Abstract
Although stimulation of dissimilatory metal reduction to promote the reductive precipitation of uranium has been shown to successfully remove uranium from some aquifer sediments, the organisms in the family Geobacteraceae that have been found to be associated with metal reduction in previous studies are not known to grow at the high salinities found in some uranium-contaminated groundwaters. Studies with a highly saline uranium-contaminated aquifer sediment demonstrated that the addition of acetate could stimulate the removal of U(VI) from the groundwater. This removal was associated with an enrichment in microorganisms most closely related to Pseudomonas and Desulfosporosinus species.
The addition of acetate to organic-poor subsurface sediments is an effective strategy for stimulating dissimilatory metal reduction in these sediments (2, 3, 14). In uranium-contaminated subsurface sediments, acetate additions result in the removal of uranium from the groundwater because dissimilatory metal-reducing microorganisms can reduce soluble U(VI) to insoluble U(IV) (6, 8). In previous studies in which dissimilatory metal reduction was stimulated with the addition of acetate, molecular analysis revealed that microorganisms in the family Geobacteraceae increased in number by orders of magnitude as metal reduction proceeded and that these organisms became the predominant members of the microbial community during metal reduction (4, 14). In a similar manner, microorganisms in the Geobacteraceae were highly enriched in the Fe(III) reduction zones of aquifers contaminated with petroleum (12) or landfill leachate (11).
In some instances, uranium-contaminated groundwater may have elevated salinities as a result of the high concentrations of acids that are used in processing uranium ores (1, 13). These salinities may far exceed those in which the microorganisms in the Geobacteraceae are known to grow (5). This raises the question of whether stimulating dissimilatory metal reduction is an appropriate strategy for removing U(VI) from highly saline groundwaters and, if so, what organisms might be responsible for U(VI) reduction.
In order to evaluate the potential for stimulating dissimilatory metal reduction in highly saline subsurface sediments, sediments were collected from a portion of the aquifer at the uranium mine tailings site in Shiprock, N.Mex. (2, 15). Sediments were from near well 854. The groundwater from the site at which these subsurface sediments were obtained contained high concentrations of Na+ (411 mM), Mg2+ (225 mM), Ca2+ (22 mM), Cl− (48 mM), SO42− (296 mM), and NO3− (28.7 mM). The sediments contained 4.5 μmol of 0.5 N HCl-extractable Fe(III) per g of sediment.
As previously described (3), sediments were collected from below the water table, shipped to the laboratory, and incubated under strict anaerobic conditions at 20°C in serum bottles under an atmosphere of N2-CO2 (93:7). Acetate was added from a concentrated anoxic stock solution. Nitrate, Fe(II), Fe(III), U(VI), and sulfate were monitored over time as previously described (3).
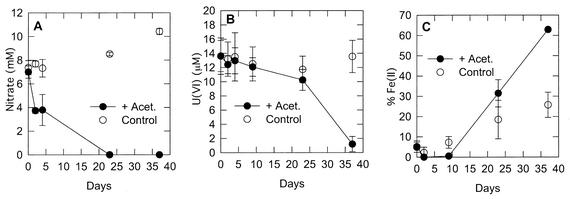
Despite the high salinity, the acetate additions stimulated the removal of nitrate from the groundwater, followed by the simultaneous accumulation of Fe(II) and the removal of U(VI), presumably as a result of Fe(III) and U(VI) reduction (Fig. 1). There was no significant loss of sulfate until after Fe(III) and U(VI) reduction was complete. There was no significant removal of nitrate or reduction of Fe(III) and U(VI) in the absence of added acetate. These results demonstrated that the addition of acetate could stimulate anaerobic respiration in these highly saline sediments and could promote the removal of contaminant U(VI).
FIG. 1.
Concentrations of dissolved nitrate and U(VI) and HCl-extractable Fe(II) in sediments from site 854 in initial incubation under anoxic conditions. The results are means of results from triplicate incubations. The error bars represent 1 standard deviation. (A) Nitrate; (B) U(VI); (C) percent Fe(II) of total iron. Acet., acetate.
In order to determine which microorganisms were associated with dissimilatory metal reduction in these sediments, DNA was extracted as previously described (4) at the start of the study as well as at day 23, in the middle of the metal reduction phase. As previously described (4), bacterial DNA was amplified with two primer sets, (i) 8F and 519R and (ii) 338F and 907R. The PCR products from the two primer sets were pooled, the clone libraries were prepared, and the inserts were amplified and digested with HhaI and MspI (4). The different band patterns were noted, and the frequency of similar patterns was scored. All amplified clones with distinct banding patterns were sequenced.
Although it was considered that Archaea might be important components of the microbial community due to the high salinity, when archaeal 16S rRNA gene sequences were amplified with PCR primers 334F and 915R, no PCR product could be detected in an agarose gel with UV illumination. When a sample from this first PCR was reamplified with primers 334F and 915R, the PCR product was recovered. However, the necessity for this second amplification suggested that the archaeal 16S rRNA gene sequences were much less numerous than the sequences of Bacteria.
Bacterial sequences were analyzed by using BLAST (National Center for Biotechnology Information) in order to find the most similar available sequences. Sequences were manually aligned with closely related 16S rRNA gene sequences from the Ribosomal Database Project and GenBank using the graphical user interface Seqlab (Wisconsin Package, version 1.0; Genetics Computer Group, Madison, Wis.). Only those sequence regions that could be aligned with confidence were included in the analyses, and gaps were treated as missing nucleotides. Phylogenetic trees were inferred from unambiguously aligned sequence data with the distance, maximum-likelihood, and maximum-parsimony tools of PAUP* (version 4.0 beta; Sinauer Associates, Sunderland, Mass.).
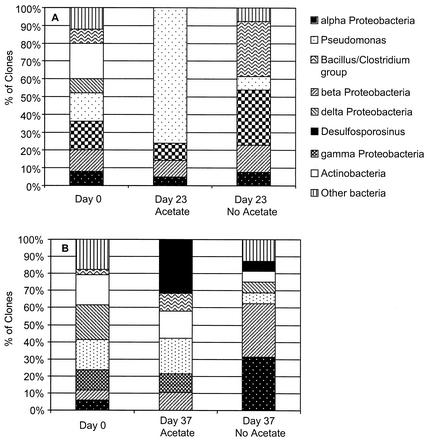
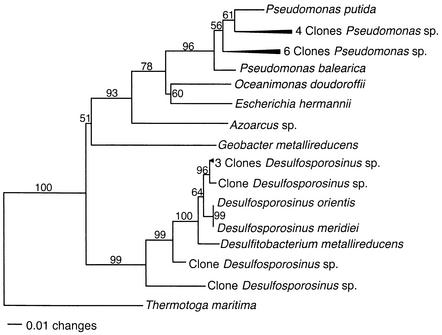
Sediments in which metal reduction was the predominant terminal electron-accepting process were highly enriched in microorganisms with 16S rRNA gene sequences closely related to known Pseudomonas species (Fig. 2A and 3). Whereas Pseudomonas 16S rRNA gene sequences accounted for 16% of the sequences at the start of the study, by day 23, 76% of the sequences represented Pseudomonas species. In contrast, only 8% of the sequences of sediments that were not amended with acetate and in which there was no reduction of nitrate or metal could be assigned to Pseudomonas. There was no apparent increase in any other group of microorganisms in the acetate-amended sediments, and no sequences of Geobacteraceae were detected. In the sediment with no acetate addition, the number of gamma proteobacteria and those of the species in the Bacillus-Clostridium group increased slightly.
FIG. 2.
Distribution of clones from the initial sediment study (A) and from the second sediment study (B).
FIG. 3.
Phylogenetic tree of Bacteria, showing the placement of Pseudomonas and Desulfosporosinus clone sequences that were enriched in acetate-amended, metal-reducing sediments.
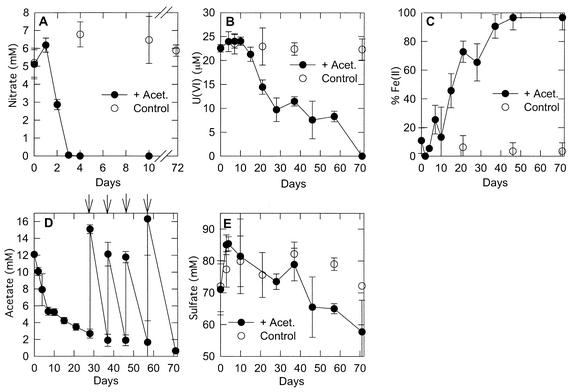
In order to evaluate the kinetics of acetate consumption in the sediments, a similar study was conducted with a subsample of the same sediments 1 year later. This time, the acetate concentrations were monitored over time with high-performance liquid chromatography as previously described (7). In this study, acetate was added back to the sediment as it became depleted (Fig. 4). As was observed in the initial study with freshly collected sediments, the addition of acetate stimulated the reduction of nitrate and then of Fe(III) and U(VI) (Fig. 4). However, in contrast to the results of the initial study, there clearly was a reduction in sulfate prior to the completion of U(VI) reduction.
FIG. 4.
Incubation of site 854 sediments with multiple acetate additions. The results are means of results from triplicate incubations. The error bars represent 1 standard deviation. Arrows in panel D indicate the readdition of acetate (Acet.). (A) Nitrate; (B) U(VI); (C) percent Fe(II) of total iron; (D) acetate; (E) sulfate.
Although the sediments had been stored aerobically for a year prior to the second study, the composition of the microbial community at the start of the second study was quite similar to that in the freshly collected sediments. This similarity is attributed to the facts that the sediments are aerobic in situ and that without the addition of an exogenous electron donor, microbial metabolism remains slow. An analysis of the microbial community on day 37, when metal reduction was the predominant terminal electron-accepting process, prior to detectable sulfate reduction, indicated that there was only a slight increase in the Pseudomonas sequences that were so highly enriched during metal reduction in the acetate-amended freshly collected sediments. More obvious was an enrichment in the 16S rRNA gene sequences that were closely related to Desulfosporosinus (Fig. 2B and 3). None of these sequences were detected at time zero, but they accounted for 32% of the clone sequences on day 37. In the sediments not amended with acetate, only 6% of the sequences were Desulfosporosinus species. As in the initial study, no sequences of Geobacteraceae were detected in any of the sediment treatments.
The differences in the microbial communities in the two incubations may be due to a variety of factors. In the second incubation, significantly more of the available Fe(III) had been reduced when a sediment sample was taken for community analyses. Furthermore, it is possible that spore-forming Desulfosporosinus species were favored over Pseudomonas species after storage of the sediment for 1 year or that the sequential addition of acetate to the sediments favored the initiation of sulfate reduction and the growth of Desulfosporosinus species.
These results demonstrate that it may be possible to bioremediate highly saline, uranium-contaminated aquifers with the simple addition of an electron donor such as acetate. However, the organisms involved in metal reduction in such aquifers are not likely to be the Geobacteraceae that have previously been implicated in metal reduction in aquifers with freshwater and marine-like salinities. The salinity of site 854 is 10-fold higher than typical marine salinities, which probably accounts for the lack of Geobacteraceae at this site. Attempts to isolate some of the Pseudomonas and Desulfosporosinus species which might be responsible for metal reduction in the site 854 sediments have as yet been unsuccessful. However, there are strains of Pseudomonas (9, 16) and Desulfosporosinus (10) that can grow at high salinities, and organisms in these genera are known to be capable of Fe(III) reduction (5, 10). Thus, further attempts to isolate the salt-tolerant metal-reducing organisms and study their physiology seem warranted in order to provide insights into the physiological factors controlling the rate and extent of metal reduction in highly saline subsurface environments.
Acknowledgments
We thank Robert Anderson for assistance with sediment sampling and Dawn Holmes for assistance with molecular analyses. This work was supported by the U.S. Department of Energy NABIR program (grants DE-FG02-00ER62985 and DE-FG02-97ER62475).
REFERENCES
- 1.Cerda, J., S. Gonzalez, J. Rios, and T. Quintana. 1993. Uranium concentrates bioproduction in Spain: a case study. FEMS Microbiol. Rev. 11:253-260. [Google Scholar]
- 2.Finneran, K., R. Anderson, K. Nevin, and D. Lovley. 2002. Potential for bioremediation of uranium-contaminated aquifers with microbial U(VI) reduction. Soil Sediment Contam. 11:339-357. [Google Scholar]
- 3.Finneran, K., M. Housewright, and D. Lovley. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4:510-516. [DOI] [PubMed] [Google Scholar]
- 4.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovley, D. 2000. Fe(III) and Mn(IV)-reducing prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 6.Lovley, D. R. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Ind. Microbiol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 7.Lovley, D. R., and E. J. P. Phillips. 1989. Requirement for a microbial consortium to completely oxidize glucose in Fe(III)-reducing sediments. Appl. Environ. Microbiol. 55:3234-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 9.Nishimori, E., K. Kita-Tsukamoto, and H. Wakabayashi. 2000. Pseudomonas plecoglossicida sp. nov., the causative agent of bacterial haemorrhagic ascites of ayu, Plecoglossus altivelis. Int. J. Syst. Evol. Microbiol. 50:83-89. [DOI] [PubMed] [Google Scholar]
- 10.Robertson, W., J. Bowman, P. Franzmann, and B. Mee. 2001. Desulfosporosinus meridiei sp. nov., a spore-forming sulfate-reducing bacterium isolated from gasolene-contaminated groundwater. Int. J. Syst. Evol. Microbiol. 51:133-140. [DOI] [PubMed] [Google Scholar]
- 11.Röling, W. F. M., B. M. van Breukelen, M. Braster, B. Lin, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schippers, A., R. Hallmann, S. Wentzein, and W. Sand. 1995. Microbial diversity in uranium mine waste heaps. Appl. Environ. Microbiol. 61:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snoeyenbos-West, O., K. Nevin, R. Anderson, and D. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 15.UMTRA Project. 1995. UMTRA Project water sampling and analysis plan, Shiprock, New Mexico. Document no. DOE/AL/62350-78. Office of Environmental Management, U.S. Department of Energy, Washington, D.C.
- 16.Yumoto, I., T. Kusano, T. Shingyo, Y. Nodasaka, H. Matsuyama, and H. Okuyama. 2001. Assignment of Pseudomonas sp. strain E-3 to Pseudomonas psychrophila sp. nov., a new facultatively psychrophilic bacterium. Extremophiles 5:343-349. [DOI] [PubMed] [Google Scholar]