Abstract
In this study, a lab-scale rotating biological contactor (RBC) treating a synthetic NH4+ wastewater devoid of organic carbon and showing high N losses was examined for several important physiological and microbial characteristics. The RBC biofilm removed 89% ± 5% of the influent N at the highest surface load of approximately 8.3 g of N m−2 day−1, with N2 as the main end product. In batch tests, the RBC biomass showed good aerobic and anoxic ammonium oxidation (147.8 ± 7.6 and 76.5 ± 6.4 mg of NH4+-N g of volatile suspended solids [VSS]−1 day−1, respectively) and almost no nitrite oxidation (< 1 mg of N g of VSS−1 day−1). The diversity of aerobic ammonia-oxidizing bacteria (AAOB) and planctomycetes in the biofilm was characterized by cloning and sequencing of PCR-amplified partial 16S rRNA genes. Phylogenetic analysis of the clones revealed that the AAOB community was fairly homogeneous and was dominated by Nitrosomonas-like species. Close relatives of the known anaerobic ammonia-oxidizing bacterium (AnAOB) Kuenenia stuttgartiensis dominated the planctomycete community and were most probably responsible for anoxic ammonium oxidation in the RBC. Use of a less specific planctomycete primer set, not amplifying the AnAOB, showed a high diversity among other planctomycetes, with representatives of all known groups present in the biofilm. The spatial organization of the biofilm was characterized using fluorescence in situ hybridization (FISH) with confocal scanning laser microscopy (CSLM). The latter showed that AAOB occurred side by side with putative AnAOB (cells hybridizing with probe PLA46 and AMX820/KST1275) throughout the biofilm, while other planctomycetes hybridizing with probe PLA886 (not detecting the known AnAOB) were present as very conspicuous spherical structures. This study reveals that long-term operation of a lab-scale RBC on a synthetic NH4+ wastewater devoid of organic carbon yields a stable biofilm in which two bacterial groups, thought to be jointly responsible for the high autotrophic N removal, occur side by side throughout the biofilm.
Sustainable wastewater treatment systems are being developed that minimize energy consumption, CO2 emission, and sludge production. However, these systems typically yield effluents rich in ammonium-nitrogen (NH4+-N) and poor in biodegradable organic carbon, thereby making them less suitable for biological N removal through the conventional nitrification-denitrification sequence.
Different N removal processes that could be successfully integrated in a sustainable wastewater treatment system are being studied. The Sharon process (single-reactor high-activity ammonium removal over nitrite) (15) uses the principle that at higher temperatures (30 to 35°C), pH 7 to 8, and a cell residence time of 1 day, aerobic ammonia-oxidizing bacteria (AAOB) are able to maintain themselves in the system while nitrite-oxidizing bacteria (NOB) are washed out. Given the reaction stoichiometry of the two groups of nitrifying bacteria (equations 1 and 2), this process yields nitrite (NO2−) instead of nitrate (NO3−), needing less energy for aeration and less carbon source for subsequent denitrification:
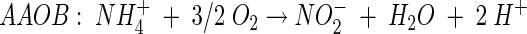 |
(1) |
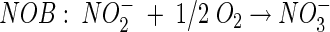 |
(2) |
Combination of the Sharon process with the newly discovered anaerobic ammonium oxidation (Anammox) process (equation 3), which involves the autotrophic, anoxic oxidation of NH4+ to N2 with NO2− as the electron acceptor (43), can bring about completely autotrophic N removal (41):
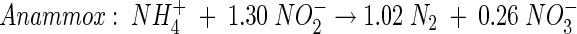 |
(3) |
The bacteria involved in the Anammox process are anaerobic ammonia-oxidizing bacteria (AnAOB) belonging to a new, deep-branching group within the Planctomycetales (Candidatus Brocadia anammoxidans [43]) and showing less than 80% sequence similarity to previously recognized planctomycetes. More recently, a one-reactor combination of partial nitrification to NO2− and Anammox was proposed (completely autotrophic nitrogen removal over nitrite [CANON], equation 4), described mainly for suspended systems (41, 44) but also modeled as a biofilm process (14):
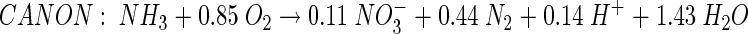 |
(4) |
In parallel with the research on Sharon and Anammox, several reports of unexplained N losses in full-scale nitrifying biofilm reactors have been published (16, 20, 40). The process has been called aerobic/anoxic deammonification (18). Autotrophic N-removing biofilm reactors have been examined at the compositional and ecophysiological level (10, 17, 18) and have been subjected to mathematical modeling (25). A second type of planctomycete AnAOB (Candidatus Kuenenia stuttgartiensis [38]), only distantly (<91% sequence similarity) related to the original anaerobic AOB, has been detected. By using specific PCR primers and fluorescence in situ hybridization (FISH) probes developed in earlier studies (33, 38, 43), several close relatives of K. stuttgartiensis (>98% sequence similarity) have subsequently been detected in different full-scale and lab-scale biofilm systems treating primarily raw leachates (10, 19).
The principle of autotrophic N removal under O2 limitation has been examined in our laboratory and termed oxygen-limited autotrophic nitrification-denitrification (OLAND) (28). AAOB are of particular importance in this process because it is postulated that these bacteria play an active role in the anoxic part of the oxygen-limited biofilm, next to the known AnAOB, which differentiates the OLAND principle from the CANON process. Removals of up to 50 mg of N liter−1 day−1 were possible using an actively nitrifying enrichment culture (ammonium binding inoculum liquid [ABIL]; Avecom, Beernem, Belgium) under O2 limitation, and the end product was mainly N2 (28). After these experiments, a lab-scale rotating biological contactor (RBC) was started using ABIL as an inoculum (36). In a second phase, granular anaerobic sludge from a full-scale upflow anaerobic sludge blanket reactor treating potato-processing wastewater (Primeur, Waregem, Belgium) was added to this reactor as a second biocatalyst inoculum. High N removals were achieved afterward, which were shown to be due to a coupling of NH4+ and NO2− to N2 under anoxic conditions (37). In this paper, we describe several important physiological and microbial characteristics of the RBC biomass that developed from these two different kinds of biocatalyst after long-term operation on a synthetic NH4+ wastewater devoid of organic carbon.
MATERIALS AND METHODS
Lab-scale RBC reactor.
The reactor configuration used in these experiments has been fully described previously (36). It has a total liquid volume of 44 liters, and 40 polyvinyl chloride disks, with a total surface area of 6.3 m2, resulting in 50% submersion of the disk surface. Low-level aeration, required for optimal OLAND conditions, was provided by gentle rotation of all disks along a horizontal shaft (2.5 rpm). The influent flow rate during the whole period described in this paper was set at 62.3 liters day−1 (hydraulic residence time, ca. 17 h). Detailed information on the reactor performance prior to the period described in this paper is published elsewhere (36, 37).
Chemical analyses.
The concentrations of NH4+-N (measured as total ammoniacel N) and total Kjeldahl-N were determined by the Kjeldahl distillation method (7). Influent and effluent nitrate, nitrite, phosphate, and sulfate concentrations were determined using a DX 600 ion chromatograph (Dionex, Sunnyvale, Calif.) equipped with a conductivity detector. Operational parameters were as follows: column AS9-HC; eluent, 9 mM Na2CO3; flow, 1 ml min−1; sample loop, 200 μl. The pH was determined potentiometrically with a portable digital pH meter (Knick portamess 751). The dissolved-oxygen (DO) concentration was measured with a portable digital DO meter (Endress-Hauser COM 381) with a detection limit of 0.015 mg of O2 liter−1. Calibration and cleaning were done every 2 to 3 weeks as specified by the manufacturer. Chemical oxygen demand and volatile and total suspended solids (VSS and TSS) were determined by standard methods (13).
Quantification of gaseous N production by the RBC reactor.
Gaseous N losses were measured after closing the RBC reactor with an airtight Plexiglas cap on day 644. Headspace air (calculated volume, 151 liters) was mixed with fans and refreshed with atmospheric air at a dilution rate of 0.57 liter min−1. N2O and NO accumulation were monitored continuously during 8-h periods before removing the Plexiglas cap to avoid anoxic conditions. To estimate the N2 production, enclosed headspace air (already containing about 79% N2) was purged with a 21% O2-79% Ar mixture at the beginning of the experiment. N2 accumulation was monitored for 2 h by discontinuous sampling (every 10 min) of the headspace gas. For NO and N2O, a mass balance [dC/dt = (Q/V) × (Cair − C) + P, where C and Cair are the concentration of NO or N2O in the reactor headspace and air, respectively, Q is 0.57 liter min−1, V is 151 liters, and P is the production rate of NO or N2O] was calculated; a second-order equation was fitted to the sampling points and integrated between 0 and 8 h. From this, the production rate, P, could thus be calculated as the only remaining unknown parameter. The same calculation method was used for N2 production, but using a first-order best fit due to the absence of a dilution rate (Q = 0) in this mass balance.
The nitrous oxide (N2O) concentration was determined using a photoacoustic detector connected to an infrared gas analyser (type 1302; Brüel & Kjær). The nitric oxide (NO) concentration was determined using a continuous-flow chemiluminiscence analyser (CLD 77 AM; Eco Physics). The dinitrogen (N2) concentration was determined with a gas chromatograph (CP 9000; Chrompack), equipped with a CTR I column and a thermal conductivity detector.
Aerobic and anoxic batch experiments with RBC biomass.
The aerobic batch experiments were carried out in 500-ml Erlenmeyer flasks. Non-O2-limiting conditions (DO concentration, ≥6 mg liter−1) were used to measure the nitrification capacity of the RBC biofilm, similar to activity measurements of AAOB and NOB in other O2-limited systems (41). To minimize anaerobic ammonia-oxidizing and denitrification activity, biomass was preaerated. Allylthiourea was used (10 mg liter−1) to inhibit nitrification activity in some batch tests to measure NH4+ production through cell decay. All Erlenmeyer flasks contained 25 mM KH2PO4-25 mM K2HPO4 as a buffer, 6 mM NaHCO3 and 2 ml of trace-element solution liter−1 (28). The initial pH was 7.15 ± 0.03. This test was carried out at a temperature (T) of 29 ± 2°C. Prior to sampling, aeration to the flasks was stopped for about 5 min for biomass sedimentation. The mixed liquor (5 ml) was sampled, filtered through a 0.40-μm-pore-size filter, and analyzed for NH4+-N, NO2−-N, and NO3−-N. Before biomass sedimentation, pH, DO concentration, and T were recorded. The TSS and VSS of all flasks were also determined.
The anaerobic batch experiments were carried out to measure the anaerobic ammonia-oxidizing activity of the biomass in 120-ml serum bottles containing 80 ml of mixed liquor (40-ml headspace). The mixed liquor contained 6 mM NaHCO3, 0.4 mM KH2PO4, and 2 ml of a trace-element solution liter−1. Biomass was added, and the serum bottles were closed with butyl rubber stoppers and aluminum caps (initial pH 7.6). Anaerobic conditions were obtained by applying 20 cycles of sequential headspace vacuum removal/replacement with Ar (700-kPa overpressure). Substrate was added to a final concentration of 100 mg of NH4+-N and NO2−-N liter−1, and biomass was incubated at 29 ± 2°C. Samples (5 ml) were taken at regular intervals, filtered through a 0.40-μm-pore-size filter, and analyzed for NH4+-N, NO2−-N, and NO3−-N. Samples of the headspace (1 ml) were also analyzed for N2O and N2. In addition, a set of anoxic-activity experiments was performed with increased PO43− concentrations (1.8, 3.6, and 9.2 mM) and with 10 mg of allylthiourea liter−1 (in triplicate).
To compare batch kinetic experiments with reactor kinetics, the amount of VSS on the rotating disks was estimated at the time of the batch experiments by measuring the VSS of five independent, representative samples of 100 cm2 of biofilm.
DNA extraction and purification, PCR, and DGGE.
Total DNA was extracted from the biofilm (5) and purified using Wizard PCR prep columns (Promega, Madison, Wis.). To increase the sensitivity and to facilitate the denaturing gradient gel electrophoresis (DGGE) by analyzing fragments of the same length, a nested PCR technique was applied as described previously (6). For first-round PCR amplification of 16S rRNA genes of members of the β subdivision of AAOB, the CTO189fAB plus CTO189fC and CTO653r primer set was used (27). During the second round of PCR, the obtained fragments were reamplified by using primers P338f and P518r (34). In a first planctomycete (PLA)-targeted PCR round, primers PLA46f (5′-GGATTAGGCATGCAAGTC-3′) and PLA886r (5′-GCCTTGCGACCATACTCCC-3′) were used, which are broadly PLA targeted and were used under conditions that would permit amplification of the known AnAOB, having mismatches at positions 9 and 14 of primer PLA886r (A versus G). During the second round of PCR, the obtained PLA fragments were reamplified by using primers P338f and P518r (34). However, these primers did not allow us to retrieve sequences with high homology to known AnAOB. Hence, a more rigorous second PCR-based identification was attempted that used only primers with perfect complementarity to all known AnAOB: a PLA-specific sequence PLA40f (5′-CGGCRTGGATTAGGCATG-3′) and the eubacterially conserved primer P518r (34). For the second round of PCR, a 50:50 mixture of P338f and P338-IIf (5′-ACACCTACGGGTGGCTGC-3′) with a 40-bp GC clamp as forward primers was used with P518r, to overcome a possible anomaly due to mismatches of certain PLA with P338f as described previously (8). Specific primers and PCR conditions for the different bacterial groups were used as listed in Table 1.
TABLE 1.
PCR primers, targets, and PCR conditions
| Target | Primers | PCR conditions
|
Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of cycles | Denaturation
|
Annealing
|
Elongation
|
||||||
| Temp (°C) | Time (min) | Temp (°C) | Time (min) | Temp (°C) | Time (min) | ||||
| β ammonia oxidizers (AAOB) | |||||||||
| First PCR round | CTO189fAB-CTO189fC and CTO653r | 35 | 94 | 1 | 57 | 1 | 72 | 2 | 27 |
| Second PCR round | P338,a P518r | 30 | 95 | 1 | 53 | 1 | 72 | 2 | 34 |
| Planctomycetales | First primer set | ||||||||
| First PCR round | PLA46f and PLA886rb | 30 | 95 | 1 | 56 | 1 | 72 | 2 | 38 |
| Second PCR round | P338f,a P518r | 30 | 95 | 1 | 53 | 1 | 72 | 2 | 34 |
| Planctomycetales | Second primer set | ||||||||
| First PCR round | PLA40f and P518r | 30 | 94 | 0.5 | 60 | 1 | 72 | 2 | 9 |
| Second PCR round | P338f and P338-IIf,a,c P518r | 30 | 95 | 1 | 53 | 1 | 72 | 2 | 34 |
A GC-clamp of 40 bp (5) was added for DGGE analysis.
Derived from probes developed by Neef et al. (33); Schmid et al. (38) did not use PLA886r. This primer is complementary to the oligonucleotide probe PLA886 constructed by Neef et al. (33).
A 50:50 mixture of both forward primers was used; the P338-IIf primer was developed from the oligonucleotide probe constructed as EUB338-II by Daims et al. (8).
DGGE was performed with the D Gene System (Bio-Rad, Hercules, Calif.) by using standard methods (32). The products of the second round of PCR were loaded onto 8% (wt/vol) polyacrylamide gels in 1 × TAE (20 mM Tris, 10 mM acetate, 0.5 mM EDTA [pH 7.4]). The polyacrylamide gels were made with a denaturing gradient ranging from 45 to 60%. The electrophoresis was run for 16 h at 60°C and 37 V. After electrophoresis, the gels were soaked for 5 min in fixation buffer (10% ethanol, 0.5% acetic acid) and then for 20 min in SYBR Green I nucleic acid gel stain (1:10,000 dilution; FMC BioProducts, Rockland, Maine). The stained gel was immediately photographed on a UV transillumination table with a video camera module (Vilbert Lourmat, Marne-la Vallé, France).
Cloning of the 16S rRNA genes and sequencing of the most dominant species.
A library of AAOB and PLA 16S rRNA gene clones was constructed using amplicons from the first round of PCR and the TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) as specified by the manufacturer. Transformant colonies were lysed by boiling (10 min), and the presence of amplicons in the cloning vector was verified by PCR with M13 primers. Diversity in the clone library was verified by restriction fragment length polymorphism (RFLP) of approximately 100 clones with HaeIII (3 h at 37°C), visualized on an 8% acrylamide gel (6 h for 45 V; poststained with SYBR Green I). Clones were ordered in different RFLP groups, and representatives of each (for both PLA and AAOB libraries) were selected and sequenced (IIT Biotech Bioservice, Bielefeld, Germany). The presence of chimerae in the obtained sequences was determined by using the CHECK_CHIMERA program from the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu) (29). Analyses of DNA sequences and homology searches were completed with standard DNA sequencing programs and the BLAST server of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) using the BLAST algorithm for the comparison of a nucleotide query sequence against a nucleotide sequence database (blastn). The diversity of the clone library was assessed by estimating clonal richness as a function of increasing clone numbers (EstimateS version 6.0b1, copyright R. K. Colwell [http://viceroy.eeb.uconn.edu/estimates]) by using the ChaoII index, which represents a suitable parameter for estimating microbial richness (21). Phylogenetic trees were constructed using the PHYLIP interface from the RDP. Sequences were first aligned using the sequence alignment software provided by the RDP. A similarity matrix was calculated by the Jukes-Cantor method, using a transition/transversion of 0.5 as recommended in PHYLIP. Phylogenetic trees were constructed using the neighbor-joining method with Thermotoga maritima as the outgroup.
FISH.
The probe sequences used, the target sites, the target organisms, and the optimal formamide concentrations in the hybridization buffers are reported in Table 2. Probes were purchased as Cy3-, Cy5-, and 5,6-carboxyfluorescein-N-hydroxysuccinimide (FLUO)-labeled derivatives (Eurogentec, Seraing, Belgium). Hybridizations were performed on 4% (wt/vol) paraformaldehyde-fixed biofilm samples immobilized on gelatin-coated microscopic slides as described previously (1). Sequential hybridization was applied for probes requiring different stringency (45). Hybridization buffer (9 μl) was mixed with probe working solutions (1 μl of each probe; 30 ng μl−1 for Cy3- and Cy5-labeled probes, and 50 ng μl−1 for FLUO-labeled probes) and competitor probes (1 μl of each probe, when required) and spotted into the corresponding well of the microwell slide.
TABLE 2.
Oligonucleotide probes used for CSLM-FISH in this study, probe sequence, target sites, target organisms, and formamide concentration in the hybridization buffer
| Probe name | Labeling dye | Stringencya (% [vol/vol] formamide) | Target siteb (16S rRNA positions) | Target organism(s) | Reference(s) |
|---|---|---|---|---|---|
| PLA886 and competitor | FLUO/Cy5 | 35 | 886-904 | Most Pirellula, Planctomyces, Isosphaera, and Gemmata spp. | 33 |
| PLA46 | Cy5 | 25 | 46-63 | Gemmata, Isosphaera, Pirellula, and Planctomyces sp. and the Anammox branch (Brocadia and Kuenenia spp.) | 33, 38 |
| NSO190 | Cy3 | 35c | 190-208 | Ammonia-oxidizing β-Proteobacteria | 30 |
| AMX820 | Cy3 | 25 | 820-841 | Brocadia anammoxidans | 42 |
| KST1275 | Cy3 | 25 | 1275-1292 | Kuenenia stuttgartiensis | 38 |
The formamide concentration used during hybridization.
Escherichia coli numbering.
Although the original publication on the use of NSO190 indicated the use of 55% FA in FISH applications, we experimentally confirmed that at 35% half of the probe signal was removed with Nitrosomonas europaea as the test organism, consistent with other publications describing the use of NSO190 at much lower stringencies than originally published (4, 26).
Microscopy.
Hybridized biofilm samples were inspected by confocal scanning laser microscopy (CSLM) (Leitz DM RBE microscope; true confocal system TCS SP with a multiband confocal imaging spectrophotometer [Leica Microsystems, Heidelberg, Germany]) equipped with He and Ar/Ne lasers (set at approximately 50% of maximum intensity). Excitation wavelengths were set at 488, 543, and 633 nm for FLUO, Cy3, and Cy5, respectively, and fluorescence signals were collected, after passage through a 488/543/633 filter in three different photomultiplier tubes with collection ranges of 500 to 525, 550 to 575, and 645 to 750 nm for FLUO, Cy3, and Cy5, respectively. Image stacks were collected with z sections at 1- to 2-μm intervals, with four scans per section. Images were captured, quantified, and rendered using the PowerScan and Power3D features of the standard manufacturer software (Leica confocal software TCS NT/SP).
Nucleotide sequence accession numbers.
16S rRNA gene fragment sequences from this study have been deposited in the GenBank database, the AAOB clones AOB1 to AOB6 under accession numbers AF525950 to AF525955 and the PLA clones PLA1 to PLA14 and PLA15 to PLA18 under accession numbers AF525956 to AF525969 and AY167665 to AY167668, respectively.
RESULTS
Chemical profile of the performance of the RBC reactor.
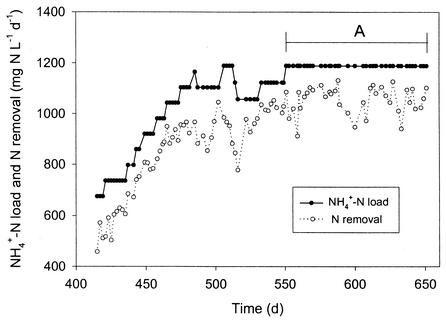
A 236-day performance period of the RBC fed with NH4+-rich wastewater devoid of organic carbon (starting 615 days after reactor startup and 415 days after addition of granular anaerobic sludge) is shown in Fig. 1. The N load consisted of NH4+-N (synthetic wastewater), while the N removal was calculated as the difference between the amount of NH4+-N in the feed and the sum of all inorganic N species (NH4+-N, NO2−-N, and NO3−-N) in the effluent. This deficit was in part due to conversion to gaseous N products like NO, N2O, and N2 and in part due to N uptake for cell synthesis. During the 236 days described here, an average removal of 86% ± 6% was achieved at loading rates ranging from 675 to 1,189 mg of N liter−1 day−1 (or 4,714 to 8,304 mg of N m−2 day−1). More detailed information on the performance and removal characteristics is given in Table 3 for a 100-day period (indicated as period A in Fig. 1) with constant loading. A stable and high removal efficiency of 89% ± 5% was achieved during this period. The bulk T, pH, and DO concentration in the reactor were 29.1 ± 1.8°C, 7.85 ± 0.15, and 0.57 ± 0.12 mg of O2 liter−1, respectively. The estimated total amount of biomass on the reactor disk surface at the end of the experimental period was 259.6 ± 17.8 g of VSS. Based on this estimation and the results in Table 3, the total (sum of aerobic and anoxic) in-reactor specific ammonia oxidation rate at that time was 192.4 ± 16.0 mg of N g of VSS−1 day−1.
FIG. 1.
Nitrogen loading and removal in the RBC reactor during a 236-day period. A constant loading of 1,189 mg of N liter−1 day−1 (or 8,307 mg of N m−2 of disk surface area day−1) was applied to the reactor during a 100-day period (period A).
TABLE 3.
Reactor performance during a 100-day period of constant loadinga
| Effluent concn (mg liter−1) | NH4+-N oxidationb
|
N removal rates
|
|||||
|---|---|---|---|---|---|---|---|
| NH4+-N | NO2−-N | NO3−-N | mg liter−1 day−1 | mg m−2 day−1 | % | mg liter−1 day−1 | mg m−2 day−1 |
| 38 ± 38 | 8 ± 6 | 47 ± 11 | 1,135 ± 55 | 7,931 ± 376 | 89 ± 5 | 1,058 ± 60 | 7,390 ± 418 |
The 100-day period corresponds to that labeled A in Fig. 1. This period was characterized by constant loading (1,189 mg of N liter−1 day−1 or 8,307 mg of N m−2 day−1). Values are given as mean ± standard deviation (n = 37). The influent NH4+-N concentration was 840 mg of N liter−1, and the flow rate was ca. 62 liters day−1.
Calculated as the sum of aerobic and anaerobic ammonium oxidation.
Measurement of the gas phase of the RBC reactor.
On day 644, possible gaseous N losses, specifically NH3, NO, N2O, and N2, from the reactor were quantified. At a total reactor load of 52.32 g of N day−1, under steady-state performance, and with an average removal efficiency of 89% (N loss of 46.56 g of N day−1), no NH3 was detected. N2O production accounted for 1.43 g of N day−1 (3.1% of the N loss), while NO production was at 149 μg of N day−1 (0.0003%). Calculation of the N2 production rate proved to be more difficult, probably due to the relatively short measuring interval of 2 h (this was chosen so as not to deplete O2 and thus significantly change the aeration profile of the reactor) and possible interference of atmospheric N2 during sampling and sample injection. As a result, the N2 production estimate exceeded the N input. We conclude that the remaining 97% of the gaseous N losses were most probably N2. Based on the results presented in Table 3 and the gas measurements, a reactor N balance for the inorganic N species is as follows (charges, H, and O not balanced):
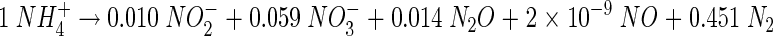 |
(5) |
Aerobic and anoxic batch experiments with RBC biomass.
To test the hypothesis that the biofilm removes N through a combination of both aerobic and anoxic ammonium oxidation, batch experiments were carried out to quantify both processes.
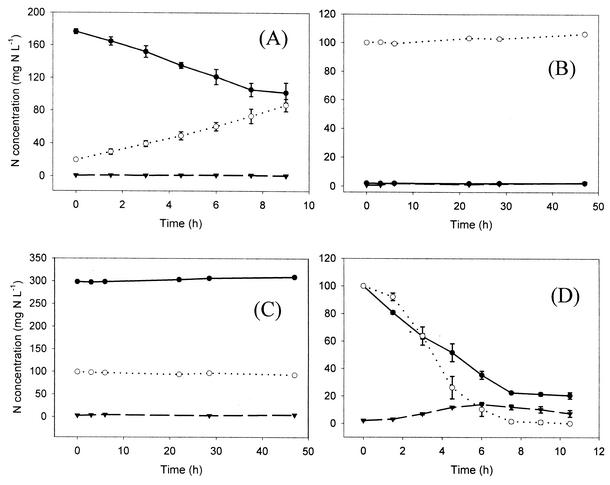
The aerobic batch experiments were carried out to determine the short-term (48-h) NH4+ and NO2− oxidation potential of the AAOB and NOB community in the biofilm, respectively. Results are shown for tests with RBC biomass supplemented with (i) NH4+, (ii) NO2−, and (iii) NH4+ + NO2− + ATU (Fig. 2A to C). The aerobic NH4+ oxidation potential of the biofilm, producing NO2− as the only oxidized N compound, was quantified as 147.8 ± 7.6 mg of N g of VSS−1 day−1 (Fig. 2A). To verify the absence of NO2−-oxidizing activity, biofilm samples were aerated in the presence of NO2− as the only N source and NO3− formation was found to be extremely low (<1 mg g of VSS−1 day−1) (Fig. 2B). To check whether N could be removed aerobically, biomass was aerated in the presence of NH4+ and NO2−, with ATU (10 mg liter−1) as a nitrification inhibitor to prevent NO2− formation by AAOB. This concentration of ATU was first shown not to inhibit the anoxic ammonium oxidation in an anoxic batch test, with NH4+ and NO2− being removed simultaneously, similar to the control experiment. No decrease in either NH4+ or NO2− concentration during 48 h of aeration was observed; the NH4+ concentration even increased slightly, probably due to biomass decomposition during the test (Fig. 2C). This increase was also seen in a test where biomass was inoculated with ATU in the absence of NH4+ or NO2− (data not shown). The rate of NH4+ production in these tests was about 1.7 mg of NH4+-N g of VSS−1 day−1.
FIG. 2.
Batch test results for RBC biomass under different conditions: NH4+/O2 (A), NO2−/O2 (B), NH4+/NO2−/O2 + ATU (C), and NH4+/NO2− in the absence of O2 (D). Average NH4+-N (solid circles), NO2−-N (open circles), and NO3−-N (solid triangles) levels are shown, along with standard deviations (n = 3). For the aerated tests, the DO concentration was ≥6 mg liter−1 and the pH was around 7; for the anoxic test, flasks were made anoxic by 20 cycles of 2 min of Ar overpressure and vacuum; the pH was around 8.
When the same test was carried out under anoxic conditions, simultaneous removal of both NH4+ and NO2− with concomitant NO3− production was measured (Fig. 2D), suggesting anaerobic ammonium oxidation. In the headspace, 99.7% of the N loss was found as N2-N and 0.3% was found as N2O-N. The latter was produced during the first hour of the experiment; no N2O formation was measured during the rest of the test. The maximum specific anoxic ammonia oxidation rate was 76.5 ± 6.4 mg of N g of VSS−1 day−1. The anoxic N removal stoichiometry of the process based on this batch test is presented in equation 6 (charges, H, and O not balanced):
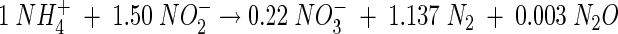 |
(6) |
Based on these aerobic and anoxic activity measurements, the maximal total specific ammonia oxidation rate of the RBC biomass was thus 224.3 ± 9.9 mg of N g of VSS−1 day−1. In batch tests, phosphate was shown to partially inhibit the AnAOB responsible for the anoxic ammonium oxidation in the RBC biofilm. Anoxic activity decreased to 63% of the normal activity at 1.8 mM PO43− and further to 20% at 3.6 mM PO43−. At 9.2 mM PO43−, no further decrease was observed (80% inhibition).
Clone library of the RBC biofilm samples.
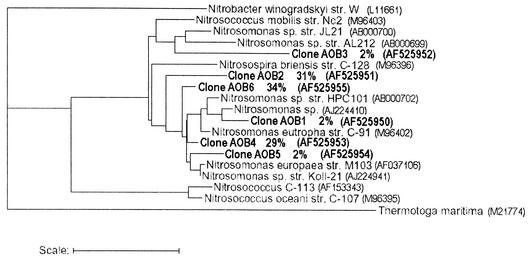
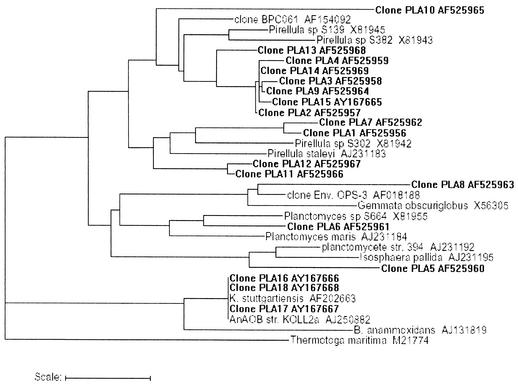
After PCR amplification of the biofilm community DNA with primers specific for AAOB and PLA, DGGE analysis was carried out. The high diversity of PLA based on this analysis (data not shown) prompted us to develop a clone library. Partial 16S rRNA gene sequences of AAOB and PLA, representative of different RFLP groups, were obtained. For the specific CTO primer set used in this study, 94 clones were analyzed that fell into six HaeIII RFLP groups. Representatives of each class were sequenced (465 bp), and all clones were highly similar to different Nitrosomonas sp. (Fig. 3). For the PLA, two different primer sets were used. For the first (PLA46f and PLA886r), 48 clones were analyzed that fell into 18 HaeIII RFLP groups. Rarefaction analysis of this clone library revealed that the ChaoII index reached a plateau of 60 distinct clones after 32 clones sampled and then remained stable. Thus, the number of analyzed clones represents the diversity in the planctomycete community with the primers used. None of the clone sequences displayed a chimera pattern. Of the 18 partial 16S rRNA gene fragments (one of each group) sequenced (±800 bp), 14 were clear PLA sequences (Fig. 4, PLA1 to PLA14). The other four clones could not be affiliated with the PLA and were most similar to an uncultured eubacterial clone (AF446328), an uncultured bacterial clone (AF418968), an Antarctic bacterial clone (AF173819), and a Sulfobacillus partial strain (AJ306699). However, these four groups of clones made up only about 6% of the total amount of clones, and so notwithstanding the “nonstringent” conditions of this primer set, 94% PLA sequences were retrieved. The phylogenetic analysis (Fig. 4) showed that the clones retrieved with these primers roughly clustered in all four classical branches of the PLA, with the largest fraction of the clones belonging to the Pirellula branch. The diversity within this group is also quite large, with some clones tending to form separate clusters within the group. However, the second primer set (PLA40f and P518r), which targets both the classical PLA groups and the newly discovered Anammox PLA branch, provided more complete information on the PLA sequence diversity and showed the dominant presence (about 90% of a total of 40 clones analyzed) of close relatives of the known AnAOB PLA K. stuttgartiensis in the biofilm (Fig. 4, PLA16 to PLA18). Each of these clones showed a similarity of more than 98% to K. stuttgartiensis based on partial 16S rRNA gene sequence comparison (530 bp). The remaining 10% of the clones (Fig. 4, PLA15) were affiliated with the Pirellula branch of the Planctomycetales.
FIG. 3.
Phylogenetic tree based on partial AAOB 16S rRNA gene sequences retrieved from the biofilm (indicated in bold, numbered as clones AOB1 to AOB6, with the percentage of clones having the same sequence indicated), some of their closest relatives, and sequences obtained from the RDP (29). T. maritima was used as an outgroup. Scale bar, 10 inferred nucleotide substitutions per 100 nucleotides. GenBank accession numbers are also indicated.
FIG. 4.
Phylogenetic tree based on partial PLA 16S rRNA gene sequences retrieved from the biofilm with primer set PLA46f and PLA886r (numbered as clones PLA1 to PLA14) and set PLA40f and P518r (clones PLA15 to PLA18), some of their close relatives, and sequences obtained from the RDP (29). T. maritima was used as an outgroup. Scale bar, 5 inferred nucleotide substitutions per 100 nucleotides. GenBank accession numbers are also indicated.
CSLM of the RBC biofilm.
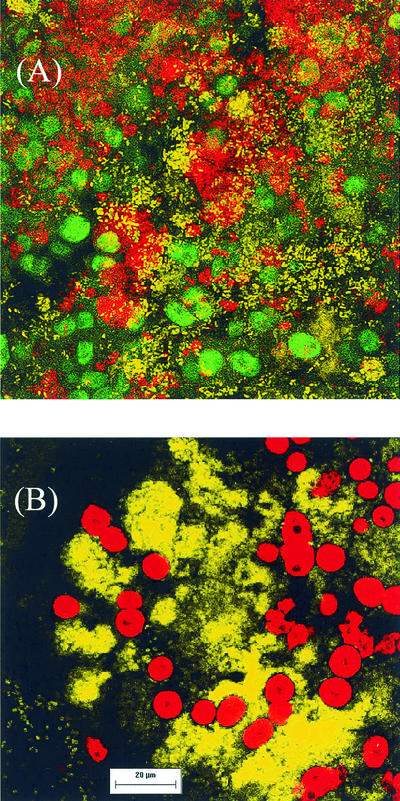
Using probe NSO190, specific for the AAOB, it was clear that the biofilm was largely a nitrifying biofilm with numerous AnAOB (hybridizing with probe PLA46) occurring side by side with the AAOB (Fig. 5A). To visualize all PLA, two previously described oligonucleotide probes, PLA46 and PLA886, were used (33). These two probes hybridized with different types of PLA, with PLA886 not targeting the AnAOB. PLA886 yielded exclusively very conspicuous, spherical fluorescent signals between 5 and 10 μm in diameter and in most cases with a dark area in the center (Fig. 5). Use of probe PLA46 and a combination of KST1275 (specific for K. stuttgartiensis) and AMX820 (specific for Brocadia anammoxidans) gave similar hybridization results, indicating that the PLA that hybridized with PLA46 consisted largely of these two organisms. However, when KST1275 and AMX820 were used in combination with PLA886, there was no overlap between the two (Fig. 5B). The same probe combination also indicated that the AnAOB are dispersed throughout the biofilm (Fig. 5B).
FIG. 5.
FISH-CSLM micrographs of the RBC biofilm. (A) Two-dimensional transparent rendering of a collection of depth scans of the RBC biofilm. Scans were taken every 1 μm along 16 μm of the z axis (depth); hybridizations were done using probe NSO190 (yellow), probe PLA46 (red), and probe PLA886 (green). (B) Single cross-sectional scan of the biofilm using probe PLA886 (red), probe KST1275 (yellow), and AMX820 (yellow). Scale bar, 20 μm. Magnification, ×630.
DISCUSSION
In this study we investigated several physiological and microbiological aspects of an autotrophic N-removing RBC biofilm almost 2 years after startup. The RBC reactor was running stable for the 236-day period described and was removing N at an average rate of 7.4 g of N m−2 day−1 (Fig. 1). This value is three to five times higher than those described for other N-removing RBC systems (17, 40) and translates into a high NH4+ surface loading rate of 8.3 g of N m−2 day−1. The N loss can be attributed to autotrophic microorganisms, because no NH3 stripping was detected and the influent synthetic wastewater was devoid of an organic carbon source. Due to the high loading and the slow rotation velocity of the disks (2.5 rpm), the DO concentration in the bulk water phase was low (0.57 mg of DO liter−1) and the biofilm on the disks was oxygen limited. The circumstances were thus ideal for the formation of a NO2−-producing biofilm (3). This is crucial for the autotrophic N removal process, because NO2− acts as the electron acceptor during anoxic oxidation of ammonium to N2. Indeed, N2 was found to be the major end product formed during N removal in the RBC (computed at 97%), although N2O was also detected (approximately 3%). N2O formation could be due to incomplete denitrification by resident heterotrophic bacteria (potentially using electron donors derived from biomass decay) or formation by nitrifiers under oxygen-limited conditions (2, 12, 22, 24, 35). During the period with constant loading, the effluent of the reactor contained on average 47 mg of NO3−-N liter, which was about 6% of the total amount of NH4+-N removed (Table 3). This NO3− production could be due to the activity of NOB or to the occurrence of a process in the biofilm similar to the Anammox process. NO3− production described for the CANON process is 11% (equation 4) of the total amount of NH4+ removed (41), almost double the value reported here (6% [Table 3 and equation 5]). Hence, it is likely that some NO3− reduction occurred simultaneously, also influencing the N balance in the RBC reactor. A mature biofilm under a high NH4+ loading rate most probably consists of three major groups of bacteria: the AAOB, NOB, and AnAOB (the last belonging to the PLA) (14, 25, 44). These three groups compete for several substrates in the biofilm: the AAOB and NOB compete for O2, the AnAOB and the NOB compete for NO2−, and the AAOB and AnAOB compete for NH4+. A first direct indication of the activity of these three groups in the biofilm was achieved by some additional batch tests.
In a first series of batch tests, biofilm biomass was aerated to examine its NH4+ and NO2− oxidation potential. The presence and activity of AAOB in the biofilm was obvious (Fig. 2A). However, despite the initial presence of NOB based on high NO3− production during periods before the one described in this paper (36, 37), the long-term (more than 600 days of operation) oxygen-limited conditions in the RBC reactor established a biofilm in which the NOB were apparently outcompeted (Fig. 2B). Negligible NOB activity was measured (even after 2 days of aeration, less than 1 mg of NO2−-N was oxidized g of VSS−1 day−1), which could give rise to a maximum of 4 mg of NO3−-N liter−1 in the reactor effluent (259.6 g of VSS on the disks). The anoxic ammonium oxidation activity (76.5 mg of NH4+-N g of VSS−1 day−1 [equation 6]) would yield about 70 mg of NO3−-N liter of effluent−1, significantly higher than the NOB contribution. A possible explanation for the apparent absence of NOB could be that in an oxygen-limited biofilm system, these bacteria tend to grow in the outermost part of the biofilm because of their lower O2 affinity than that of the AAOB. Due to the dynamic behavior (sloughing and regrowth of the outer layers) of a biofilm and the lack of available O2 at higher N loads, NOB were probably unable to maintain themselves in large numbers in the biofilm and most probably washed out. A biofilm sample supplied with NH4+ and NO2− under aerated conditions did not show any N removal, indicating that the N removal process in the RBC occurred via anoxic ammonium oxidation, which was also deduced from earlier 15N-labeling experiments (37). Indeed, 76.5 ± 6.4 mg of N g of VSS−1 day−1 was removed, a similar value to that reported for the Anammox process, with the higher NO2− and N2 levels per unit NH4+ possibly caused by some activity of nitrifiers and heterotrophic denitrifiers (equation 6). Inhibition experiments indicated that the AnAOB in the biofilm were sensitive to elevated PO43− concentrations, similar to the archetype B. anammoxidans (23) but contradictory to other reports (10). This sensitivity could be due to higher autooxidation rates of hydrazine, one of the Anammox intermediates, at higher PO43− concentrations (31). Based on the results of these tests and the reactor performance, it was concluded that two major groups of bacteria were active and responsible for the autotrophic N removal in the RBC biofilm: the AAOB and AnAOB, consistent with similar biofilm studies (19). The maximal specific total ammonia oxidation rate determined in batch tests (224.3 ± 9.9 mg of N g of VSS−1 day−1) was slightly higher than that estimated for the reactor (192.4 ± 16.0 mg of N g of VSS−1 day−1), most probably as a result of more optimal batch conditions and/or variations in the estimation of the biomass on the RBC disks.
At present, only two microorganisms that perform anaerobic ammonium oxidation are fully characterized: Candidatus B. anammoxidans and Candidatus K. stuttgartiensis, both belonging to a deep-branching lineage within the order Planctomycetales (38). Although these organisms have similar physiological and morphological features, their 16S rRNA sequence similarity is below 91% (38). This could imply that there is diversity in microorganisms (belonging to the Planctomycetales) capable of doing the same process. Considering this and the specific startup procedure of this reactor, which was different from those described in literature, the diversity of AAOB and PLA in the RBC biofilm was examined. The AAOB clones were dominated by close relatives of Nitrosomonas europaea and N. eutropha and showed little diversity. However, the clones obtained with the PLA primer set PLA46f and PLA886r displayed a high diversity (Fig. 4, PLA1 to PLA14), with representatives of each of the four major groups of Planctomycetales(Pirellula, Planctomyces, Gemmata, and Isosphaera) present in the biofilm. This diversity in PLA signals (Fig. 4) is much wider than that observed by others (38). Their presence in a mainly autotrophic RBC biofilm under oxygen limitation is not expected due to the aerobic, organoheterotrophic nature of most PLA (11). However, some Pirellula species are known to be facultative nitrate reducers (11). Although the studied RBC is mainly an autotrophic system, some electron donors derived from decaying biomass may be available, permitting the growth of heterotrophic organisms. Use of a second, less biased primer set, PLA40f and P518r, specific for all groups of PLA including the Anammox branch, revealed the dominant presence of presumed AnAOB in the biofilm. Three clones (PLA16 to PLA18), making up 90% of the clones analyzed, were highly similar (>98%) to K. stuttgartiensis. The remaining 10% of the clones were shown to belong to another group of planctomycetes (Pirellula, PLA15). The overall microbiology of this autotrophic N-removing reactor highly resembles that of similar systems (10, 19), despite the different bacterial inocula and type of wastewater (synthetic wastewater versus leachate).
Using combinations of group-specific FISH probes for AAOB (NSO190) and PLA in general (PLA46 and PLA886) and probes for the detection of known AnAOB (such as KST1275 and AMX820), we were able to visualize the spatial organization of the bacterial groups thought to be responsible for the N removal in the RBC reactor. Overall, the FISH analysis confirmed what was shown in the physiological and microbial characterization. CSLM revealed that the biofilm was largely a nitrifying community, with AnAOB spread throughout the biofilm and with conspicuous signals of microorganisms hybridizing with probe PLA886 (Fig. 5). Due to low signal resolution, we could not determine if the latter were large groups of cells or cell clusters. Whether these microorganisms play an active role in the N removal process could not be assessed, but their active presence relative to the AAOB and AnAOB, as shown in Fig. 5, certainly supports the assumption that they do. Besides the presence of these typical PLA886 signals, there is the question of the activity and the role of the nitrifiers in the deeper regions of the biofilm. The AAOB were shown to be active at around the same depth as the AnAOB (Fig. 5A), possibly having two functions: either they consume O2 (thus producing NO2−), protecting the AnAOB and supplying substrate, or they have some kind of anoxic metabolism. Our finding of AAOB next to AnAOB in presumably anoxic parts of the biofilm is consistent with an anoxic trickling filter where AAOB of the β subclass of the Proteobacteria made up about half of the AnAOB biomass (38). Their presence in the biofilm in close proximity to the AnAOB and their documented anoxic ammonia oxidation features in a mixed community with AnAOB in the presence of oxidized N species (39) certainly warrant further investigation of a possible anoxic role for these bacteria in the N removal process in the oxygen-limited RBC biofilm.
Conclusions.
A highly loaded RBC reactor showing a nitrogen loss of approximately 7.4 g of N m−2 day−1 was shown to be the habitat of a fairly homogeneous group of autotrophic AAOB and a group of close relatives to the known AnAOB K. stuttgartiensis. Due to their known respective aerobic and anoxic ammonia oxidation potential, these two groups are considered to be jointly responsible for the high N loss. However, activity of anoxic nitrifiers and some heterotrophs cannot be ruled out, and a small but diverse group of other PLA with a so far unknown role in the biofilm was also detected. NOB are considered to be present only in very small numbers based on physiological activity measurements. We postulated that the AnAOB are responsible for the autotrophic N removal process and that the AAOB might play a significant role in this process, based on their side-by-side spatial organization with the AnAOB in the biofilm.
Acknowledgments
This research was funded by the Flemish Institute for the Improvement of Scientific-Technological Research in Industry (IWT). During the time of this study, B. F. Smets was supported by a Senior Postdoctoral Fellowship of the Fund for Scientific Research, Flanders (FWO-Vlaanderen).
We thank Katrien Crul, Farida Doulami, and Han Vervaeren for help with the molecular and FISH work and Hendrik Nollet, Geert Lissens, and Wim De Windt for critical readings of the manuscript. We also thank Bjarke Bak Christensen of the Danish Veterinary and Food Administration (Søborg, Denmark) for access to the confocal scanning laser microscope.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in-situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, I. C., and J. S. Levine. 1986. Relative rates of nitric oxide and nitrous oxide production by nitrifiers, denitrifiers, and nitrate respirers. Appl. Environ. Microbiol. 51:938-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernet, N., P. Dangcong, J. P. Delgenes, and R. Moletta. 2001. Nitrification at low oxygen concentration in biofilm reactor. J. Environ. Eng. 127:266-271. [Google Scholar]
- 4.Biesterfeld, S., L. Figueroa, M. Hernandez, and P. Russell. 2001. Quantification of nitrifying bacterial populations in a full-scale nitrifying trickling filter using fluorescent in-situ hybridization. Water Environ. Res. 73:329-338. [DOI] [PubMed] [Google Scholar]
- 5.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2000. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, 12gfp. Appl. Environ. Microbiol. 66:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2001. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl. Environ. Microbiol. 67:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bremner, J. M., and R. D. Keeney. 1965. Steam distillation methods for determination of ammonium, nitrate and glycine. Anal. Chem. Acta 32:485-495. [Google Scholar]
- 8.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: cevelopment and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 9.Derakshani, M., T. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egli, K., U. Fanger, P. J. J. Alvarez, H. Siegrist, J. R. van der Meer, and A. J. B. Zehnder. 2001. Enrichment and characterization of an anammox bacterium from a rotating biological contactor treating ammonium-rich leachate. Arch. Microbiol. 175:198-207. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst, J. A. 1995. The planctomycetes—emerging models for microbial ecology, evolution and cell biology. Microbiology 141:1493-1506. [DOI] [PubMed] [Google Scholar]
- 12.Goreau, T. J., W. A. Kaplan, S. C. Wofsy, M. B. McElroy, F. W. Valois, and S. W. Watson. 1980. Production of NO2− and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl. Environ. Microbiol. 40:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg, A. E., L. S. Clesceri, and A. D. Eaton (ed.). 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, American Water Works Association, Water Environment Federation, Washington, D.C.
- 14.Hao, X. D., J. J. Heijnen, and M. C. M. van Loosdrecht. 2002. Sensitivity analysis of a biofilm model describing a one-stage completely autotrophic nitrogen removal (CANON) process. Biotechnol. Bioeng. 77:266-277. [DOI] [PubMed] [Google Scholar]
- 15.Hellinga, C., A. Schellen, J. W. Mulder, M. C. M. van Loosdrecht, and J. J. Heijnen. 1998. The SHARON process: an innovative method for nitrogen removal from ammonium-rich wastewater. Water Sci. Technol. 37:135-142. [Google Scholar]
- 16.Helmer, C., and S. Kunst. 1998. Simultaneous nitrification/denitrification in an aerobic biofilm system. Water Sci. Technol. 37:183-187. [Google Scholar]
- 17.Helmer, C., S. Kunst, S. Juretschko, M. C. Schmid, K.-H. Schleifer, and M. Wagner. 1999. Nitrogen loss in a nitrifying biofilm system. Water Sci. Technol. 39:13-21. [Google Scholar]
- 18.Helmer, C., C. Tromm, A. Hippen, K. H. Rosenwinkel, C. F. Seyfried, and S. Kunst. 2001. Single stage biological nitrogen removal by nitritation and anaerobic ammonium oxidation in biofilm systems. Water Sci. Technol. 43:311-320. [PubMed] [Google Scholar]
- 19.Helmer-Madhok, C., M. Schmid, E. Filipov, T. Gaul, A. Hippen, K. H. Rosenwinkel, C. F. Seyfried, M. Wagner, and S. Kunst. 2002. Deammonification in biofilm systems: population structure and function. Water Sci. Technol. 46:223-231. [PubMed] [Google Scholar]
- 20.Hippen, A., K. H. Rosenwinkel, G. Baumgarten, and C. F. Seyfried. 1997. Aerobic deammonification: a new experience in the treatment of wastewaters. Water Sci. Technol. 35:111-120. [Google Scholar]
- 21.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itokawa, H., K. Hanaki, and T. Matsuo. 2001. Nitrous oxide production in high-loading biological nitrogen removal process under low COD/N ratio condition. Water Res. 35:657-664. [DOI] [PubMed] [Google Scholar]
- 23.Jetten, M. S. M., M. Strous, K. T. van de Pas-Schoonen, J. Schalk, U. van Dongen, A. A. van de Graaf, S. Logemann, G. Muyzer, M. C. M. van Loosdrecht, and J. G. Kuenen. 1998. The anaerobic oxidation of ammonium. FEMS Microbiol. Rev. 22:421-437. [DOI] [PubMed] [Google Scholar]
- 24.Kester, R. A., W. deBoer, and H. J. Laanbroek. 1997. Production of NO and N2O by pure cultures of nitrifying and denitrifying bacteria during changes in aeration. Appl. Environ. Microbiol. 63:3872-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch, G., K. Egli, J. R. Van der Meer, and H. Siegrist. 2000. Mathematical modeling of autotrophic denitrification in a nitrifying biofilm of a rotating biological contactor. Water Sci. Technol. 41:191-198. [Google Scholar]
- 26.Konuma, S., H. Satoh, T. Mino, and T. Matsuo. 2001. Comparison of enumeration methods for ammonia-oxidizing bacteria. Water Sci. Technol. 43:107-114. [PubMed] [Google Scholar]
- 27.Kowalchuk, G. A., P. L. E. Bodelier, G. H. J. Heilig, J. R. Stephen, and H. J. Laanbroek. 1998. Community analysis of ammonia-oxidising bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridisation. FEMS Microbiol. Ecol. 27:339-350. [Google Scholar]
- 28.Kuai, L., and W. Verstraete. 1998. Ammonium removal by the Oxygen-Limited Autotrophic Nitrification-Denitrification system. Appl. Environ. Microbiol. 64:4500-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maidak, B. L., J. R. Cole, C. T. Parker, G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moliner, A. M., and J. J. Street. 1989. Decomposition of hydrazine in aqueous solutions. J. Environ. Qual. 18:483-487. [Google Scholar]
- 32.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of Polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neef, A., R. Amann, H. Schlesner, and K. H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 34.Ovreas, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poth, M., and D. D. Focht. 1985.15N kinetic analysis of N2O production by Nitrosomonas europaea: an examination of nitrifier denitrification. Appl. Environ. Microbiol. 49:1134-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pynaert, K., R. Sprengers, J. Laenen, and W. Verstraete. 2002. Oxygen-limited nitrification and denitrification in a lab-scale rotating biological contactor. Environ. Technol. 23:353-362. [DOI] [PubMed] [Google Scholar]
- 37.Pynaert, K., S. Wyffels, R. Sprengers, P. Boeckx, O. Van Cleemput, and W. Verstraete. 2002. Oxygen-limited nitrogen removal in a lab-scale rotating biological contractor treating an ammonium-rich wastewater. Water Sci. Technol. 45:357-363. [PubMed] [Google Scholar]
- 38.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K. H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt, I., C. Hermelink, K. van de Pas-Schoonen, M. Strous, H. J. Op den Camp, J. G. Kuenen, and M. S. M. Jetten. 2002. Anaerobic ammonia oxidation in the presence of nitrogen oxides (NOx) by two different lithotrophs. Appl. Environ. Microbiol. 68:5351-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegrist, H., S. Reithaar, G. Koch, and P. Lais. 1998. Nitrogen loss in a nitrifying rotating contactor treating ammonium-rich wastewater without organic carbon. Water Sci. Technol. 38:241-248. [Google Scholar]
- 41.Sliekers, A. O., N. Derwort, J. L. C. Gomez, M. Strous, J. G. Kuenen, and M. S. M. Jetten. 2002. Completely autotrophic nitrogen removal over nitrite in one single reactor. Water Res. 36:2475-2482. [DOI] [PubMed] [Google Scholar]
- 42.Strous, M. 2000. Microbiology of anaerobic ammonium oxidation. Ph.D. thesis. Technical University Delft, Delft, The Netherlands.
- 43.Strous, M., J. A. Fuerst, E. H. M. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. M. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 44.Third, K. A., A. O. Sliekers, J. G. Kuenen, and M. S. M. Jetten. 2001. The CANON system (completely autotrophic nitrogen removal over nitrite) under ammonium limitation: interaction and competition between three groups of bacteria. Syst. Appl. Microbiol. 24:588-596. [DOI] [PubMed] [Google Scholar]
- 45.Wagner, M., R. Amann, P. Kampfer, B. Assmus, A. Hartmann, P. Hutzler, N. Springer, and K. H. Schleifer. 1994. Identification and in-situ detection of gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405-417. [Google Scholar]