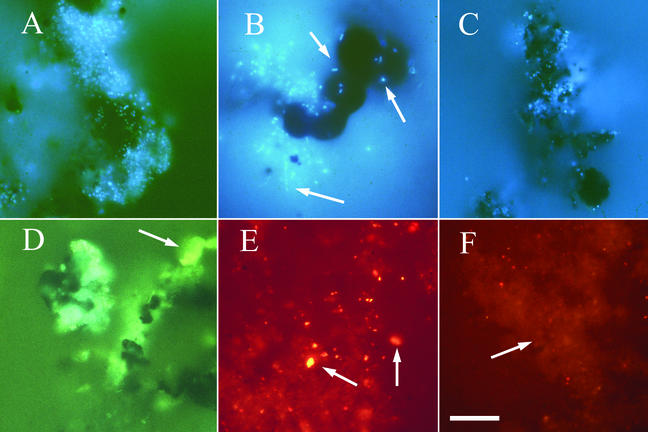
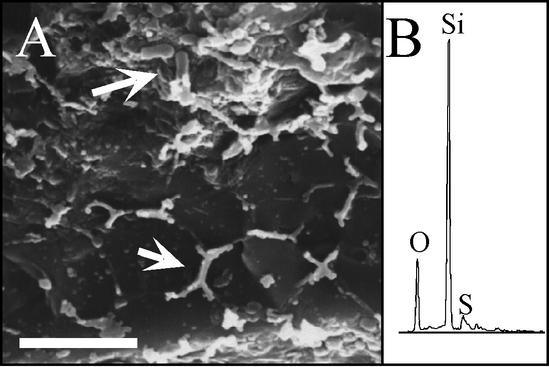
Microscopic observation of chimney material revealed that microorganisms were attached to mineral surfaces, forming multicellular biofilms and cell clusters (Fig. 3A to C). The most extensive microbial communities were in Z1 to Z3 of the chimney. The biofilms in Z1 consisted primarily of filaments and rods. In Z2, the biofilms were more than 10 μm thick and contained filaments, short rods, and cocci (Fig. 3B). SEM-EDS analysis showed that many organisms from this zone were coated with silica (Fig. 4). Morphologies similar to those determined by SEM were also stained with DAPI, suggesting that the structures observed were microorganisms. Attached microbial communities in Z3 were sporadically distributed and consisted almost entirely of small cocci (diameters, ∼1 μm) aggregated into biofilms and microcolonies (diameters, 5 to 10 μm) on mineral surfaces (Fig. 3A and C). An extremely patchy cell distribution was observed in Z4, with attached microorganisms occurring as individual cocci or as microcolonies consisting of a few cells. However, the low biomass and coconcentration and autofluorescence of small minerals upon extraction interfered with detailed analysis of these samples.
FIG. 3.
Epifluorescence photomicrographs of fluorescently stained microbial communities inhabiting the walls of Finn. (A to C) Images obtained by DAPI staining of attached microbial communities. (D to F) Images obtained by FISH to mineral surfaces and cell extracts. Biofilms were a common feature in Z2 and Z3 (A) and in some cases were more than 10 μm thick. Dispersed cell clusters (B) and microcolonies (C) were also observed within Z2 and Z3, respectively. Attached communities were composed of cells with diverse morphologies, including rods, cocci, and filaments (arrows in panel B). (D) Whole-cell hybridization of a microbial biofilm attached to unextracted rock material from Z3 with ARC915 (green fluorescence). Background fluorescence of the mineral substratum (yellow-green) is indicated by an arrow. (E) FISH to a cell extract from Z2, showing the nonspecific binding of probe ARC915 (red fluorescence) to small mineral particles (indicated by arrows). In this experiment we did not use the blocking agents bovine serum albumin and poly(A), which were typically used in FISH experiments in this study. (F) FISH to a cell extract from Z3 with the crenarchaea-specific probe CREN499 (red fluorescence). The diffuse fluorescence of nonspecific probe binding is indicated by an arrow, in contrast to the point sources of fluorescence (red dots), which were counterstained with DAPI and were counted as hybridized cells. All images were captured with a 35-mm microscope camera, scanned, and processed in Adobe Photoshop, version 6.0. (Adobe). Scale bar = 15 μm.
FIG. 4.
(A) Cryo-SEM image of rod-shaped microbial cells (arrows) attached to a fractured mineral surface within Z2 of Finn. Similar morphologies were observed by epifluorescence microscopy. (B) EDS spectra indicated that cells were coated with silica (SiO2). Scale bar = 10 μm.
Biomass estimates.
PLFA analysis and cell extraction and enumeration indicated that there was widespread distribution of microbial biomass within Finn (Table 2). Between 4 and 2.3 × 104 pmol of PLFA/g (dry weight) of sulfide was detected in Z1 and Z2, which corresponds to a cell density of 7 × 104 to 4.6 × 108 cells/g (dry weight) based upon a conversion factor of 2.5 × 104 cells/pmol of PLFA, as described by Balkwill et al. (7). In Z1 the PLFA were composed of monoenoic (indicative of gram-negative bacteria), normal saturated, and microeukaryotic lipids. Z2 contained a more diverse range of lipid types, including predominantly monoenoic and normal saturated PLFA, as well as terminally and mid-branched saturated PLFA (indicative of sulfate-reducing bacteria) and branched monoenoic PLFA (indicative of obligate anaerobes, including sulfate- and iron-reducing bacteria). Ether lipids were analyzed and detected only in Z1 and Z2 and were composed entirely of diether lipids in both cases. The cell densities obtained by epifluorescence microscopy were between 1.5 × 105 and 3.9 × 106 cells/g (dry weight) in the outer portion of the chimney (Z1). The cell density increased to 7.2 × 107 to 2.9 × 108 cells/g (dry weight) in Z2. Z3 contained between 2.5 × 106 and 7.9 × 107 cells/g (dry weight). Within Z4 the cell abundance varied significantly between samples, but the average was 2.1 × 105 cells/g (dry weight).
FISH.
A high percentage of cells in Z1 and Z2 were stained with the probes used in this study (>85% of the DAPI-stained cells hybridized when ARC915 and EUB338 were used simultaneously). The hybridized cells in Z1 consisted of both eubacteria (53%) and archaea (33%) (Table 2). FISH indicated that organisms in Z2 also consisted of eubacteria and archaea, although archaea made up a far greater percentage of the hybridized community in this region than in Z1 (26% eubacteria versus 65% archaea). In Z3 and Z4 nearly all of the cells that hybridized were archaea, although in Z4 <25% of cells hybridized with the rRNA-targeted probes. FISH analysis with subdomain probes revealed that members of the Euryarchaeota were a predominant part of the hybridized archaeal community in Z1 (48%). In Z2 to Z4, members of the Crenarchaeota made up a larger portion of the hybridized archaea (48 to 69%). Cells that were stained with the TC probe (TC589) accounted for a consistent but low (<3%) portion of the hybridized cells. Cells that stained with the MC probe cocktail (MC688 plus MC1109) exhibited peak abundance (4 to 5%) within Z2 and Z3 of the chimney (Table 2).
Analysis of archaeal 16S rRNA genes.
DNA was successfully extracted from one rock sample from Z1 and two samples each from Z2 (Z2a and Z2b) and Z3 (Z3a and Z3b), and five clone libraries of archaeal 16S rRNA genes were constructed from these samples. Partial sequence analysis of 228 clones revealed 64 different phylotypes based upon 97% sequence similarity of approximately 600 homologous bases (Table 3). The clone sequences correspond closely to the sequences of known organisms cultured from hydrothermal vents and to the sequences of many groups of unknown microorganisms found in terrestrial and marine environments (1, 8, 27, 28, 36, 47, 49-51, 56) (Table 3 and Fig. 5). However, some sequences were not closely related to previously characterized microorganisms and may represent novel phylogenetic diversity beyond the species level.
TABLE 3.
Summary of archaeal 16S rRNA clone sequences from Finn Z1 to Z3
| Phylogenetic group | Clone(s)a | Closest match (accession no.)b | Similarityc | No. of clonesd
|
||||
|---|---|---|---|---|---|---|---|---|
| Z1a | Z2a | Z2b | Z3a | Z3b | ||||
| Korarchaeota | FZ2bA214 | pOWA133 (AB007303) | 0.94 | 1 | ||||
| Crenarchaeota | ||||||||
| Marine group I | FZ1aA3, FZ1aA5, FZ1aA6, FZ1aA14, FZ1aA67, FZ1aA159, FZ1aA160, FZ1aA163, FZ2aA59, FZ2bA18, FZ2bA16, FZ2bA30, FZ2bA64, FZ2bA101, FZ2bA206 | JTB167 (AB015275) | 0.94-0.97 | 17 | 6 | 20 | 1 | 9 |
| UCI | FZ1aA18, FZ1aA60, FZ2bA20, FZ3aA1, FZ3aA20, FZ3bA114 | SUBT-14 (AF361211) | 0.84-0.92 | 6 | 15 | 1 | 22 | 29 |
| UCIIa | FZ2bA12, FZ2bA54, FZ2bA108 | 20a-6 (AJ299151) | 0.91-0.93 | 11 | ||||
| UCIIb | FZ2aA56, FZ2bA4 | 20a-6 (AJ299151) | 0.93-0.94 | 1 | 1 | |||
| UCIII | FZ2bA61, FZ3aA130 | pISA9 (AB019732) | 0.84-0.88 | 1 | 1 | |||
| Thermoproteales | FZ2aA55 | GBa2r048 (AF419633) | 0.93 | 1 | ||||
| UCIV | FZ2bA53, FZ2bA211, FZ2bA221 | pJP89 (L25305) | 0.87-0.89 | 4 | ||||
| Euryarchaeota | ||||||||
| UEI | FZ1aA12, FZ1aA168, FZ2aA72, FZ2bA36, FZ2bA106, FZ2bA167, FZ3bA23 | CRA13-11 (AF119124) | 0.84-0.96 | 20 | 6 | 11 | 3 | 4 |
| UEII | FZ1aA158, FZ2aA51, FZ2bA26, FZ2bA42, FZ2bA63, FZ2bA121, FZ2bA105 | G26_C49 (AF356632) | 0.78-0.83 | 4 | 4 | 9 | ||
| UEIIIa | FZ1aA166, FZ2bA110 | DOUR010 (AF201364) | 0.73-0.76 | 1 | 1 | |||
| UEIIIb | FZ2bA11, FZ2bA69 | pMC2A17 (AB019747) | 0.72-0.75 | 2 | ||||
| UEIV | FZ2aA5, FZ2bA3, FZ2bA112, FZ2bA217 | pMC2A24 (AB019736) | 0.83-0.87 | 1 | 3 | |||
| Methanosarcinales | FZ2bA17, FZ2bA209 | ARB5 (AF293016) | 0.85-0.93 | 4 | ||||
| Methanococcales | FZ3aA10 | 33-P62-A98 (AF355922) | 0.96 | 1 | ||||
| Palaeococcus type | FZ2bA58, FZ2bA113 | GR-1 (AF069725) | 0.94-0.95 | 3 | ||||
| Thermococcales | FZ3aA26, FZ3aA31, FZ3bA104 | T. celer (M21529) | 0.93-0.99 | 2 | 1 | |||
| Archaeoglobales | FZ2aA25 | pMC2A228 (AB019735) | 0.98 | 1 | ||||
Representative 16S rRNA sequence(s) used in phylogenetic analyses, based upon 97% similarity grouping.
Clone name (GenBank accession number) based upon the results of a BLAST search (NCBI).
Based upon the results from a similarity matrix of unambiguously aligned nucleotides. A value of 1.00 indicates that sequences are identical.
The total numbers of clones analyzed for Z1a, Z2a, Z2b, Z3a, and Z3b were 48, 35, 72, 30, and 43, respectively. The numbers of different phylotypes were 21, 16, 35, 11, and 7, respectively.
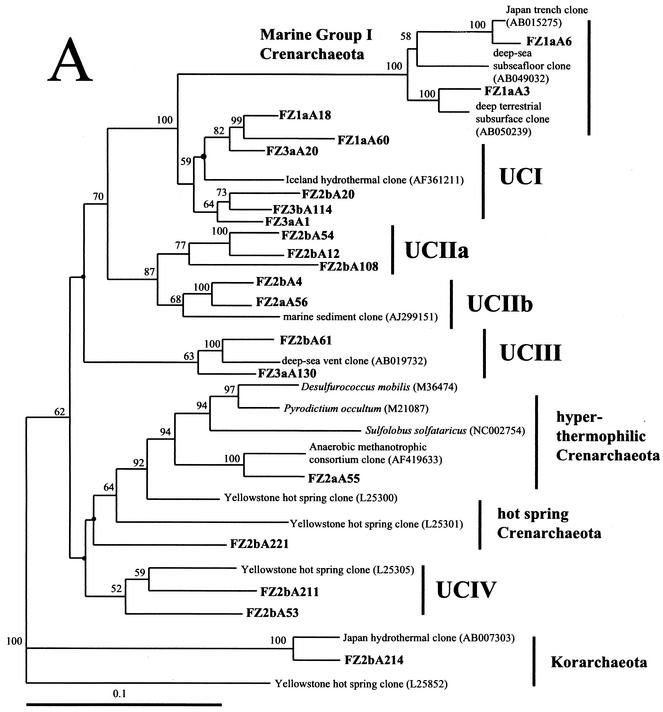
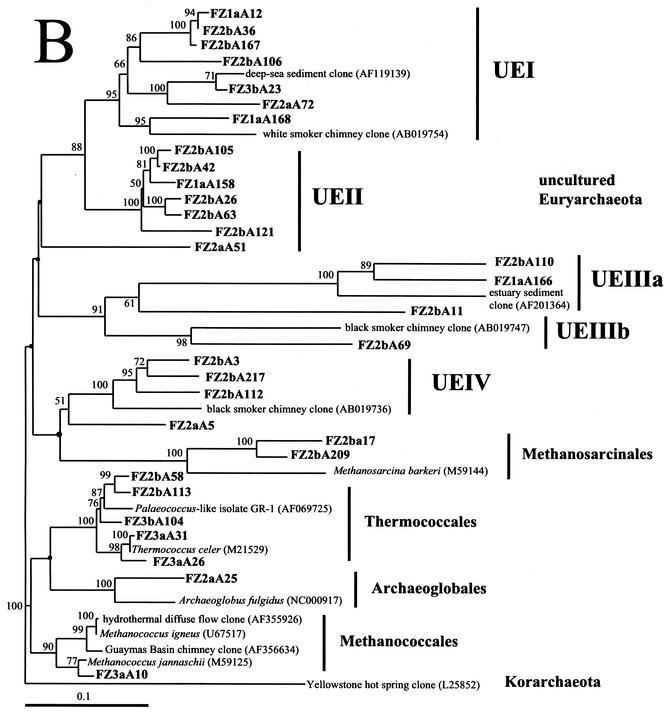
FIG. 5.
Phylogenetic trees based upon 16S rRNA gene sequences of archaea, including clones obtained from Z1 to Z3 of the black smoker chimney Finn. Each of the trees was inferred by phylogenetic analysis of approximately 600 homologous positions of sequences by using the Fitch-Margolis distance-based method. The numbers in parentheses are the GenBank accession numbers for sequences obtained from NCBI. Percentages of 100 bootstrap resamplings greater than 50% are indicated adjacent to the corresponding nodes. The bootstrap values were less than 50% for the branch points marked with solid circles and no numerical values. Scale bars = 0.1 expected nucleotide change per sequence position. (A) Tree depicting the phylogenetic diversity of Crenarchaeota in Z1 to Z3 of Finn. Some clone phylotypes in the marine group I Crenarchaeota were omitted for simplicity. (B) Tree indicating the phylogenetic relationships of Euryarchaeota in Z1 to Z3 of Finn. FZ1a is from Z1 of Finn, FZ2a and FZ2b are from Z2 of Finn, and FZ3a and FZ3b are from Z3 of Finn. Corresponding data are presented in Table 3.
Cloning of the archaeal 16S rRNA gene from Z1 revealed a total of 21 unique phylotypes among the 48 clones analyzed (Table 3). A majority of these clones were related to uncultured organisms in marine group I of the Crenarchaeota or to uncultured members of the Euryarachaeota from benthic marine environments (groups UEI and UEII). Six sequences from the Z1 library were similar to an uncultured crenarchaeal group (group UCI) obtained from subterranean hot springs (36). Two clone libraries were constructed from separate DNA extractions of Z2 (Z2a and Z2b). Thirty-five and 72 clones were analyzed from libraries Z2a and Z2b, respectively (Table 3). Some of the crenarchaeal sequences obtained from Z2 were related to marine group I, like sequences found in Z1. A high level of UCI sequences was found in the Z2a library (15 of 23 Crenarchaeota sequenced), in addition to a single sequence (FZ2aA55) related to the Thermoproteales. The Z2b library contained several unique phylotypes, including sequences related to uncultured hot spring, subsurface, and benthic marine microorganisms (groups UCII to UCIV) and one clone (FZ2bA214) similar to a korarchaeal sequence from a Japanese hydrothermal environment (8, 36, 49). Among the Euryarchaeota, some of the sequences identified in the Z2a and Z2b libraries were similar to the uncultured euryarchaeal groups found in Z1 (groups UEI, UEII, and UEIIIa). However, we found some groups that were not present among the sequences from the Z1 library, including uncultured Euryarchaeota (groups UEIIIb and UEIV), methanogens (Methanosarcinales), and sequences closely related to groups of hyperthermophilic Euryarchaeota (Thermococcales, Archaeoglobales) (Table 3). The Z3 clone libraries (Z3a and Z3b) were dominated by a cluster of related crenarchaeal phylotypes (group UCI), which accounted for 49 of the 73 clones sequenced (Table 3). Sequences related to the orders Thermococcales and Methanococcales were also found in the Z3 libraries.
Fifty-three marine group I sequences from Z1 to Z3 of Finn were closely related (94 to 97%) to sequences of uncultured microorganisms obtained from marine environments. Group UCI contained the single most abundant phylotype within the clone libraries and accounted for 71 of the 228 archaeal clones sequenced (Table 3). These clones were 84 to 92% related to SUBT-14, a sequence obtained from a low-salinity, subterranean hot spring in Iceland (36) (Fig. 5A). UCIIa, UCIIb, UCIII, and UCIV clones were present only in the Z2 and Z3 archaeal libraries and were related primarily to sequences of uncultured organisms obtained from terrestrial and marine hot spring environments in Yellowstone National Park, Iceland, and Japan (8, 36, 49) (Fig. 5A). A single sequence (FZ2aA55) from the UCIV cluster was related to the hyperthermophilic crenarchaeal group Thermoproteales. Finally, a clone from the Z2b library (FZ2bA214) was closely related (94%) to a korarchaeal sequence obtained from a hydrothermal environment at the Suiyo Seamount near Japan (49).
Among the Euryarchaeota, UEI and UEII sequences were abundant in the Z1 and Z2 clone libraries, and UEI sequences were present in the Z3 clone libraries (Table 3). UEI clones were related primarily to sequences from benthic marine environments (1, 56), whereas UEII sequences formed a distinct group (Fig. 5B). UEII sequences were only 78 to 83% similar to previously published sequences based upon similarity matrices (Table 3). UEIII sequences formed two distinct clusters (clusters UEIIIa and UEIIIb) (Fig. 5B). UEIIIa was most closely related to sequences obtained from marine sediments (1), although the levels of similarity between sequences were low (73 to 76%). UEIIIb encompassed sequences from hydrothermal chimney environments and contained only clones from Z2 (50, 51). UEIV was comprised of sequences from diverse deep-sea hydrothermal vent environments and formed a cluster with clones related to the methanogenic group Methanosarcinales (27, 50). A methanogen-related clone (FZ3aA10) that was similar to the Methanococcales was also detected in the Z3a library. Finally, the Z2 and Z3 clone libraries contained sequences related to cultured strains of Thermococcales and Archaeoglobales, many of which have been isolated from deep-sea hydrothermal vent environments (Table 3). Three clones from Z2 were 94 to 95% similar to cultured strains of Palaeococcus, an anaerobic, hyperthermophilic, iron-reducing heterotroph (52). Three Thermococcales sequences were found in the Z3 clone libraries, and they included a sequence that was 93% similar to the sequence of an Fe(III)-reducing hyperthermophilic isolate from the East Pacific Rise (44) and two sequences related to Thermococcus celer. One sequence from Z2 (FZ2aA25) was closely related to an Archaeoglobus sequence (98%) obtained from a deep-sea black smoker chimney (50).
DISCUSSION
Thermal and chemical gradients within the walls of active hydrothermal chimneys can select for and sustain organisms adapted to specific environmental conditions. On this basis, the compositions of microbial communities within different microenvironments, such as the exterior and interior of a sulfide chimney, are expected to be vastly different. This rationale was used to investigate the diversity of microbial communities within defined mineralogical zones of the 302°C hydrothermal chimney designated Finn. Microscopic observation of mineral surfaces and cell extracts from Finn revealed intact microorganisms within all zones of the chimney wall. Overall, cell abundance was greatest in the three outermost zones of the chimney wall (Z1 to Z3; 1.9 × 106 to 1.7 × 108 cells/g of sulfide). These results compare favorably with other measurements of microbial abundance in the middle and exterior regions of small active chimney structures (24, 26, 50, 51) and likely reflect the relatively moderate temperatures and diverse and abundant nutrient and energy sources in these regions (30). Multispecies biofilms composed of eubacteria and archaea were abundant in Z1 and Z2 of Finn. In contrast, the FISH-stained microbial communities in Z3 and Z4 were nearly entirely small archaeal cocci. Archaeal 16S rRNA sequences from Finn revealed diverse phylotypes within the chimney, ranging from marine group I Crenarchaeota and benthic Euryarchaeota near the exterior of the chimney to predominantly uncultured Crenarchaeota and hyperthermophilic Euryarchaeota near the interior of the chimney. Surprisingly, intact cells were also detected in Z4 in material less than 1 to 4 cm away from the conduit venting 302°C fluids.
Z1: chimney exterior.
Z1 encompassed the outer 2 to 3 cm of the chimney wall and was characterized by an abundance of oxidized sulfide minerals and the presence of macrofauna. Metal sulfides exposed to cold, oxygenated seawater rapidly undergo oxidation in the deep sea and may support unique microbial communities adapted to this favorable metabolic couple (6, 19). Macrofauna likely influence microbial communities in Z1 directly through symbioses and excreted organic material and indirectly as flow channels (fossilized tube structures) that allow perfusion of seawater into the chimney wall. The direct contact of Z1 with 2°C seawater, the presence of living and partially fossilized macrofauna, and the abundance of oxidized minerals result in a low-temperature, organic matter-rich, oxygenated environment.
The microbial communities in Z1 (and to a lesser extent in Z2) contained abundant eubacteria and archaea, as reflected by both FISH data (Table 2) and lipid analysis results. Previous studies have also found that eubacteria occur at similar if not higher levels than archaea in the exterior portions of sulfide chimneys (24, 25, 51). Diverse eubacterial species are known to be associated with chimneys, including animal symbionts, thermophilic hydrogen oxidizers, and members of the ɛ subclass of the class Proteobacteria (25, 42). The exterior and outermost walls of sulfide chimneys, percolating diffuse fluids, provide low-temperature (2 to 30°C), energy-rich environments where oxygen-respiring microorganisms and macrofauna and their associated symbionts can flourish (34, 38, 43). At higher temperatures (>60°C) in reducing areas of the chimney wall, thermophilic and hyperthermophilic, anaerobic and microaerophilic genera may be important components of the microbial community and carry out processes such as hydrogen oxidation and sulfur reduction (25, 33, 42).
A number of 16S rRNA sequences found in Finn can be used to carefully infer environmental conditions within the chimney wall, either through their relationship to cultured microorganisms or through their similarity to sequences obtained from other defined environments. For example, many of the crenarchaeal sequences from Z1 fall into the marine group I cluster, which encompasses sequences from pelagic and benthic marine environments (1, 56). A second component of the Z1 clone libraries is sequences related to uncultured Euryarchaeota found in benthic marine and estuarine environments (1, 50, 56). In the context of a vent chimney, the presence of these clusters likely reflects infiltration of seawater through exterior surfaces of the chimney wall. These results indicate that there are psychrophilic to mesophilic, aerobic habitats near the exterior of the chimney, which support a microbial population similar to the populations in benthic deep-sea environments.
Z2: silicified pyrite plus zinc sulfide.
The greatest biomass (>108 cells/g) was found 2 to 10 cm inside the chimney walls in a porous iron- and zinc sulfide-rich zone characterized by abundant late deposition of amorphous silica (Z2). The enhanced deposition of amorphous silica in Z2 required conditions that favored conductive cooling of reduced hydrothermal fluids coinciding with an absence of seawater mixing (23, 54, 55). However, the localized presence of barite, anhydrite, and clay minerals within the chimney wall indicates that at least episodically, seawater circulation occurred. These observations indicate that there is a dynamic and complex environment in Z2 characterized by mixing of warm or hot reduced fluids and oxidizing seawater.
The microbial community in Z2 consisted of both eubacteria and archaea, like the community in Z1; however, the archaeal component shifted from predominantly Euryarchaeota in Z1 to Crenarchaeota in Z2 to Z4 (Table 2). This may reflect variations in environmental conditions between zones which promote the growth of different groups of microorganisms. However, without additional information about the uncharacterized species that inhabit the chimney wall it is difficult to infer thermal and chemical information from the microbiological data alone. The Z2 archaeal clone libraries contained several sequences similar to the sequences of cultured organisms, including Archaeoglobus (98% similar) and Palaeococcus (94 to 95%). Cultured strains of these microorganisms are anaerobic and hyperthermophilic (optimal growth occurs at >80°C), which indicates that there are hot, reducing environments that are 2 to 10 cm inside the chimney wall (30, 52). Few Thermococcales sequences have been recovered from hydrothermal vent environments despite their ubiquity in culture-based studies (27, 42, 51). However, Reysenbach et al. (42) found abundant Thermococcus and Archaeoglobus sequences in analyses of surfaces colonized during the deployment of an in situ growth chamber on an actively venting chimney. Z2 clone libraries also contained sequences related to anaerobic, methanogenic archaea belonging to the order Methanosarcinales, the closest relatives of which have been found on the surfaces of iron and manganese micronodules in association with metal-oxidizing bacterial communities (47). Mineral surfaces similar to the micronodules are abundant on and around hydrothermal vents in the deep sea (29, 30). The occurrence of these groups of organisms suggests that there was surface association of multispecies microbial communities within the exterior zones of Finn. Additionally, a number of crenarchaeal sequences from Z2 of Finn are similar to sequences obtained from terrestrial and marine hydrothermal environments, such as Iceland, Yellowstone National Park, Guaymas Basin, and the Okinawa Trough (8, 36, 51, 53). Although very few of these organisms have been cultured, their occurrence in thermophilic to hyperthermophilic source habitats supports the inference that high-temperature regions are present within the chimney wall, which is consistent with the mineralogy and high venting temperatures.
We hypothesized that the interface of mixing between highly reducing hydrothermal fluids and oxidizing seawater was most intense in Z2 and supported an abundant and highly active microbial community (>85% of the cells in Z2 hybridized as determined by FISH, indicating that they contained abundant ribosomes [5]). Microenvironments created by mixing between end member fluids provide habitats that can support a rich diversity of microorganisms, as shown by 16S rRNA sequences that range from marine group I Crenarchaeota to hyperthermophilic Euryarchaeota. The organization of microorganisms from Z2 into thick biofilms may have facilitated the resistance of these organisms to extreme environmental changes, such as oxygen stress or high temperature, and may also have provided conditions that favored increasing physiological diversity within the microbial community (9, 32).
Z3: porous pyrite plus zinc sulfide.
The mineralogy of Z3 was characterized by a predominantly pyrite and zinc sulfide matrix with chalcopyrite-lined fractures and local patches of anhydrite. In general, Z3 lacked abundant amorphous silica in its interstices and likely sustained higher temperatures than Z2 because silica deposition is temperature dependent (23). Chalcopyrite-lined fractures in Z3 indicate that there were breakouts of high-temperature fluids from the central conduit. Furthermore, localized anhydrite deposition in Z3 and Z4 required intermittent circulation of seawater and temperatures in excess of 150°C, as anhydrite dissolves at temperatures below this value (11, 23). The presence of anhydrite rather than clay minerals in this zone indicates that little mixing took place between hydrothermal fluids and seawater during the depositional episodes (23, 54, 55). Together, these observations describe a high-temperature anoxic environment subject to transient breakouts of hydrothermal fluids from the central conduit and at other times episodic inflow of seawater.
In contrast to the microbial populations in the exterior regions of the chimney, the Z3 and Z4 microbial communities were nearly all archaea, which formed biofilms (Fig. 3A) and microcolonies (Fig. 3C) on mineral surfaces. Crenarchaeota comprised most of the FISH-staining archaeal population from Z3, although hyperthermophilic Euryarchaeota were also detected. Cells that were stained with TC and MC probes comprised 2 to 5% of the population in Z2 and Z3 (Table 2). Analyses of 16S rRNA sequences from Z3 clone libraries revealed the prevalence of a single crenarchaeal cluster (cluster UCI) (Table 3), which is related to uncultured Crenarchaeota obtained from deep subsurface hydrothermal environments (36). Sequences related to hyperthermophilic Thermococcales and Methanococcales species were also found in the Z3 clone libraries. Thermococcales and Methanococcales have commonly been cultured from sulfide chimneys (30) and were also cultured in this study (data not shown), although their abundance in situ has been poorly constrained. All of the known species of these groups are anaerobic, and all of the members of the Thermococcales that have been described are hyperthermophilic (30). Thus, the presence of sequences related to Thermococcales and Methanococcales and the presence of cells that were stained with the TC and MC probes indicate that there were hot, reducing environments within Z3 of Finn. Further development and application of molecular technologies and culturing methods to vent samples are necessary to increase our understanding of the ecology of these commonly encountered groups and the many uncultured phylotypes that populate 16S rRNA clone libraries from chimney habitats. Due to the location of Z3 within the chimney wall and its mineralogy, either Z3 had to be intensively cooled by fluid circulation to life-sustaining temperatures or the organisms in Z3 had to possess novel characteristics that allowed them to survive at temperatures well in excess of 100°C.
Z4: chalcopyrite plus zinc sulfide.
The mineralogy in Z4 was characterized by a thick (0.2- to 10-cm) chalcopyrite layer lining the inner surface of the vent conduit. The chalcopyrite rind exhibited very low porosity and was characterized by subparallel microscopic fractures that bound some mineral grains. It is likely that fluid transport across this boundary was controlled by these fine-scale fracture networks. However, major pulses of end member hydrothermal fluids into the outer portions of the wall also occurred through large-scale fractures. The highly conductive nature of the monominerallic layer of chalcopyrite, coupled with the presence of anhydrite bounding the chalcopyrite rind, required prolonged temperatures in excess of 150°C (11) and an upper temperature boundary constrained by the 302°C venting fluid. In the interior of the chimney (Z4) organisms were detected both in cell extracts and by direct examination of mineral surfaces, although few cells hybridized with the oligonucleotide probes used in this study. This may have been due to either the impermeability of the organisms to the probe, the absence of a target sequence, or the inaccessibility or lack of sufficient ribosomes (5, 46). In addition to the high temperature, the variable permeability of the chalcopyrite lining in Z4 may have been sufficient to inhibit the growth of significant biomass as exposure to nutrient sources was probably quite low. Detection of lipid biomarkers and intact cells in material adjacent to hot hydrothermal fluids is not unprecedented. Hedrick et al. (26) found archaeal di- and tetraether lipids in a hot flange adjacent to 350°C fluids, and intact cells have been observed in interior portions of small high-temperature sulfide chimneys (24, 51). The presence of intact cells in Z4, the mineralogical region directly adjacent to the venting 302°C fluids, indicates that microorganisms from this region may have adaptive strategies that allow them to survive at extremely high temperatures.
Survival at extreme temperatures?
The presence of intact microorganisms in regions of Finn where the temperature must have been well in excess of 100°C was not expected. Our data indicate that microorganisms can survive and possibly grow under the extreme conditions present in active chimneys. Two observations from this study provide some insight into the microbial ecology of such environments. First, the communities that inhabited interior zones of the sulfide chimney resemble known groups of organisms in that they were stained with DNA and RNA probes. 16S rRNA sequences from Z3, together with FISH data, indicated that uncultured groups of Crenarchaeota were prevalent in interior zones of Finn. Examining the physiology of these organisms in greater detail in either culture-based or molecular studies should increase our understanding of the microbial ecology in chimney environments. Second, a key observation based on samples obtained within the walls of Finn was the association of multispecies microbial communities with mineral surfaces. Microbial biofilms may influence the microenvironments that organisms encounter in unique ways, which allow the organisms to survive despite extreme environmental stresses (9). The occurrence of intact microorganisms attached to mineral surfaces in Finn implies that surface association may have important implications for microbial growth and survival at extremely high temperatures.
Ongoing analyses of several large sulfide structures from the Edifice Rex project are providing multifaceted maps of the vent chimneys and are allowing integration of multiple data sets into a coherent biogeochemical model. Our findings provide a key piece of information for the modeling effort as they show that the distribution and composition of microbial communities vary significantly in different regions of an actively venting sulfide structure. Once the data set is complete, the conditions that supported the uncultured microbial communities in Finn should be more completely described and aid our attempts to culture and characterize these organisms. Conversely, if well constrained, the metabolic diversity of specific microbial communities and biochemical and biogeochemical markers may be used as indicators of the conditions present within a given vent environment (48). The challenge of correlating in situ temperatures inside sulfide chimneys with microbiology was an obvious limitation of the present study and resulted from both technical difficulty and the dynamic conditions of such environments. With this in mind, our group is currently developing new methods to obtain coregistered measurements of temperature, chemistry, and microbial growth within active hydrothermal vent chimneys in situ.
Acknowledgments
This work was supported by NSF-LEXEN (grant OCE9816491), NSF (grant OCE0096510), NSF-IGERT (grant DGE-9870713), Washington Sea Grant (grant NA76RG0119), and the NASA Astrobiology Institute through the Carnegie Geophysical Institute.
We thank the participants in the Edifice Rex Sulfide Recovery Project, including the crews of the R/V Tully, R/V Thompson, R/V Atlantis, ROV ROPOS, and ROV JASON, for their assistance with site characterization and sample retrieval. We are grateful to Ed Mathez and the American Museum of Natural History for facilitating the sulfide recovery, David Mogk at Montana State University for assistance with SEM, and Mitchell Elend for help with preparing figures. We also thank Julie Huber, Jon Kaye, Mausmi Mehta, Sheryl Bolton, and Diane Nielsen for their thoughtful insights and helpful discussions.
REFERENCES
- 1.Abreu, C., G. Jurgens, P. D. Marco, A. Saano, and A. A. Bordalo. 2001. Crenarchaeota and Euryarchaeota in temperate estuarine sediments. J. Appl. Microbiol. 90:713-718. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amend, J. P., and E. Shock. 2001. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 25:175-243. [DOI] [PubMed] [Google Scholar]
- 7.Balkwill, D. L., F. R. Leach, J. T. Wilson, J. F. McNabb, and D. C. White. 1988. Equivalence of microbial biomass measures based on membrane lipid and cell wall components, adenosine triphosphate and direct counts in subsurface aquifer sediments. Microb. Ecol. 16:73-84. [DOI] [PubMed] [Google Scholar]
- 8.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beveridge, T. J., S. A. Makin, J. L. Kadurugamuwa, and Z. Li. 1997. Interactions between biofilms and the environment. FEMS Microbiol. Rev. 20:291-303. [DOI] [PubMed] [Google Scholar]
- 10.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 11.Blount, C. W., and F. W. Dickson. 1969. The solubility of anhydrite (CaSO4) in NaCl-H2O from 100 to 450 degrees C and 1 to 1000 bars. Geochim. Cosmochim. Acta 33:227-245. [Google Scholar]
- 12.Bond, P. L., and J. F. Banfield. 2001. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb. Ecol. 41:149-161. [DOI] [PubMed] [Google Scholar]
- 13.Burgraff, S., T. Mayer, R. Amann, S. Schadhauser, C. R. Woese, and K. O. Stetter. 1994. Identifying members of the domain Archaea with rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 60:3112-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaney, J. R., D. S. Kelley, E. A. Mathez, D. R. Yoerger, J. Baross, M. Schrenk, M. K. Tivey, J. Kaye, and V. Robigou. 2001. “Edifice Rex” sulfide recovery project: analysis of a submarine hydrothermal microbial habitat. EOS Trans. 82:67-73. [Google Scholar]
- 15.Delaney, J. R., D. S. Kelley, M. D. Lilley, D. A. Butterfield, J. A. Baross, W. S. D. Wilcock, R. W. Embley, and M. Summit. 1998. The quantum event of oceanic crustal accretion: impacts of diking at mid-ocean ridges. Science 281:222-230. [PubMed] [Google Scholar]
- 16.Delong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deming, J. W., A.-L. Reysenbach, S. A. Macko, and C. R. Smith. 1997. Evidence for the microbial basis of a chemoautotrophic invertebrate community at a whale fall on the deep seafloor: bone-colonizing bacteria and invertebrate endosymbionts. Microsc. Res. Tech. 37:162-170. [DOI] [PubMed] [Google Scholar]
- 18.Deming, J. W., and J. A. Baross. 1993. Deep-sea smokers: windows to a subsurface biosphere? Geochim. Cosmochim. Acta 57:3219-3230. [DOI] [PubMed] [Google Scholar]
- 19.Eberhard, C., C. O. Wirsen, and H. Jannasch. 1995. Oxidation of polymetal sulfides by chemolithoautotrophic bacteria from deep-sea hydrothermal vents. Geomicrobiol. J. 13:145-164. [Google Scholar]
- 20.Edwards, K. J., P. L. Bond, T. M. Gihring, and J. F. Banfield. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287:1796-1799. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 23.Hannington, M. D., I. R. Jonasson, P. M. Herzig, and S. Peterson. 1995. Physical and chemical processes of seafloor mineralization at mid-ocean ridges, p. 115-157. In S. E. Humphris, R. A. Zierenberg, L. S. Mullineaux, and R. E. Thomson (ed.), Seafloor hydrothermal systems: physical, chemical, biological, and geological interactions. Geophysical monograph 91. American Geophysical Union, Washington, D.C.
- 24.Harmsen, H. J. M., D. Prieur, and C. Jeanthon. 1997. Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl. Environ. Microbiol. 63:2876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmsen, H. J. M., D. Prieur, and C. Jeanthon. 1997. Group-specific 16S rRNA-targeted oligonucleotide probes to identify thermophilic bacteria in marine hydrothermal vents. Appl. Environ. Microbiol. 63:4061-4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedrick, D. B., R. D. Pledger, D. C. White, and J. A. Baross. 1992. In-situ microbial ecology of hydrothermal vent sediments. FEMS Microbiol. Ecol. 101:1-10. [Google Scholar]
- 27.Huber, J. A., D. A. Butterfield, and J. A. Baross. 2002. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingaki, F., K. Takai, T. Komatsu, T. Kanamatsu, and K. Horikoshi. 2001. Archaeology of Archaea: geomicrobiological record of Pleistocene thermal events concealed in a deep-sea subseafloor environment. Extremophiles 5:385-392. [DOI] [PubMed] [Google Scholar]
- 29.Juniper, S. K., and B. M. Tebo. 1995. Microbe-metal interactions and mineral deposition at hydrothermal vents, p. 219-253. In D. S. Karl (ed.), The microbiology of deep-sea hydrothermal vents. CRC Press, Boca Raton, Fla.
- 30.Kelley, D. S., J. A. Baross, and J. R. Delaney. 2002. Volcanoes, fluids, and life at mid-ocean ridge spreading centers. Annu. Rev. Earth Planet. Sci. 30:385-491. [Google Scholar]
- 31.Kelley, D. S., J. R. Delaney, and D. R. Yoerger. 2001. Geology and venting characteristics of the Mothra Hydrothermal Field, Endeavour Segment, Juan de Fuca Ridge. Geology 29:959-962. [Google Scholar]
- 32.LaPaglia, C., and P. L. Hartzell. 1997. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl. Environ. Microbiol. 63:3158-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.L'-Haridon, S., V. Cilia, P. Messner, G. Raguenes, A. Gambacorta, U. B. Sleytr, D. Prieur, and C. Jeanthon. 1998. Desulfurobacterium thermolithotrophum gen. nov., sp. nov., a novel autotrophic, sulphur-reducing bacterium isolated from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 48:701-711. [DOI] [PubMed] [Google Scholar]
- 34.Longnecker, K., and A.-L. Reysenbach. 2001. Expansion of the geographic distribution of a novel lineage of ɛ-proteobacteria to a hydrothermal vent site on the southern East Pacific Rise. FEMS Microbiol. Ecol. 35:287-293. [DOI] [PubMed] [Google Scholar]
- 35.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marteinsson, V. T., H. Sigurbjörg, C. F. V. Hobel, H. Kristmannsdóttir, G. O. Hreggvidsson, and J. K. Kristjánsson. 2001. Phylogenetic diversity analysis of subterranean hot springs in Iceland. Appl. Environ. Microbiol. 67:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maruyama, A., and M. Sunamura. 2000. Simultaneous direct counting of total and specific microbial cells in seawater using a deep-sea microbe as a target. Appl. Environ. Microbiol. 66:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCollom, T. M., and E. L. Shock. 1997. Geochemical constraints on chemolithoautotrophic metabolism by microorganisms in seafloor hydrothermal systems. Geochim. Cosmochim. Acta 61:4375-4391. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen, D. C., D. S. Kelley, J. R. Delaney, and M. O. Schrenk. 2002. Investigation of steep thermal and chemical gradients in submarine microbial habitats. Astrobiology 2:552. [Google Scholar]
- 40.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 41.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reysenbach, A.-L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarrazin, J., S. K. Juniper, G. Massoth, and P. Legendre. 1999. Physical and chemical factors influencing species distribution on hydrothermal sulfide edifices of the Juan de Fuca Ridge, northeast Pacific. Mar. Ecol. Prog. Ser. 190:89-112. [Google Scholar]
- 44.Slobodkin, A., B. Campbell, S. C. Cary, E. Bonch-Osmolovskaya, and C. Jeanthon. 2001. Evidence for the presence of thermophilic Fe(III)-reducing microorganisms in deep-sea hydrothermal vents at 13°N (East Pacific Rise). FEMS Microbiol. Ecol. 36:235-243. [DOI] [PubMed] [Google Scholar]
- 45.Smith, D. C., A. J. Spivack, M. R. Fisk, S. A. Haveman, H. Staudigel, and Ocean Drilling Program Leg 185 Shipboard Scientific Party. 2000. Tracer-based estimates of drilling-induced microbial contamination of deep sea crust. Geomicrobiol. J. 17:207-219. [Google Scholar]
- 46.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 47.Stein, L. Y., M. T. LaDuc, T. J. Grundl, and K. H. Nealson. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 3:10-18. [DOI] [PubMed] [Google Scholar]
- 48.Summit, M., and J. A. Baross. 2001. A novel microbial habitat in the mid-ocean ridge subseafloor. Proc. Natl. Acad. Sci. USA 98:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takai, K., and Y. Sako. 1999. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol. Ecol. 28:177-188. [Google Scholar]
- 50.Takai, K., and K. Horikoshi. 1999. Genetic diversity of Archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takai, K., T. Komatsu, F. Inagaki, and K. Horikoshi. 2001. Distribution of Archaea in a black smoker chimney structure. Appl. Environ. Microbiol. 67:3618-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takai, K., A. Sugai, T. Itoh, and K. Horikoshi. 2000. Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int. J. Syst. E vol. Microbiol. 50:489-500. [DOI] [PubMed] [Google Scholar]
- 53.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. de Vera Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tivey, M. K. 1995. The influence of hydrothermal fluid composition and advection rates on black smoker chimney mineralogy: insights from modeling transport and reaction. Geochim. Cosmochim. Acta 59:1933-1949. [Google Scholar]
- 55.Tivey, M. K., and R. E. McDuff. 1990. Mineral precipitation in the walls of black smoker chimneys: a quantitative model of transport and chemical reaction. J. Geophys. Res. 95:12617-12637. [Google Scholar]
- 56.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A.-L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White, D. C., W. M. Davis, J. S. Nickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 40:201-211. [DOI] [PubMed] [Google Scholar]