Abstract
To investigate the impact of genetically modified, antibiotic-producing rhizobacteria on the indigenous microbial community, Pseudomonas putida WCS358r and two transgenic derivatives were introduced as a seed coating into the rhizosphere of wheat in two consecutive years (1999 and 2000) in the same field plots. The two genetically modified microorganisms (GMMs), WCS358r::phz and WCS358r::phl, constitutively produced phenazine-1-carboxylic acid (PCA) and 2,4-diacetylphloroglucinol (DAPG), respectively. The level of introduced bacteria in all treatments decreased from 107 CFU per g of roots soon after sowing to less than 102 CFU per g after harvest 132 days after sowing. The phz and phl genes remained stable in the chromosome of WCS358r. The amount of PCA produced in the wheat rhizosphere by WCS358r::phz was about 40 ng/g of roots after the first application in 1999. The DAPG-producing GMMs caused a transient shift in the indigenous bacterial and fungal microflora in 1999, as determined by amplified ribosomal DNA restriction analysis. However, after the second application of the GMMs in 2000, no shifts in the bacterial or fungal microflora were detected. To evaluate the importance of the effects induced by the GMMs, these effects were compared with those induced by crop rotation by planting wheat in 1999 followed by potatoes in 2000. No effect of rotation on the microbial community structure was detected. In 2000 all bacteria had a positive effect on plant growth, supposedly due to suppression of deleterious microorganisms. Our research suggests that the natural variability of microbial communities can surpass the effects of GMMs.
Antagonistic bacteria can suppress plant diseases caused by microbial pathogens (52, 55). A major mechanism of control of soilborne plant pathogens by fluorescent pseudomonads is the production of antibiotics (14, 19, 30, 55). Many naturally occurring antibiotics, such as 2,4-diacetylphloroglucinol (DAPG), phenazine, pyoluteorin, and pyrrolnitrin, have been identified (20, 52). Application of biocontrol agents to suppress soilborne diseases is often unsuccessful because of inconsistent performance under field conditions. The explanations for this inconsistency include variable plant root colonization by the biocontrol agent, insufficient concentration of the antibiotic, instability or degradation of the antibiotic, and genetic diversity of the pathogen (31, 53). One approach to overcome some of these difficulties is to genetically modify biocontrol strains for enhanced and/or constitutive biosynthesis of antibiotics.
Before genetically modified microorganisms (GMMs) can be commercially used as biocontrol agents, field studies must be performed to obtain information about their possible impact on nontarget organisms. So far, effects of introduced wild-type strains and GMMs on the soil ecosystem have been studied mainly in microcosms (21, 46). In most field studies attention has been focused primarily on the fate of the introduced GMMs (49, 51). In only a few studies have the workers described effects on the indigenous microbial community (10, 40). In particular, GMMs with an enhanced capacity to produce antibiotic compounds are likely to affect nontarget microorganisms. Glandorf et al. (18) demonstrated that introduction of the root-colonizing bacterium Pseudomonas putida WCS358r, genetically modified to produce the antimicrobial compound phenazine-1-carboxylic acid (PCA), caused a differential but transient shift in the fungal rhizosphere microflora of wheat plants compared to the effect of the parental strain.
In the present study strain WCS358r, modified to constitutively produce PCA or DAPG, was introduced in two consecutive years into a field to monitor possible long-term effects on the rhizosphere microflora. The objectives were to determine whether the effects on the microbial community observed by Glandorf et al. (18) are specific for PCA or apply also to another antibiotic. To simulate commercial application protocols, we introduced WCS358r and genetically modified derivatives of this strain into the same field plots in two consecutive years in order to investigate whether a second application of the strains resulted in intensified effects on the microflora. Population dynamics and dispersal of the introduced bacteria were also studied.
A general method of studying effects on the microbial community is plate count enumeration of culturable microorganisms from rhizosphere samples. Because only 0.1 to 1% of the total microflora can be cultured (2, 24), molecular techniques based on direct extraction of nucleic acid from samples are more suitable (23, 39, 44). Here, we used PCR-based amplified ribosomal DNA (rDNA) restriction analysis (ARDRA) to detect restriction fragment length polymorphisms of the microbial rRNA genes. The impact of the GMMs on the microflora was compared with the effects resulting from a common agricultural practice, crop rotation, a strategy that is often used for control of soilborne plant diseases (23). In our study microbial communities in field plots cultured with wheat for two consecutive years were compared with the microbial communities in plots planted with wheat in the first year and with potato in the second year.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used are listed in Table 1. WCS358r, a spontaneous rifampin-resistant mutant of plant-growth-promoting P. putida strain WCS358, was used as the wild type and has been described previously (3, 15, 16). A constitutively PCA-producing derivative contained the phzABCDEFG gene cluster on the disarmed mini-Tn5 transposon-based vector pUTKm (22) inserted into the chromosome. This strain, WCS358r::phz, was identical to the strain designated GMM 8 used in the study of Glandorf et al. (18). A DAPG-producing GMM was constructed by inserting the phlFACBDE genes from Pseudomonas fluorescens Q2-87 (4), a naturally DAPG-producing strain, into the chromosome of WCS358r, and it was designated WCS358r::phl. The disarmed mini-Tn5 transposon including the nptII gene coding for kanamycin resistance contained in pUTKm was used as the vector. In this construct the phlF repressor gene is disrupted, resulting in constitutive production of DAPG. WCS358r was cultured on King's medium B (KB) agar (26) supplemented with 150 μg of rifampin ml−1. The transconjugants were cultured on KB agar in the presence of 150 μg of rifampin ml−1 and 30 μg of kanamycin ml−1. All bacteria were incubated at 28°C for 2 days.
TABLE 1.
Bacterial strains used in this study
Seed treatment.
Wheat seeds (Triticum aestivum cv. Baldus) were treated with 1:1 mixtures of washed bacterial suspensions (WCS358r, WCS358r::phz, WCS358r::phl) and 3% methylcellulose, as described by Glandorf et al. (18). For the control treatment the bacterial suspension was replaced by 10 mM MgSO4. Coated seeds were air dried overnight and sown the next day. Coating resulted in approximately 107 CFU per seed, as determined by plate count enumeration.
Experimental field.
Experiments were performed in 1999 and 2000 in an experimental field located near the Botanical Garden of Utrecht University. The field had a planting history of grass, and the soil consists of clay (12%) and has an organic matter content of 4% and a pH (KCl) of 5.0 (18). In 2000 the plots were fertilized 1 day before sowing (35 g of P2O5 per m2, 57 g of K2O per m2, 8 g of NO3− per m2).
The field was divided into two halves, each containing 18 1-m2 plots. For each half a random block design with six treatments, each with three replicates, was used. The plots were separated by unplanted, 50-cm-wide buffer zones, and the field was surrounded by an additional buffer zone that was 1.8 m wide. The site was surrounded by straw mats and a fence and was covered with a net to prevent rabbits and birds from entering.
Wheat seeds were sown and potatoes were planted in April and harvested in August. The following treatments were used: seeds coated with either WCS358r, WCS358r::phz, WCS358r::phl, or a 1:1 mixture of WCS358r::phz and WCS358r::phl, a control in which uninoculated seeds were used, and a rotation treatment in which uninoculated wheat seeds were used in 1999 and nontreated potatoes (Solanum tuberosum L. cv. Modesta) were used in 2000. In each plot 70 g of wheat seeds was sown in 11 rows that were 1 m long at a depth of 2 to 3 cm. In the rotation plots in the second year 40 potatoes were planted in eight rows, each containing five seed tubers. Plant and rhizosphere soil samples were taken during the growing season at five to eight different times.
Population dynamics of the introduced strains.
On each sampling date during the growing season, plant roots with adhering soil were harvested from three randomly selected spots within each plot. Prior to dilution plating, samples were prepared as described by Glandorf et al. (18). Population densities of WCS358r and the PCA- and DAPG-producing GMMs were determined by plating on KB+ (15) supplemented with 150 μg of rifampin ml−1 (KB+rif). The numbers of rifampin-resistant CFU were determined after incubation for 2 days at 28°C. Wild-type organisms and GMMs were also differentiated by fluorescence under UV light (366 nm). PCA and DAPG are UV-absorbing compounds (34), and production of these compounds results in weaker fluorescence of the GMMs on KB+ agar compared to the fluorescence of the wild type. Single colonies of the GMMs grown on KB+rif were also transferred to KB+rif containing kanamycin (KB+rif/km) to verify the presence of the gene cluster.
Soil samples to check for dispersal of GMMs were collected between the plots and in the surrounding buffer zones twice in both seasons.
Probes and colony hybridization.
To distinguish between WCS358r::phz and WCS358r::phl in the rhizosphere of plants treated with the mixture of GMMs, specific probes were developed. Escherichia coli 1936 JLS containing the phz gene locus was grown on Luria-Bertani agar (37) supplemented with kanamycin (30 μg ml−1) for 1 day at 37°C. P. fluorescens Q2-87, containing the phl gene locus, was grown on KB agar plates for 2 days at 28°C. Colonies were lysed and diluted 1:50, and 1 μl was used as template DNA for PCR. Primers Phl2a (5′-GAGGACGTCGAAGACCACCA-3′) and Phl2b (5′-ACCGCAGCATCGTGTATGAG-3′) (36) amplify a 745-bp fragment in the sequence of the phlD gene. Primers Phen1 (5′-CCCCTGTTGACAATTAATCATCGG-3′) and Phen2 (5′-ACCTTGACGTTGTACCATTCCCAA-3′) target sites in the sequences of the phzA and phzB genes and amplify a 1,014-kb fragment. Both primer sets were synthesized by Eurogentec, Maastricht, The Netherlands. Each 50-μl reaction mixture contained 5 μl of diluted heat-lysed cells, PCR buffer (Amersham Pharmacia Biotech), each deoxynucleoside triphosphate at a concentration of 200 μM, each phl or phz primer at a concentration of 200 μM, and 1.5 U of Taq DNA polymerase (Amersham Biosciences, Roosendaal, The Netherlands). The PCR program consisted of 4 min at 92°C and 30 cycles of 92°C for 30 s, 56°C for 30 s, and 72°C for 60 s. The sizes of the PCR products were checked on a 0.7% agarose gel, and the products were excised from the gel and purified with a QIAEX II agarose extraction kit (Qiagen, Hilden, Germany) used according to the manufacturer's protocol. The probes were then labeled with alkaline phosphatase from the Gene Images AlkPhos direct labeling and detection system (Amersham Biosciences) according to the protocol provided by the manufacturer.
To determine the relative numbers of the PCA- and DAPG-producing GMMs in the treatments that received the mixture of the two GMMs, at each sampling time 120 bacterial colonies were randomly selected from KB+rif/km plates containing colonies from plants treated with the mixture. The colonies were transferred to Hybond-N+ nylon membranes (Amersham Pharmacia Biotech), the bacterial cells were lysed, and the cell debris was washed off by standard methods (37). The DNA was fixed to the filters by exposure to UV light (365 nm) for 2 min (Hoefer Scientific Instruments, San Francisco, Calif.). Hybridization with the Gene Images AlkPhos direct labeling and detection system (Amersham Pharmacia Biotech) and detection with the CDP-Star chemiluminescent detection reagent (Amersham Biosciences) were performed as described in the supplier's protocol.
PCA extraction from the wheat rhizosphere.
To determine whether PCA was produced by WCS358r::phz in the rhizosphere, samples were collected 18 days after sowing. Samples from three replicate plots were pooled, yielding two replicates per treatment. The procedure was carried out as described by Bonsall et al. (7) and modified by Glandorf et al. (18). The detection limit of PCA in roots or in soil was 15 ng per sample.
Enumeration of the culturable microflora.
Selected groups of bacteria and fungi in the wheat rhizosphere were quantified by plating on different media. The aerobic heterotrophic bacterial population was determined on tryptic soy agar (Difco Laboratories, Detroit, Mich.) containing 100 μg of cycloheximide ml−1 after incubation at 20°C for 8 days. To determine the total number of Bacillus spores, dilutions of rhizosphere samples were incubated for 15 min at 80°C to inactivate vegetative cells prior to plating on tryptic soy agar and incubation at 28°C for 2 days. Plate count enumeration of pseudomonads was performed on KB+ agar after incubation at 28°C for 2 days. The size of the predominant fungal community was estimated on 0.25× potato dextrose agar (PDA) (Difco) containing 2 μl of Triton X-100 ml−1 and 200 μg of aureomycin ml−1. Fusarium spp. were determined on Komada's agar (27); fungi belonging to the order Mucorales were identified on PDA supplemented with 50 μg of benomyl ml−1 (6), and Trichoderma species were identified on 0.25× PDA and on Trichoderma selective medium (12). All fungi were incubated at 20°C for 5 days.
Determination of the compositions of predominant fungal and bacterial communities by ARDRA.
A cultivation-independent approach based on amplification of the small-subunit rRNA was used to compare the compositions of the bacterial and fungal communities for different treatments. Three replicates from each treatment were pooled to produce two replicate samples for analysis. Samples were treated as described previously (18), and total DNA was extracted by using a bead beater as described previously (43). PCR with the DNA extracts were performed with fungus- and bacterium-specific primers. Primers EF3 (5′-TCCTCTAAATGACCAAGTTTG-3′) and EF4 (5′-GGAAGGG[G/A]TGT-ATTTATTAG-3′) (18, 44) amplify a 1.4-kb DNA fragment of fungal 18S rDNA. Eubacterial primers 338F (5′ACTCCTA-CGGG[A/G][G/C]GCAGC-3′) (1) and 1492R (5′-GGTTACCTTGTTACGACTT-3′ (13) were used to amplify a 1.1-kb fragment of bacterial 16S rDNA. The PCR conditions used with primers EF3 and EF4 were as follows: 5 min at 94°C for one cycle, 1 min at 94°C, 1 min at 48°C, and 3 min at 72°C for 40 cycles, and 10 min at 72°C. The PCR conditions used with primers 338F and 1492R differed only in the annealing temperature, which was 60°C.
Fungal PCR products were digested with TaqI, and bacterial PCR products were digested with HinfI. The samples were loaded on precast polyacrylamide gels (GeneGelExcel 12.5; Amersham Pharmacia Biotech), and bands were separated with a GenePhor horizontal electrophoresis unit (Amersham Pharmacia Biotech) and silver stained with a Hoefer automated gel stainer by using a DNA silver staining kit (Amersham Pharmacia Biotech). Gel images were digitalized by using the GeneGenius bioimaging system (SYNGENE, Maidenhead, United Kingdom). The Dice coefficient was used to calculate the similarities of the banding patterns. The algorithm used for clustering the resulting DNA patterns was the unweighted pair group method using arithmetic averages (Bionumerics program, version 2.0; Applied Maths, Sint-Martens-Latem, Belgium). Clusters were defined by a cutoff similarity value of 60%.
Plant growth.
Plant growth was determined by measuring the heights and fresh and dry weights of 10 to 15 plants per plot on each sampling date. Plant dry weight was determined after plant shoots were dried at 70°C for 1 to 3 days. After harvest, 20-ear and 100-seed weights were determined for each plot.
NPA.
To determine the effects of the GMMs on the nitrifying microorganisms, we determined the nitrifying potential activity (NPA) during both seasons. At each sampling date 10 g of root-free rhizosphere soil from each plot was suspended in 25 ml of assay medium (5, 45), and the mixture was shaken at 200 rpm for 48 h at 25°C. Samples (1 ml) were withdrawn and centrifuged for 5 min at 13,000 × g. Then 0.5 ml of each supernatant was mixed with an equal amount of 2 M KCl to stop the nitrification reaction. Samples were stored at −20°C until analysis of NO3− with an autoanalyzer (SanPlus System, interface 8708/16; Skalar).
Statistical analysis.
All data except those obtained with ARDRA were statistically analyzed by using the SPSS software package (version 10.0). Results for the populations of WCS358r, the GMMs derived from this strain, the culturable microflora, and plant yield were determined for each sampling date by one-way analysis of variance (ANOVA), followed by a Bonferroni correction. A significant interaction between time and treatment was analyzed with repeated-measurement ANOVA. Before ANOVA was performed, the homogeneity of variances and normal distribution were verified. In cases of heterogeneity of variances or nonnormal distribution, the Kruskal-Wallis nonparametric test was used. For the ARDRA dendrograms the Dice similarity indices were calculated based on a comparison between the banding patterns (Bionumerics program, version 2.0; Applied Maths) and were statistically analyzed by using a permutation test with random sampling. In all cases the confidence interval used was 95%.
RESULTS
Construction of WCS358r::phl.
The rhizobacterium P. putida WCS358r was genetically modified by insertion of the phlFACBDE genes from P. fluorescens Q2-87 (4), encoding production of DAPG. The resulting strain, WCS358r::phl, contained one copy of the gene cluster in the chromosome and produced the same amount of DAPG in vitro as the donor strain produced (data not shown).
Population dynamics of P. putida WCS358r and GMMs derived from it.
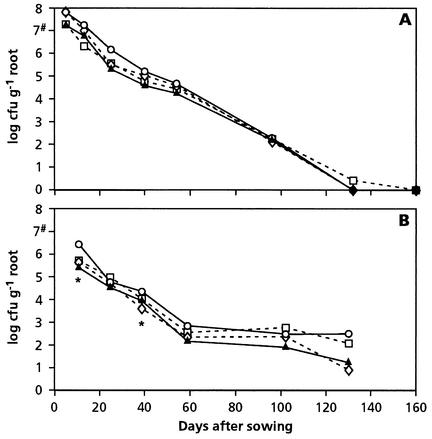
After inoculated seeds were planted in 1999, the rhizosphere populations of both wild-type strain WCS358r and the GMMs decreased from over 107 CFU/g of roots at 5 days after sowing to less than 102 CFU/g of roots at 100 days after sowing (Fig. 1A). In the year 2000 the number of cells dropped more rapidly, from about 106 CFU/g of roots at 11 days after sowing to 102 to 103 CFU/g of roots at 60 days (Fig. 1B). During the next 40 days the cell numbers remained relatively constant, after which an additional but variable decrease was observed at 130 days after sowing. No differences between treatments were apparent, except at two times in the second year. Eleven days after sowing the number of CFU of the GMM strains was significantly lower than the number of CFU of the parental strain (for WCS358r::phz, P = 0.028; for WCS358r::phl, P = 0.049; for WCS358r::phz plus WCS358r::phl, P = 0.004). At 39 days the number of CFU of WCS358r::phz was significantly lower than the number of CFU of the parental strain (P = 0.018).
FIG. 1.
Population dynamics of WCS358r (○), its PCA- and DAPG-producing derivatives WCS358r::phz (⋄) and WCS358r::phl (□), and a combination of the two derivatives (▴) in the wheat rhizosphere in 1999 (A) and 2000 (B). The numbers of CFU were determined on KB+rif. An asterisk indicates that there was a significant difference between the parent strain and WCS358r::phz, WCS358r::phl, or a mixture of the two GMMs 11 days after sowing (P = 0.028, P = 0.049, and P = 0.004, respectively) and between the parent strain and WCS358r::phz 39 days after sowing in 2000 (P = 0.018). The number sign indicates the number of CFU per seed at the beginning of the field experiment.
To investigate if the effect of one antibiotic is enhanced by the presence of a second antibiotic, we applied a mixture of the PCA- and the DAPG-producing GMMs at a 1:1 ratio onto the wheat seeds. During the season the proportions of both GMMs in the plots were calculated from colony hybridization data (Table 2). In 1999 the percentage of the DAPG-producing GMM decreased to about 30% during the growing season. In 2000 the percentage of the DAPG-producing GMM in the combination treatment varied between 37% at the beginning and 67% during the field trial. The latter results were compared to results obtained by plate count enumeration. Because of their different morphologies WCS358r::phz and WCS358r::phl could be easily distinguished on KB+rif/km agar plates. The colonies of WCS358r::phl appeared later and were smaller. Using these two independent assays, we obtained similar results for the average percentage of the DAPG-producing GMM in 2000 (44% ± 26% as determined by plate count enumeration and 53% ± 15% as determined by colony hybridization analysis).
TABLE 2.
Percentages of rifampin-resistant CFU of the DAPG-producing GMM isolated from the rhizospheres of plantsa
| No. of days after sowing | % of rifampin-resistant CFU
|
||
|---|---|---|---|
| 1999 colony blots | 2000 colony blots | 2000 plate counts | |
| 0 | NDb | 37 ± 8c | 17 ± 11 |
| 13 or 11d | 52 ± 11 | ND | 16 ± 10 |
| 25 | 30 ± 5 | 67 ± 20 | 59 ± 19 |
| 39 | 36 ± 6 | 64 ± 40 | 73 ± 20 |
| 59 or 54e | 31 ± 8 | 45 ± 7 | 54 ± 46 |
| Mean | 37 ± 10 | 53 ± 15 | 44 ± 26 |
Seeds were coated with a 1:1 mixture of the PCA- and DAPG-producing GMMs. Colony hybridization was performed on five sampling dates during the field trials in 1999 and 2000. A total of 120 colonies were tested on each sampling date. Plate counting to morphologically distinguish PCA-producing GMMs from DAPG-producing GMMs was performed only in 2000.
ND, not determined.
Mean ± standard deviation.
Thirteen days after sowing in 1999 and 11 days after sowing in 2000.
Fifty-nine days after sowing in 1999 and 54 days after sowing in 2000.
In the control treatment, no CFU were detected on KB+rif. Early in the season single colonies of rifampin- and kanamycin-resistant strains were found outside the plots into which they were introduced (data not shown), indicating that there was dispersal of the introduced bacteria. Later in the season neither the wild type nor the GMMs were detected outside the plots.
Stability of the phz and phl genes.
Under field conditions, the stability of the mini-Tn5 transposon in the GMMs, which, besides the phz and phl genes, contains the nptII gene coding for kanamycin resistance, was examined by determining kanamycin resistance in the GMMs. At each sampling date rhizosphere samples were plated on KB+rif agar with and without kanamycin, after which the numbers of colonies on the two media were compared. No significant differences between the numbers of colonies on the two media were observed in either season. This indicates that the constructs containing the phl or phz genes remained stable in the chromosome of WCS358r.
Detection of PCA in the rhizosphere.
Roots with adhering soil were sampled 18 days after sowing, and PCA was extracted and analyzed by reversed-phase high-performance liquid chromatography. In 1999 40 and 37 ng of PCA per g of roots were detected in the field plots into which the PCA-producing GMM was introduced alone and in combination with the DAPG-producing GMM, respectively (data not shown). No PCA was detected in the other plots. In 2000 PCA was detected only in the plots into which both GMMs were introduced. Quantification was not possible because the amounts were near the detection limit (47).
Effects of the GMMs on the indigenous culturable microflora.
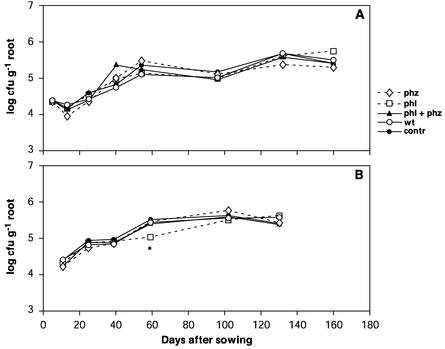
The effects of WCS358r and the GMMs on the numbers of component groups of the culturable indigenous microflora were determined by plating rhizosphere samples on selective media. Besides aerobic heterotrophic bacteria, fluorescent pseudomonads, actinomycetes, Bacillus spp. spores, filamentous fungi, and Fusarium spp., in 1999 members of the order Mucorales and Trichoderma species were also included. With a single exception in neither season was a significant effect of the introduced bacteria on fungal or bacterial communities detected. The single exception concerned the filamentous fungi at 59 days after sowing in 2000, as shown in Fig. 2. There was a significant difference between the counts of filamentous fungi from the rhizospheres of the control plants and the counts of these organisms from plants treated with the DAPG-producing GMM (P = 0.032, as determined by one-way ANOVA). In both seasons the population of filamentous fungi increased from 104 CFU per g of roots to about 5 × 105 CFU per g of roots after harvest.
FIG. 2.
Population dynamics for total filamentous fungi in the rhizospheres of wheat plants treated with WCS358r (wt) (○), its PCA- and DAPG-producing derivatives WCS358r::phz (phz) (⋄) and WCS358r::phl (phl) (□), and a combination of the two derivatives (phl + phz) (▴) in the 1999 (A) and 2000 (B) field seasons. In the control treatment (contr) (•) seeds were coated with methylcellulose (no bacteria). Numbers of CFU were determined on PDA. The asterisk indicates that there was a significant difference between the control treatment and the treatment with WCS358r::phl (P = 0.032).
Effects of the GMMs on the predominant fungal and bacterial microflora.
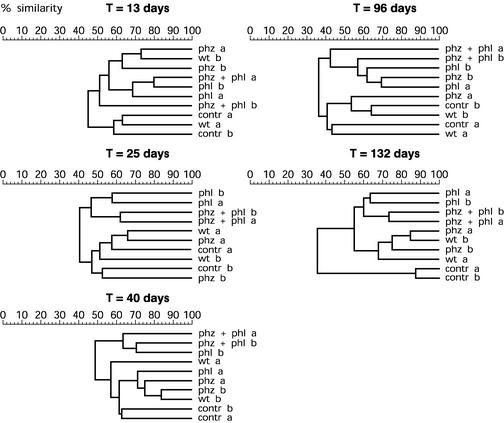
Total DNA from rhizosphere samples from each plot was extracted and analyzed by ARDRA. The field was laid out in such a way that three replicates of each treatment were located in the left half of the field, and the other three replicates were located in the right half. For analysis samples of the three plots from each half were pooled, resulting in two replicates per treatment. A particular partitioning for all dendrograms was obtained by cutting the dendrograms at a similarity index of 60%. Figure 3 shows the dendrograms based on the ARDRA patterns of the fungal community in the wheat rhizosphere in 1999. Throughout the season one distinct cluster was apparent, which contained fungal communities from plants treated with WCS358r::phl, either alone or in combination with WCS358r::phz. Other clusters contained fungal communities from plants treated with the control, WCS358r, and WCS358r::phz. At 25 days samples from plants treated with WCS358r::phl and with WCS358r::phz plus WCS358r::phl clustered separately at the 60% similarity cutoff value, although at the 48% breakpoint they clustered together, apart from all other treatments. At 96 days the cluster contained one rhizosphere sample from the WSC358r::phz treatment. The distinct clustering was most evident at the end of the season, at 132 days. Only at that time did the two control samples cluster together with a similarity of more than 80%. The clustering at 132 days suggests that the WCS358r and WCS358r::phz treatments also affected the fungal community.
FIG. 3.
Dendrograms based on the percentages of similarity of rhizosphere fungal communities of field-grown wheat plants in 1999 that were not treated (contr) or were treated with P. putida WCS358r (wt), with WCS358r::phz (phz), with WCS358r::phl (phl), or with a combination of the two GMMs (phz + phl). The similarities are based on ARDRA patterns generated from amplified 18S rDNA digested with TaqI. Samples were taken 13, 25, 40, 96, and 132 days after sowing. Samples from three plots were pooled, resulting in two replicates per treatment from the left half (a) and right half (b) of the field. The levels of similarity are shown above each dendrogram.
In the bacterial microflora the transgenic WCS358r::phl strain also caused the same distinct clustering that it caused in the fungal communities 13 and 25 days after sowing (data not shown). Later in the season the banding pattern affected by the DAPG-producing GMMs, alone or in combination, clustered in more than one group. However, at 132 days the samples treated with the DAPG-producing GMMs clustered together, including one replicate that received the WCS358r::phz treatment. The dendrogram of the bacterial community showed larger differences than the dendrogram of the fungal community showed (data not shown). The similarity indices within treatments were compared with the average similarity indices between treatments. To do this, we determined all pairwise similarity indices and compared the average within-treatment similarities with the average pairwise similarities between different treatments, using a permutation test with random sampling. Significant differences were detected for the fungal community at 25 days (P = 0.014) and 132 days (P = 0.004) and for the bacterial community at 25 days (P = 0.008) (data not shown).
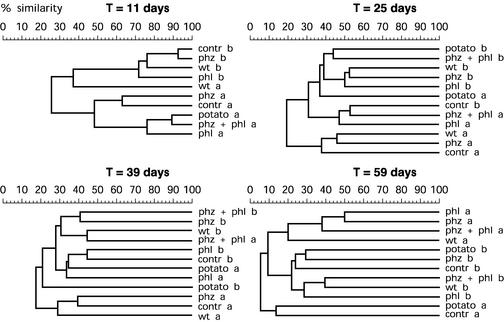
In the field experiment in 2000 rhizosphere samples from the six treatments grouped in a different way than the samples grouped in 1999. At the beginning of the field experiment both the fungal community (Fig. 4) and the bacterial community (data not shown) clustered mainly according to their positions in the field (rhizosphere samples from the left half or the right half of the field) regardless of the treatment. This was most evident for the fungal community (Fig. 4) and the bacterial community (data not shown) 11 days after sowing. During the growing season the differences between the fungal communities in the plots increased, and the communities clustered at the 60% level in eight groups, whereas the bacterial communities clustered in three, six, and five groups at 25, 39, and 59 days, respectively (data not shown). Each group contained mostly rhizosphere samples from either the left or the right half of the experimental field. The heterogeneity between samples for the fungal and bacterial communities was high, comparable with that found in 1999 for the bacterial community. No effect of the introduced strains was detected.
FIG. 4.
Dendrograms based on the percentages of similarity of rhizosphere fungal communities of field-grown wheat plants in 2000 that were not treated (contr) or were treated with P. putida WCS358r (wt), with WCS358r::phz (phz), with WCS358r::phl (phl), or with a combination of the two GMMs (phz + phl). The similarities are based on ARDRA patterns generated from amplified 18S rDNA digested with TaqI. Samples were taken 11, 25, 39, and 59 days after sowing. Samples from three plots were pooled, resulting in two replicates per treatment from the left half (a) and the right half (b) of the field. The levels of similarity are shown above each dendrogram.
In 2000 potatoes were planted in the rotation plots. We anticipated that different crops would support different microbial communities, as described in other studies (33). However, ARDRA, as shown in Fig. 4, did not reveal a distinct clustering of the rotation plots. At no time did similarity indices analyzed with the permutation test reveal significant differences.
Effects on NPA.
The nitrifying bacteria, such as Nitrosomonas and Nitrobacter, are an important group of soil microorganisms, which oxidize ammonium to nitrite or nitrate (2). One way to study the nitrifying activity in soil is to measure the NPA (5). The NPA was determined on each sampling date. In 1999 the field was not fertilized, and the NPA remained between 0.4 and 2.6 nmol of NO3− g (dry weight) of soil−1 h−1 during the 54 days after sowing (data not shown). In 2000 the field was fertilized, and the NPA for all treatments varied between 2 and 8 nmol of NO3− g (dry weight) of soil−1 h−1 before it decreased to 0.5 nmol of NO3− g (dry weight) of soil−1 h−1 at 59 days after sowing (data not shown). Neither in 1999 nor in 2000 did introduction of WCS358r or the GMMs significantly affect the soil nitrifying potential (P > 0.05) (data not shown).
Plant yield.
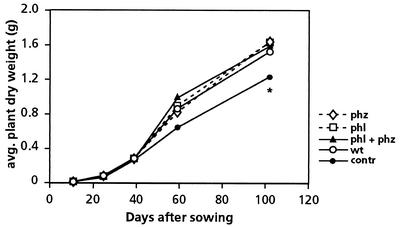
To determine if the introduced bacteria affected plant development, the fresh and dry weights of 10 to 15 shoots from each plot per sampling date were determined. In addition, after harvest 100-seed and 20-ear weights were determined. In 1999 the introduced bacteria had no effect on plant growth and yield (data not shown). However, in 2000 all introduced bacteria had a positive effect on the shoot fresh and dry weights. Dry weight was increased by 41% by all bacterial treatments compared to the control dry weight at harvest, 102 days after sowing (P < 0.04) (Fig. 5). The 20-ear weight increased by 14 to 21%, and there was a significant difference between the control plants and the GMM-treated plants (P < 0.04); the 100-seed weight increased by 5 to 8%, and there was a significant difference between control and bacterium-treated plants (P < 0.035) except for the WCS358r::phz-treated plants (P = 0.131) (data not shown). However, no significant difference in plant growth yield between the plots that received the parent strain and the plots that received the GMM derivatives was detected.
FIG. 5.
Dry weight per shoot for wheat plants treated in 2000 with P. putida WCS358r (wt) (○), with the PCA- and DAPG-producing derivatives WCS358r::phz (phz) (⋄) and WCS358r::phl (phl) (□), and with a combination of the two derivatives (phl + phz)(▴). In the control treatment (contr) (•) seeds were coated with methylcellulose (no bacteria). The values are means for 10 plants per treatment and sampling date. The asterisk indicates that there was a significant difference between the control plants and the bacterium-treated plants (P < 0.04).
DISCUSSION
P. putida WCS358r is a well-studied plant growth-promoting rhizobacterium that suppresses Fusarium wilt in carnation and radish based on the production of the fluorescent siderophore pseudobactin 358 (11, 35). In Arabidopsis thaliana it suppresses Fusarium wilt by induction of systemic resistance (50). Glandorf et al. (18) modified WCS358r with the phz gene locus, which resulted in constitutive production of PCA. WCS358r and two PCA-producing derivatives were introduced onto wheat seeds by a single application in two separate field experiments in 1997 and 1998. In both years a transient effect on the indigenous fungal microflora was detected (18).
In our field study we released twice into the same field WCS358r and its PCA-producing derivative WCS358r::phz, as well as a DAPG-producing derivative, WCS358r::phl, alone and in combination with WCS358r::phz. We simulated commercial application protocols by applying the strains as seed coatings in two consecutive years. Moreover, a single application of a biological control agent is unlikely to be fully effective in controlling disease (9). Production of PCA or DAPG is an important trait in biological control agents and can lead to suppression of plant root diseases (55). A single copy of a disarmed Tn5 transposon containing the genes for the production of DAPG was inserted into the chromosome of WCS358r, resulting in WCS358r::phl. Chromosomal insertions are more stable and are less frequently subject to horizontal gene transfer than extrachromosomal elements (41). Thus, the possibility that the Tn5 transposon that includes the DAPG gene cluster is transferred to other bacteria is minimized, an important factor when microorganisms are released into the environment.
After introduction of the wild type and the GMMs into the same field in 1999 and 2000, the numbers of cells of the inoculated bacteria dropped more rapidly during the first 60 days in 2000 than in 1999.
Successful colonization is a process that depends, among other things, on environmental conditions, such as weather. There was no major difference in the average temperature in the 2 years studied (14.2°C in 1999, 14.7°C in 2000). However, a total rainfall of 85 mm was registered at the nearby Royal Netherlands Meteorological Institute in May 2000, compared to only 52 mm in May 1999. In both years the population dynamics in the wheat rhizosphere revealed no significant differences between the wild type and the GMMs. This indicates that the GMMs were as fit as the wild type under the conditions tested. These results confirm previous observations that chromosomal insertion of the phz genes into Pseudomonas has no negative effect on the fitness of the organism in the wheat rhizosphere (18, 48). In two strains, P. fluorescens 2-79 and Pseudomonas chlororaphis 30-84, production of phenazine antibiotics contributes to the ability of the organisms to survive in and colonize the wheat rhizosphere in competition with the indigenous soil microflora (31). However, genetic modification to produce antibiotics can also lead either to reduced ecological fitness or to defects in competitive or survival ability (9).
Since DAPG is also effective against bacteria (25), effects on the fungal microflora (18) and also effects on the bacterial microflora were expected. Introduction of WCS358r, WCS358r::phz, and WCS358r::phl had no effect on the numbers of culturable bacteria and fungi after either the first introduction or the second introduction. In vitro assays indicated that compared to WCS358r, the PCA- and DAPG-producing GMMs had an increased inhibitory effect against saprophytic fungi isolated from the same field soil (data not shown). This finding confirms that the results obtained under laboratory conditions can differ from those obtained under field conditions due to complex environmental conditions (42).
In 1999, ARDRA revealed that introduction of the DAPG-producing GMM, alone or in combination with the PCA-producing GMM, affected the composition of the rhizosphere microflora. It is reasonable to expect that repeated introduction of the bacterial inoculants increase their effect. In 2000, however, rhizosphere samples clustered according to their positions in the experimental field (the left or right half of the field) independent of the seed treatment. It is possible that changes in environmental conditions caused a difference between the left and right halves of the experimental field, resulting in the distinct clustering in the second year, and that effects of the bacteria did not exceed this variability. Despite the fact that PCA was produced in the rhizosphere of wheat, there was no effect of the PCA-producing GMM. Glandorf et al. (18), however, found that the same strain, WCS358r::phz (GMM 8), after a single introduction did affect the fungal community. In our study the transient effect on the fungal and bacterial microflora resulted from introduction of the DAPG-producing GMM.
A primary benefit of crop rotation relies on the activity of the indigenous microorganisms in the soil, which deplete the energy sources for pathogenic fungi after crops are harvested (8). A 1-year rotation is sufficient, for instance, to control root pathogens of barley and wheat (8). Miethling et al. (32) compared the effects of crop specificity, soil origin, and a bacterial inoculant on the establishment of microbial communities. With the methods which they used, community-level physiological profile, fatty acid analysis, and temperature gradient gel electrophoresis, they showed that crop species had the most pronounced effect on the composition of rhizosphere microorganisms. Tomato and flax had a selective influence on populations of fluorescent pseudomonads, as demonstrated by Lemanceau et al. (29). In this study phenotypic and taxonomic characteristics of type strains and wild-type isolates from soil and plants were compared. The bacteria were grouped on the basis of their enzyme activities and the ability to utilize 147 substrates.
Comparison of lipopolysaccharide and cell envelope protein patterns of pseudomonads isolated from potato, grass, and wheat revealed 30 distinct patterns for each crop, and the majority of these patterns were not observed for the other crops (17). Differences in the root exudates of different plant species and varieties are known to affect the microbial communities (28). In our field study no effect of crop rotation was detected with the methods used. The overall variability of the microbial communities of both the wheat rhizosphere and the potato rhizosphere may have been too high to detect crop specificity under the environmental conditions encountered, using ARDRA. ARDRA has been used successfully previously to detect shifts in microbial communities (18, 40). In contrast to our expectations, we could not demonstrate an enhanced effect on the microbial communities after a second introduction of the bacterial inoculants or an effect of crop rotation. This may have been due in part to the limitations of the technique which we used (DNA extraction hampered by humic acids in the soil, preferential amplification of dominant members of the microbial community, low discriminatory power of ARDRA). However, since there was a significant effect of the DAPG-producing genetically modified derivatives in the first year, it is likely that the high variability between samples in 2000 exceeded possible effects caused by the GMMs.
Only a few microbial genera are able to utilize energy derived from nitrification, a process that can be severely affected by environmental stress (2). We anticipated that the activity of nitrifying bacteria would be easily affected by the introduction of bacterial inoculants into the soil, and we therefore determined the NPA. However, in neither season did we detect an effect of the introduced microorganisms on the NPA, indicating that production of the antibiotics did not adversely affect the capacity for nitrification in the soil.
In 2000 all bacterial treatments resulted in increased plant growth compared to the control growth. Supposedly, the main mechanism in plant growth promotion by rhizobacteria is suppression of deleterious microorganisms (38). These microorganisms commonly colonize the root systems of crops such as wheat, potato, and sugar beet (14). The siderophores of the rhizobacterium WCS358r, produced under iron-limited conditions, have a higher affinity for iron than those of deleterious microorganisms have (38). Consequently, deleterious microorganisms are outcompeted by WCS358r in the rhizosphere, resulting in greater growth of plants treated with WCS358r or GMMs derived from it. However, there was no significant difference in growth between wild-type-treated plants and GMM-treated plants, suggesting that production of either PCA or DAPG is not important for the observed plant growth stimulation. This effect was detected only in the second year and not in 1999 or in the 1-year field experiments described previously (18), despite similar population dynamics of the introduced bacteria in the different years. As for the effect of the genetic modification, our study showed that the DAPG-producing GMM caused a transitory shift in the fungal and bacterial microflora. Repeated introduction of the GMMs did not lead to the expected intensified effects. So far, we have found no evidence that GMMs modified for enhanced biological control activity have a major impact on the environment. The field trial will be continued for two more years, and the effects of the GMMs will be investigated further.
Acknowledgments
This study was financed by the Dutch Ministry of Housing, Spatial Planning and the Environment.
We thank Hans van Pelt and Ientse van der Sluis for excellent technical assistance, Niko Nagelkerke for assistance with the statistical analysis, and Marjolein Kortbeek-Smithuis for editing Fig. 3 and 4. We also thank Bas Valstar, Fred Siesling, and Jeroen van Schaik (Botanical Garden, Utrecht University) for constructing and maintaining the experimental field site.
REFERENCES
- 1.Amann, R., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas, R. M., and R. Bartha. 1998. Microbial ecology: fundamentals and applications, 4th ed. Benjamin/Cunnings Science Publishing, Menlo Park, Calif.
- 3.Bakker, P. A. H. M., J. G. Lamers, A. W. Bakker, J. D. Marugg, P. J. Weisbeek, and B. Schippers. 1986. The role of siderophores in potato tuber yield increase by Pseudomonas putida in short rotation to potato. Neth. J. Plant Pathol. 92:249-256. [Google Scholar]
- 4.Bangera, G. M., and L. S. Thomashow. 1996. Characterization of a genomic locus required for synthesis of the antibiotic 2,4-diacetylphloroglucinol by biocontrol control agent Pseudomonas fluorescens Q2-87. Mol. Plant-Microbe Interact. 9:83-90. [DOI] [PubMed] [Google Scholar]
- 5.Bodelier, P. 1997. Nitrification and denitrification in the root zone of Glyceria maxima. Ph.D. thesis. University of Nijmegen, Nijmegen, The Netherlands.
- 6.Bollen, G. J. 1972. A comparison of the in vitro antifungal spectra of thiophanates and benomyl. Neth. J. Plant Pathol. 78:55-64. [Google Scholar]
- 7.Bonsall, R. F., D. M. Weller, and L. S. Thomashow. 1997. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 63:951-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook, R. J. 1992. Wheat root health management and environmental concern. Can. J. Plant Pathol. 14:76-85. [Google Scholar]
- 9.Cook, R. J. 1993. Making greater use of introduced microorganisms for biological control of plant pathogens. Annu. Rev. Phytopathol. 31:53-80. [DOI] [PubMed] [Google Scholar]
- 10.De Leij, F. A. A. M., E. J. Sutton, J. M. Whipps, J. S. Fenlon, and J. M. Lynch. 1995. Impact of field release of a genetically modified Pseudomonas fluorescens on indigenous microbial populations of wheat. Appl. Environ. Microbiol. 61:3443-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duijff, B. J., P. A. H. M. Bakker, and B. Schippers. 1994. Suppression of fusarium wilt of carnation by Pseudomonas putida WCS358 at different levels of disease incidence and iron availability. Biocontrol Sci. Technol. 4:279-288. [Google Scholar]
- 12.Elad, Y., I. Chet, and Y. Henis. 1981. A selective medium for improving quantitative isolation of Trichoderma spp. from soil. Phytoparasitica 9:59-67. [Google Scholar]
- 13.Embley, T. M., J. Smida, and E. Stackebrandt. 1988. Reverse-transcriptase sequencing of 16S ribosomal RNA from Faenia rectivirgula, Pseudonocardia hermophila and Saccharopolyspora hirsuta, 3 wall type IV actinomycetes which lack mycolic acids. J. Gen. Microbiol. 134:961-966. [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto, D. K., D. M. Weller, and L. S. Thomashow. 1995. Role of secondary metabolites in root disease suppression. ACS (Am. Chem. Soc.) Symp. Ser. 582:330-347. [Google Scholar]
- 15.Geels, F. P., and B. Schippers. 1983. Reduction of yield depression in high frequency potato cropping soil after seed tuber treatments with antagonistic fluorescent Pseudomonas spp. Phytopathol. Z. 108:207-214. [Google Scholar]
- 16.Glandorf, D. C. M., I. Brand, P. A. H. M. Bakker, and B. Schippers. 1992. Stability of rifampin resistance as a marker for root colonization studies of Pseudomonas putida in the field. Plant Soil 147:135-142. [Google Scholar]
- 17.Glandorf, D. C. M., L. G. L. Peters, I. Van der Sluis, P. A. H. M. Bakker, and B. Schippers. 1993. Crop specificity of rhizosphere pseudomonads and the involvement of root agglutinins. Soil Biol. Biochem. 25:981-989. [Google Scholar]
- 18.Glandorf, D. C. M., P. Verheggen, T. Jansen, J.-W. Jorritsma, E. Smit, P. Leeflang, K. Wernars, L. S. Thomashow, E. Laureijs, J. E. Thomas-Oates, P. A. H. M. Bakker, and L. C. van Loon. 2001. Effect of genetically modified Pseudomonas putida WCS358r on the fungal rhizosphere microflora of field-grown wheat. Appl. Environ. Microbiol. 67:3371-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas, D., C. Blumer, and C. Keel. 2000. Biocontrol ability of fluorescent pseudomonads genetically dissected: importance of positive feedback regulation. Curr. Opin. Biotechnol. 11:290-297. [DOI] [PubMed] [Google Scholar]
- 20.Handelsmann, J., and C. Stabb. 1996. Biocontrol of soilborne pathogens. Plant Cell 8:1855-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hase, C., F. Macher, Y. Moënne-Loccoz, and G. Défago. 1999. Nutrient deprivation and the subsequent survival of biocontrol Pseudomonas fluorescens CHAO in soil. Soil Biol. Biochem. 31:1181-1188. [Google Scholar]
- 22.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, G. T., N. A. Mitkowski, L. Aldrich-Wolfe, L. R. Emele, D. D. Jurkonie, A. Ficke, S. Maldonado-Ramirez, S. T. Lynch, and E. B. Nelson. 2000. Methods for assessing the composition and diversity of soil microbial communities. Appl. Soil Ecol. 15:25-36. [Google Scholar]
- 24.Hugenholtz, P., and N. R. Pace. 1996. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 14:190-197. [DOI] [PubMed] [Google Scholar]
- 25.Keel, C., U. Schnider, M. Maurhofer, C. Voisard, J. Laville, U. Burger, P. Wirthner, D. Haas, and G. Défago. 1992. Suppression of root diseases by Pseudomonas fluorescens CHAO: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant-Microbe Interact. 5:4-13. [Google Scholar]
- 26.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for demonstration of pycocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 27.Komada, H. 1975. Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soils. Rev. Plant Prot. Res. 8:114-125. [Google Scholar]
- 28.Kremer, R. J., M. F. T. Begonia, L. Stanley, and E. T. Lanham. 1990. Characterization of rhizobacteria associated with weed seedlings. Appl. Environ. Microbiol. 56:1649-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemanceau, P., T. Corberand, L. Gardan, X. Latour, G. Laguerre, J. Boeufgras, and C. Alabouvette. 1995. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl. Environ. Microbiol. 61:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurhofer, M., C. Keel, C. Voisard, D. Haas, and G. Défago. 1992. Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHAO on its disease suppressive capacity. Phytopathology 82:190-195. [Google Scholar]
- 31.Mazzola, M., J. R. Cook, L. S. Thomashow, D. M. Weller, and L. S. Pierson III 1992. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 58:2616-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miethling, R., G. Wieland, H. Backhaus, and C. C. Tebbe. 2000. Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb. Ecol. 41:43-56. [DOI] [PubMed] [Google Scholar]
- 33.Olsson, S., and S. Alström. 2000. Characterization of bacteria in soils under barley monoculture and crop rotation. Soil Biol. Biochem. 32:1443-1451. [Google Scholar]
- 34.Pierson, L. S., III, and L. S. Thomashow. 1992. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol. Plant-Microbe Interact. 5:330-339. [DOI] [PubMed] [Google Scholar]
- 35.Raaijmakers, J. M., M. Leemann, M. M. P. van Oorschot, I. van der Sluis, B. Schippers, and P. A. H. M. Bakker. 1995. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology 85:1075-1081. [Google Scholar]
- 36.Raaijmakers, J. M., D. M. Weller, and L. S. Thomashow. 1997. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 63:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Manniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Schippers, B., A. W. Bakker, and P. A. H. M. Bakker. 1987. Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices. Annu. Rev. Phytopathol. 25:339-358. [Google Scholar]
- 39.Schmidt, T. M. 1994. Fingerprinting bacterial genomes using ribosomal RNA genes and operons. Methods Mol. Cell. Biol. 5:3-12. [Google Scholar]
- 40.Schwieger, F., and C. C. Tebbe. 2000. Effect of field inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizospheres of a target plant (Medicago sativa) and a non-target plant (Chenopodium album)—linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl. Environ. Microbiol. 66:3556-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengeløv, G., K. J. Kreitensen, A. H. Sørensen, N. Kroer, and S. Sørensen. 2001. Effect of genomic location on horizontal transfer of a recombinant gene cassette between Pseudomonas strains in the rhizosphere and spermosphere of barley seedlings. Curr. Microbiol. 42:160-167. [DOI] [PubMed] [Google Scholar]
- 42.Sharifi-Tehrani, A., M. Zala, A. Natsch, Y. Moënne-Loccoz, and G. Défago. 1998. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified16S rDNA. Eur. J. Plant Pathol. 104:631-643. [Google Scholar]
- 43.Smalla, K., N. Cresswell, L. C. Mendonca-Hagler, A. Wolters, and J. D. Van Elsas. 1993. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J. Appl. Bacteriol. 74:78-85. [Google Scholar]
- 44.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stienstra, A. W., P. Klein Gunnewiek, and H. J. Laanbroek. 1994. Repression of nitrification in soils under a climax grassland vegetation. FEMS Microbiol. Ecol. 14:45-52. [Google Scholar]
- 46.Thirup, L., F. Ekelund, K. Johnsen, and C. S. Jacobsen. 2000. Population dynamics of the fast-growing sub-populations of Pseudomonas and total bacteria, and their protozoan grazers, revealed by fenpropimorph treatment. Soil Biol. Biochem. 32:1615-1623. [Google Scholar]
- 47.Thomashow, L. S., R. F. Bonsall, and D. M. Weller. 1997. Antibiotic production by soil and rhizosphere microbes in situ, p. 493-499. In C. J. Hurst, G. R. Knudsen, M. J. McInery, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 48.Timms-Wilson, T. M., R. J. Ellis, A. Renwick, D. J. Rhodes, D. V. Mavrodi, D. M. Weller, L. S. Thomashow, and M. J. Bailey. 2000. Chromosomal insertion of phenazine-1-carboxylic acid biosynthetic pathway enhances efficacy of damping-off disease control by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 13:1293-1300. [DOI] [PubMed] [Google Scholar]
- 49.Van Overbeek, L. S., J. A. Van Veen, and J. D. Van Elsas. 1997. Induced reporter gene activity, enhanced stress resistance, and competitive ability of a genetically modified Pseudomonas fluorescens strain released into the field. Appl. Environ. Microbiol. 63:1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Wees, S. C. M., C. M. J. Pieterse, A. Trijssenaar, Y. A. M. Van't Westende, F. Hartog, and L. C. Van Loon. 1997. Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol. Plant-Microbe Interact. 10:716-724. [DOI] [PubMed] [Google Scholar]
- 51.Völksch, B., and R. May. 2000. Biological control of Pseudomonas syringae pv. glycinea by epiphytic bacteria under field conditions. Microb. Ecol. 41:132-139. [DOI] [PubMed] [Google Scholar]
- 52.Walsh, U. F., J. P. Morrissey, and F. O'Gara. 2001. Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr. Opin. Biotechnol. 12:289-295. [DOI] [PubMed] [Google Scholar]
- 53.Weller, D. M. 1988. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26:379-407. [Google Scholar]
- 54.Weller, D. M., and R. J. Cook. 1983. Suppression of take-all of wheat by seed treatments with fluorescent pseudomonads. Phytopathology 73:463-469. [Google Scholar]
- 55.Weller, D. M., J. M. Raaijmakers, B. B. McSpadden, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]