Abstract
Although nitrification has been well studied in coniferous forests of Western North America, communities of NH3-oxidizing bacteria in these forests have not been characterized. Studies were conducted along meadow-to-forest transects at two sites (Lookout and Carpenter) in the H. J. Andrews Experimental Forest, located in the Cascade Mountains of Oregon. Soil samples taken at 10- or 20-m intervals along the transects showed that several soil properties, including net nitrogen mineralization and nitrification potential rates changed significantly between vegetation zones. Nonetheless, terminal restriction fragment length polymorphism (T-RFLP) analysis of the PCR-amplified NH3 monooxygenase subunit A gene (amoA) showed the same DNA fragments (TaqI [283 bp], CfoI [66 bp], and AluI [392 bp]) to dominate ≥45 of 47 soil samples recovered from both sites. Two fragments (491-bp AluI [AluI491] and CfoI135) were found more frequently in meadow and transition zone soil samples than in forest samples at both sites. At the Lookout site the combination AluI491-CfoI135 was found primarily in meadow samples expressing the highest N mineralization rates. Four unique amoA sequences were identified among 15 isolates recovered into pure culture from various transect locations. Six isolates possessed the most common T-RFLP amoA fingerprint of the soil samples (TaqI283-AluI392-CfoI66), and their amoA sequences shared 99.8% similarity with a cultured species, Nitrosospira sp. strain Ka4 (cluster 4). The other three amoA sequences were most similar to sequences of Nitrosospira sp. strain Nsp1 and Nitrosospira briensis (cluster 3). 16S ribosomal DNA sequence analysis confirmed the affiliation of these isolates with Nitrosospira clusters 3 and 4. Two amoA clone sequences matched T-RFLP fingerprints found in soil, but they were not found among the isolates.
For many years the process of nitrification was thought to play a minimal role in N cycling in coniferous forest ecosystems. Based on the physiological properties of a limited number of NH3-oxidizing bacteria (AOB) in pure culture, several soil factors were considered detrimental to them, including soil acidity, high C/N ratios, low N availability, and the presence of allelochemical compounds (12, 13, 22, 38, 39, 49, 54). With the advent of improved 15N tracer methods and mass spectrometric technology, N cycling processes were shown to be tightly coupled in forest soils and that both nitrification and NO3− assimilation can occur simultaneously with mineralization and ammonification (11, 15, 45, 53). These observations stimulated interest in learning more about the nature and physiological ecology of AOB in forest ecosystems (12, 14, 30, 44, 51).
In recent years, considerable progress has been made in elucidating the composition of AOB communities in soil by taking advantage of variation in gene sequences of either 16S ribosomal DNA (rDNA) or the catalytic subunit of NH3 monooxygenase, amoA (2, 19, 37, 40). Most research has focused on grasslands and agroecosystems where, in general, soil populations are dominated by the genus Nitrosospira rather than by the widely studied genus Nitrosomonas (4, 8, 24-28, 36, 47, 48). Nitrosospira can be grouped into several phylogenetically distinct clusters (3, 25), with cluster 3 being widely distributed in agricultural soils with high N availability (8, 18, 27, 28, 55). Clusters 1, 2, and 4 are more prevalent in soils that are either acidic (47, 48), retired from agricultural use (27, 28), or that were never exposed to tillage and/or applications of N fertilizer (8, 55).
The objective of the present study was to examine the nature of the AOB communities associated with soil along high-elevation (∼1,500 m) meadow-to-forest transects at two sites in the H. J. Andrews Experimental Forest of Oregon. We hypothesized that these transects would provide a gradient along which differences in vegetation composition and plant N inputs interacting with changes in soil temperature, moisture, and pH would cause changes in AOB community composition. Implicit in this hypothesis is that physiological properties vary among soil-borne AOB. For example, the response of nitrification to temperature varies widely among soils (29, 44), and the response of NH3 oxidation to temperature and pH varies considerably among strains of Nitrosospira (21). Furthermore, the response of nitrification to NH4+ concentration varies among soils (44), and AOB community composition changes in response to effluents of high NH4+ content (17, 33), whereas members of Nitrosomonas cluster 6a have a low Ks for NH4+ (46), and outcompete other AOB at low NH4+ concentrations (6, 7). In the present study we took a complementary approach to examining AOB community composition that involved generating terminal restriction fragment length polymorphism (T-RFLP) fingerprints of amoA amplified from soil DNA, followed by cloning and sequencing amoA directly from soil and from AOB isolated into pure culture.
MATERIALS AND METHODS
Site description.
The experimental sites were situated in the H. J. Andrews Experimental Forest, (44.2°N latitude and 122.2°W longitude) in the western Cascade Mountains of Oregon. This regional landscape is considered a classic example of a temperate coniferous forest biome. For the past 50 years, ongoing ecological research at the Andrews Forest has contributed to an extensive database, which can be found at the onnline (http://www.fsl.orst.edu/lter/).
Sampling Sites.
Two sites (Lookout and Carpenter) were located at elevations of ca. 1,500 m and were selected because of the close proximity of grassland meadows and coniferous forests. Lookout has a southwestern aspect with an approximately 30° slope. The majority of trees in the forest were found to be a mixture of noble fir (Abies procera), grand fir (Abies grandis), and white fir (Abies concolor) 30 to 50 years of age, with some specimens on the upper transect being ∼100 years old. Meadow vegetation was dominated by a mixture of grasses, perennial herbs, and lupine (Lupinus polyphyllus). Carpenter has a southwestern aspect with an approximately 45° slope. The majority of trees were 60- to 90-year-old noble fir, Douglas fir (Pseudotsuga menziesii), and white fir. Meadow vegetation consisted primarily of grasses, perennial herbs, and bracken fern (Pteridium aquilinium). Selected soil characteristics from both experimental sites are given in Table 1.
TABLE 1.
Soil characteristics from the Lookout and Carpenter sites
| Site | Content (g/kg)a
|
CECa (cmol/kg) | pHb | Content (mg/kg)a
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sand | Silt | Clay | C | N | Ca | K | P | |||
| Lookout meadow | 664 | 280 | 56 | 112 | 8.9 | 39.5 | 5.6 | 340 | 146 | 23 |
| Lookout forest | 760 | 199 | 41 | 150 | 9.6 | 45.1 | 5.1 | 180 | 67 | 19 |
| Carpenter meadow | 687 | 292 | 21 | 95 | 6.8 | 39.0 | 5.8 | 560 | 228 | 13 |
| Carpenter forest | 668 | 279 | 54 | 130 | 5.5 | 55.6 | 5.2 | 400 | 68 | 7 |
Content analyses were conducted on composite soil samples prepared from meadow and forest soil samples. CEC, cation-exchange chromatography.
Soil pH was measured in 2:1 (wt/vol) water-soil samples.
Field site experimental design and soil sampling.
At each experimental site, three parallel transects were positioned perpendicularly to the meadow-forest boundary. Transects were spaced 20 m apart with the exception of the furthest upslope transect at Carpenter being located 100 m from the middle transect to avoid a hiking trail. Each transect consisted of eight, evenly spaced 1-m-diameter plots; three specific to the meadow (M), three specific to the forest (F), one near the meadow-forest boundary associated with the patchy occurrence of <10-year-old trees in predominantly meadow-type vegetation (M/F), and one in the forest-meadow boundary (F/M) associated with the presence of understory woody and herbaceous vegetation with mature coniferous trees. The plots were spaced 10 m apart at Lookout and 20 m apart at Carpenter, which was necessary to account for the wider transition zone existing between meadow and forest vegetation at Carpenter. Five soil cores (15.0-cm depth) were extracted from each plot by driving sections of polyvinyl chloride pipe (6-cm inner diameter) into the ground. The humus and litter layers were removed from the top of each soil core, and the mineral soil layers were composited, transported to the laboratory, and sieved through a 4.0-mm sieve. Routine soil textural and chemical analyses were determined by standard procedures of the Soil Testing Laboratory at Oregon State University.
Nitrification potential.
A shaken soil-slurry method was used to measure the nitrification potential rates (NPRs) for all 48 soil samples within 1 week of sampling (16). Each day, eight composite samples were randomly chosen from the larger collection of samples, and triplicate 15-g subsamples were weighed into 250-ml Erlenmeyer flasks. For each sample, 100 ml of a sterile solution (pH 7.2) containing 1.5 mM NH4+ and 1.0 mM potassium phosphate buffer (K2HPO4:KH2PO4) was added, and slurries were incubated in an orbital shaker (180 rpm) at 25°C for 24 h. Each flask was sampled at 3, 5, 22, and 24 h, when 10-ml aliquots of slurry were withdrawn and centrifuged, and the supernatants were frozen until analyzed colorimetrically for NO3− with Szechrome NAS reagent (Polysciences, Inc., Warrington, Pa.). NPRs were determined by using linear regression analysis and then log transformed prior to performing analysis of variance (S+2000; Data Analysis Products Division, MathSoft, Inc., Seattle, Wash.).
Whole-soil N mineralization.
Replicate 50-g samples of field-moist soil were weighed into specimen cups and placed in wide-mouth canning jars. Water contents were adjusted to 2/3 of water-holding capacity (0.67 g of H2O g of oven-dry soil−1), and the jars were sealed and incubated at 25°C. At 5-day intervals, 5-g portions of moist soil were extracted with 40 ml of 2 M KCl and filtered, and aliquots of filtrate were analyzed for NH4+ and NO3− on an Astoria Pacific series 300 autoanalyzer (Astoria-Pacific, Inc., Clackamas, Oreg.). Representative soil samples from various transect locations were also tested for heterotrophic nitrification by determining the sensitivity of NPRs to 1-kPa acetylene (20).
MPN enumeration of AOB.
An assay for most-probable-number (MPN) enumeration of AOB was restricted to four plots along each transect representing the four vegetation zones (M, M/F, F/M, and F). Two cores were extracted from each plot and composited, and 10-fold serial dilutions were prepared to 10−7 in a general NH3 oxidizer mineral medium supplemented with 20 mM NH4+ and adjusted to pH 7.2 (43). Quadruplicate 1-ml samples from each dilution were inoculated into 9-ml portions of the same medium and incubated at 28°C in the dark. Spot checks for NO2− were performed after 1, 2, and 4 months of incubation. Estimates of the NH3-oxidizing populations were determined by coding NO2−-positive dilutions and by using MPN tables (56). At Carpenter, MPN population density estimates of NH3 oxidizers were similar in M, M/F, and F/M soil samples, ranging from 4.0 × 104 to 9.5 × 104 cells g of soil−1, and declined sharply to 500 to 2,000 cells g of soil−1 in forest samples. At Lookout, MPN estimates of NH3 oxidizer populations were generally lower than at Carpenter and often highly variable within transect positions. In M and M/F samples population densities ranged from 0.4 to 1.6 × 104 cells g−1 soil, respectively. Populations were below the limit of accurate estimation (<20 cells g of soil−1) in two of three F/M samples and two of three F samples. Values of 1600 and 1.1 × 104 cells g−1 soil were obtained in the remaining F/M and F samples, respectively.
Pure culture isolation of NH3 oxidizers.
Terminal, NO2−-positive dilutions from the MPN assay were incubated until NO2− concentrations reached >0.5 mM. Tenfold serial dilutions were made, and 1-ml aliquots were inoculated into 9 ml of fresh medium. Tubes were incubated at 28°C in the dark and checked for NO2− production until no higher dilutions tested positive. From the terminal NO2−-positive dilutions, 1-ml inocula were back-transferred to nine tubes of fresh medium and then incubated for another month. NO2−-positive samples were checked for heterotrophic contamination microscopically and by inoculation into tubes containing 9 ml of a heterotrophic contamination medium (1.5 g of glucose, 0.75 g of yeast extract, and 0.75 g of Bacto Tryptone per liter). Tubes that did not show signs of turbidity after 3 weeks incubation were subsequently inoculated into Czapek-Dox medium (34) and “medium #3” (1). Cultures are stored at room temperature in the dark and back-transferred every 2 to 3 months.
DNA extraction from soil and PCR with amoA primers.
Soil DNA was extracted by using a FastDNA spin kit (Bio 101, Inc., Carlsbad, Calif.) and amplified by PCR with amoA primers (19). The forward primer amoA-1F (5′-GGGGGTTTCTACTGGTGGT) and the reverse primer amoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC; K = G or T; S = G or C]) correspond to positions 332 to 349 and 802 to 822, respectively, of the open reading frame of the amoA gene sequence of Nitrosomonas europaea (31). For T-RFLP analysis, PCR amplification was carried out by using the forward primer (amoA-1F) 5′ labeled with the dye 6-FAM (Genset, La Jolla, Calif.) in an ABI GeneAmp DNA thermocycler (model 2400). A hot-start procedure was used to reduce nonspecific amplification. Each reaction mixture contained either 200 or 250 ng of soil template DNA in a solution containing 1× PCR buffer [75 mM Tris-HCl (pH 8.8), 20 mM (NH4)2SO4, and 0.1% (wt/vol) Tween 20], 3.0 mM MgCl2, 0.20 mM concentrations of each deoxynucleoside triphosphate, 0.16 μM concentrations of each primer, 0.04% (wt/vol) bovine serum albumin, and 2.5 U of Taq DNA polymerase (Fermentas, Inc., Hanover, Md.), topped with sterile mineral oil (Sigma, St. Louis, Mo.). Positive controls contained 20 ng of genomic DNA prepared from cultures of N. europaea ATCC 19178 and Nitrosospira sp. strain AV. Negative controls contained either no DNA or 50 ng of Escherichia coli genomic DNA as a template. The following conditions were chosen for amplification of amoA: 5 min at 94°C, followed by a pause to add Taq polymerase, and then 30 or 35 cycles (depending on whether DNA genomic concentration was 250 or 200 ng, respectively) of 60 s at 94°C (denaturation), 60 s at 60°C (annealing), and 90 s at 72°C (elongation). PCR cycling was completed with a final elongation step of 7 min at 72°C. The PCR products were electrophoresed and visualized by standard procedures (41).
T-RFLP analysis.
The fluorescence-labeled PCR products (491 bp) were gel extracted and purified by using the Wizard DNA purification system (Promega Corp., Madison, Wis.) and then quantified by using a Bio Spec-1601 DNA-protein-enzyme analyzer (Shimadzu Co., Kyoto, Japan). A portion of each DNA sample (30 ng) was digested with one of three restriction enzymes: TaqI (recognition site T/CGA), CfoI (recognition site GCG/C), and AluI (recognition site AG/CT). We added 5 U of each enzyme to a total volume of 20 μl, which included 0.1 μl of acetylated bovine serum albumin (10 μg/ml) and 1× buffer (Promega Corp.). CfoI and AluI digests were incubated at 37°C and TaqI digests at 65°C. After overnight digestion, fragments were purified and submitted for Genescan analysis at the Central Services Laboratory (Center for Gene Research and Biotechnology, Oregon State University, Corvallis). Fragments were resolved on an ABI 377 Prism DNA sequencer by using GeneScan 1000-ROX as internal size standard (ABI, Inc., Freemont, Calif.). Fragment lengths were estimated by using the local southern method in GeneScan software (v.2.1; ABI, Inc.). T-RFLP community fingerprints were determined from the presence of fragments of specific lengths and by determining the contribution of individual peak areas to the total peak area of all fragments in a sample digest. The reproducibility of T-RFLP fingerprints was confirmed by repeating the PCR amplification of soil DNA samples and processing them by T-RFLP analysis.
Cloning and sequencing.
Soil DNA was cloned from a specific Carpenter plot that contained a variety of amoA T-RFLP fragments. The PCR protocol described described above was followed with the exception that a nonlabeled forward primer was used. PCR products were purified and quantified, ligated into the pGEM-T Easy vector by using T4 ligase (Promega Corp.), and transformed into competent E. coli cells according to manufacturer's recommendations. Aliquots (100 μl) of each transformed culture were plated onto Luria-Bertani (LB)-ampicillin-IPTG (isopropyl-β-d-thiogalactopyranoside)-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) medium and then incubated at 37°C for 24 h. White colonies were streaked for isolation on LB-ampicillin plates and incubated overnight at 37°C. Single colonies were selected from each isolated clone and inoculated into vials containing 3 ml of LB-ampicillin broth. Plasmid DNA was extracted by using the QIAprep Spin Miniprep kit (Qiagen, Inc., Valencia, Calif.) and quantified. Approximately 2.0 μl of each plasmid preparation was reamplified by using the 6-FAM forward amoA primer and unlabeled reverse amoA primer. Protocols for electrophoresis, gel purification, and restriction digests were followed as described above. Purified terminal restriction fragments were submitted for GeneScan analysis. Specific clones were selected for sequencing based on the similarity of their T-RFLP patterns to those observed in the soil community profiles. Double-stranded cycle sequencing was performed by using 400 ng of plasmid DNA and 12.0 pmol of M13 forward and reverse primers while monitoring Taq dye terminator chemistry with an ABI cycle sequencer.
Genomic DNA extraction from cultures of ammonia oxidizers.
Depending on the cell density of the culture, one of two procedures was used to extract genomic DNA from isolates. Cultures that easily achieved turbidity were harvested by centrifugation (8,000 rpm for 30 min). When culture turbidity was low, cells were gently filtered onto 0.2-μm (25-mm)-pore-size polycarbonate membrane filters. Each filter was placed into a 2.0-ml Eppendorf tube with 200 μl of a 5 M guanidine thiocyanate solution (30 ml of 0.5 M EDTA, 7.5 ml of 10% N-lauroylsarkosine, and 88.6 g of guanidine isothiocyanate in 150 ml of H2O). Samples were vortexed for 1 min and centrifuged for another min at 15,000 rpm. Additional steps for genomic DNA extraction and purification were performed with the DNeasy tissue kit (Qiagen) by using a spin filter column and a series of wash solutions. As little as 200 ng of genomic DNA could be recovered from a 10-ml aliquot of culture.
16S rDNA PCR and sequencing.
Published primers for AOB were used for 16S rDNA amplification (32, 37). The forward primer (Nso190 [5′-CGATCCCCTGCTTTTCTCC]) targets positions 190 to 208 of the E. coli 16S rDNA gene, and the reverse primer (Nso1225 [5′-CGCGATTGTATTACGTGTGA]) corresponds to the E. coli 16S rDNA positions 1225 to 1244. The PCR protocol described above was used except that 2.5 mM MgCl2 and 40 ng of DNA were used in 100-μl reaction volumes. A lower and longer annealing step (55°C for 90 s) was used to optimize primer binding. Post-PCR methods for electrophoresis, gel extraction, purification, and quantification were as described above for amoA PCR products. Double-stranded cycle sequencing of the 16S rDNA fragment utilized Nso190 and Nso1225 primers for the forward and reverse reads, respectively. Approximately 700 bp could be unambiguously read from the sequence output of each strand, resulting in a 960-bp read. Both strands of the 491-bp amoA product amplified from pure cultures were also sequenced by using amoA-1F and amoA-2R.
Phylogenetic analysis.
amoA sequences were submitted to a BLAST search and sequences in the GenBank database sharing the greatest similarities were imported into the BioEdit sequence alignment editor for Windows 95/98/NT. Thirty sequences were aligned manually before using CLUSTALW for multiple sequence alignment. Masking reduced the sequences to 397 characters, and phylogenetic analysis was performed with PAUP* 4.0 (50) by using the neighbor-joining method with a Kimura two-parameter distance measure. Bootstrap support values were generated from 100 replications of the aligned sequences to determine the confidence values of tree branches.
Nucleotide sequence accession numbers.
The amoA partial sequences of the isolates and clones used in the analyses have been deposited in GenBank under accession numbers AY189141 to AY189146. The 16S rDNA partial sequences of Nitrosospira CT2F (AY189139) and Nitrosospira LT1FMd (AY189140) were also deposited in GenBank.
RESULTS
Soil chemical and physical properties.
At both sites, mineral soils (<2 mm) are sandy loam to loamy sand in texture, with the soils under forest (F) being depleted of cations, more acidic, and containing higher organic C levels than the meadow (M) soils (Table 1). Although the C/N ratios of M soils were lower than F soils at both Lookout (13 versus 16:1, respectively) and Carpenter (14 versus 24:1, respectively), site differences were apparent (Table 2). At Lookout, C:N ratios were relatively constant in both M and M/F (13:1) and F/M and F (16:1) sites. At Carpenter, the C/N ratios of organic matter increased gradually across the transect from 13:1 to 24:1.
TABLE 2.
Changes in C/N ratios and net nitrogen mineralization rates across transect locationsa
| Location | Lookout
|
Carpenter
|
||||
|---|---|---|---|---|---|---|
| Mean C/N (SEM) | Mean net Nmin (SEM)
|
C/N (SEM) | Mean net Nmin (SEM)
|
|||
| June | Nov | June | Nov | |||
| M | 12.6 (0.7)A | 2.1 (1.2)A | 2.3 (0.3)A | 13.5 (1.0)A | 2.3 (0.6)A | 1.2 (0.3)A |
| M/F | 12.8 (1.0)A | 1.5 (0.5)A | 0.8 (0.7)B | 14.8 (1.7)AB | 2.2 (0.4)A | 1.4 (0.5)A |
| F/M | 16.1 (3.8)AB | 0.7 (0.3)A | 0.2 (0.2)B | 19.9 (3.4)BC | 0.6 (0.3)B | 0.3 (0.2)B |
| F | 15.6 (1.7)B | 1.0 (0.5)A | 0.3 (0.1)B | 23.7 (2.0)C | 0.4 (0.3)B | 0.1 (0.1)B |
Location symbols are defined in Materials and Methods. Net nitrogen mineralization rates (Nmin) were measured over 10 days of incubation and are given in micrograms of N g of soil−1 day−1. Means in columns followed by the same superscript capital letter are not significantly different at P = 0.05. Nov, November.
NPRs.
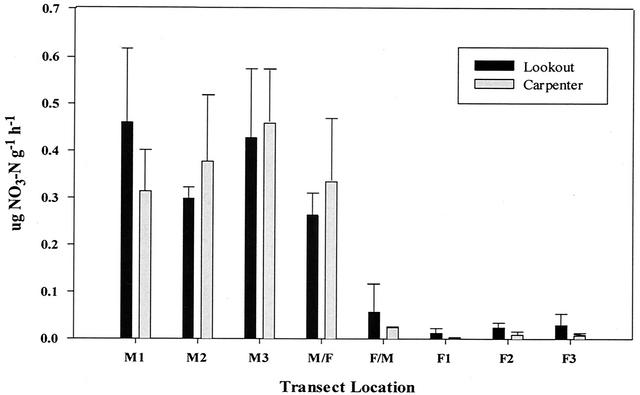
NPRs differed significantly (P < 0.05) between M and F soil samples at both sites (Fig. 1). NPRs obtained from Lookout and Carpenter M samples were ca. 19 and 64 times greater than F samples (0.39 μg of NO3−-N g of soil−1 h−1 versus 0.021 μg of NO3−-N g of soil−1 h−1 and 0.38 μg of NO3−-N g of soil−1 h−1 versus 0.006 μg of NO3−-N g of soil−1 h−1, respectively). A clear distinction could be made between NPRs of M/F and F/M at both sites (P < 0.05). At Lookout, NPRs obtained from two of the three F/M transition zone samples were below the lower limit of accurate detection (LLD, 0.003 μg of NO3−-N g of soil−1 h−1), whereas at Carpenter, NPRs obtained from the three F/M samples were similar (mean = 0.024 μg of NO3−-N g of soil−1 h−1) and ca. 7% of the mean M/F value. At Carpenter, the NPRs for three of nine F samples were <LLD, and the mean NPR of the other six samples (0.006 μg of NO3−-N g of soil−1 h−1) was ca. 25% of F/M values. At Lookout, the NPRs of two of nine F soil samples were <LLD, and the mean value of the remainder (0.021 μg of NO3−-N g of soil−1 h−1) was 3.5 times greater than the mean NPR of the F samples at Carpenter. NPRs were completely inhibited by acetylene (1 kPa) regardless of the site or the transect position (data not shown).
FIG. 1.
Profiles of NPRs across the meadow-forest transects. Values represent the means of the three transects at each site. M, M/F, F/M, and F represent meadow, meadow-forest, forest-meadow, and forest plot areas, respectively.
An examination of whole soil samples also showed that net N mineralization and nitrification differed across the transects and between the sites (Table 2). In June, N mineralization rates were higher (albeit not significantly) in M and M/F than in F/M and F samples along the transects at Lookout. In November, a significant difference was measured between M and the remaining samples from the other locations on the transect. At Carpenter there was a significant drop in net N mineralization between M/F and F/M on both sampling dates. At both sites, NH4+ did not accumulate in M and M/F samples during the incubation, whereas in F/M samples NH4+ accumulated initially and NO3− accumulation commenced only after 40 to 45 days of incubation. In F samples, NO3− appeared after 50 to 55 days at Lookout but did not appear in Carpenter samples even after 80 days of incubation (data not shown).
Molecular analysis of amoA from soil.
All but one of the 48 soil DNA samples were successfully amplified with the amoA primers and generated the expected 491-bp band size fragment of amoA. T-RFLP analysis with TaqI generated a single peak (283 bp) in all 47 soil DNA samples, suggesting that Nitrosospira- and/or Nitrosospira-like organisms predominated throughout both experimental sites (Tables 3 and 4). Based on sequence analysis, a 219-bp fragment length would have indicated the presence of Nitrosomonas species. T-RFLP analysis with AluI produced a dominant peak (392 bp) in 45 of 47 samples ( Tables 3 and 4). The 392-bp AluI fragment comprised virtually all of the AluI-digested PCR product, in 29 of 45 soil samples and never <30% of product. In addition, a full-length PCR product (491 bp) was detected in AluI-digested DNA from samples at both sites, and it was distributed primarily among M, M/F, and F/M samples at Lookout (eight of eight cases, Table 3) and at Carpenter (seven of eight cases, Table 4).
TABLE 3.
Distribution of T-RFLP fragments across transects at the Lookout site
| Location | No. of positive transectsa with restriction enzyme:
|
||||||
|---|---|---|---|---|---|---|---|
| TaqI (283 bp) |
CfoI
|
AluI
|
|||||
| 66 bp | 68 bp | 100 bp | 135 bp | 392 bp | 491 bp | ||
| M1 | 3 | 3 | 0 | 1 | 2 | 3 | 1 |
| M2 | 3 | 3 | 0 | 1 | 2 | 3 | 2 |
| M3 | 3 | 3 | 0 | 0 | 2 | 3 | 2 |
| M/F | 3 | 3 | 0 | 1 | 3 | 3 | 1 |
| F/M | 3 | 3 | 1 | 0 | 2 | 2 | 2 |
| F1 | 3 | 3 | 0 | 0 | 0 | 3 | 0 |
| F2 | 3 | 3 | 0 | 0 | 0 | 2 | 0 |
| F3 | 3 | 3 | 0 | 0 | 1 | 3 | 0 |
| Total | 24 | 24 | 1 | 3 | 12 | 22 | 8 |
There are three transects across the site; therefore, three of each specific location per site is the maximum number per location.
TABLE 4.
Distribution of T-RFLP fragments across transects at the Carpenter site
| Location | No. of positive transectsa with restriction enzyme.
|
||||||
|---|---|---|---|---|---|---|---|
| TaqI (283 bp) |
CfoI
|
AluI
|
|||||
| 66 bp | 68 bp | 100 bp | 135 bp | 392 bp | 491 bp | ||
| M1 | 3 | 3 | 0 | 0 | 2 | 3 | 1 |
| M2 | 3 | 3 | 0 | 0 | 1 | 3 | 0 |
| M3 | 3 | 3 | 2 | 0 | 2 | 3 | 2 |
| M/F | 3 | 3 | 0 | 0 | 3 | 3 | 2 |
| F/M | 3 | 3 | 0 | 2 | 2 | 3 | 2 |
| F1b | 2 | 2 | 1 | 1 | 1 | 2 | 0 |
| F2 | 3 | 3 | 0 | 0 | 0 | 3 | 0 |
| F3 | 3 | 3 | 1 | 0 | 1 | 3 | 1 |
| Total | 23 | 23 | 4 | 3 | 12 | 23 | 8 |
There are three transects across the site; therefore, three of each specific location per site is the maximum number per location.
PCR products of amoA were not successfully produced from one F1 soil sample.
T-RFLP analysis of CfoI digests produced a consistently prominent peak (66 bp) in all 47 samples (Tables 3 and 4). Three additional fragments (68, 100, and 135 bp) were observed in CfoI digests of 5, 6, and 24 soil DNA samples, respectively (Tables 3 and 4). The CfoI 135-bp (CfoI135) fragment was distributed primarily among M and M/F samples at Lookout (9 of 12 cases, Table 3) and at Carpenter (8 of 12 cases, Table 4), and yet its relative abundance was quite variable, ranging between 6 and 49% of the total CfoI products. Although found in F samples, the 135-bp fragment was always a minor product; it ranged between 3 and 18% at Lookout and was <5% in the Carpenter samples. In 13 of 24 cases, the 135-bp CfoI fragment was associated with the unrestricted full-length AluI fragment, and this combination occurred almost exclusively in M and M/F soil samples at the Lookout and Carpenter sites (12 of 13 cases). Furthermore, at Lookout the AluI491-CfoI135 combination was found in five of six soil samples expressing net N mineralization rates of ≥1.9 μg of N g−1 day−1 and was present in only one of six cases in which the rate was ≤1.9 (the overall range of N mineralization rates at Lookout was 0.5 to 3.9 μg of N g−1 day−1). There was no obvious association between the AluI491-CfoI135 fragment combination and high N mineralization rates at Carpenter. The 100-bp CfoI fragment appeared in M and M/F soils at Lookout and in F/M and F soils at Carpenter. The relative peak areas of the 68- and 100-bp fragments averaged <14% of the total peak areas in the 11 samples in which they were found.
Cloning and sequencing amoA from isolates and soil.
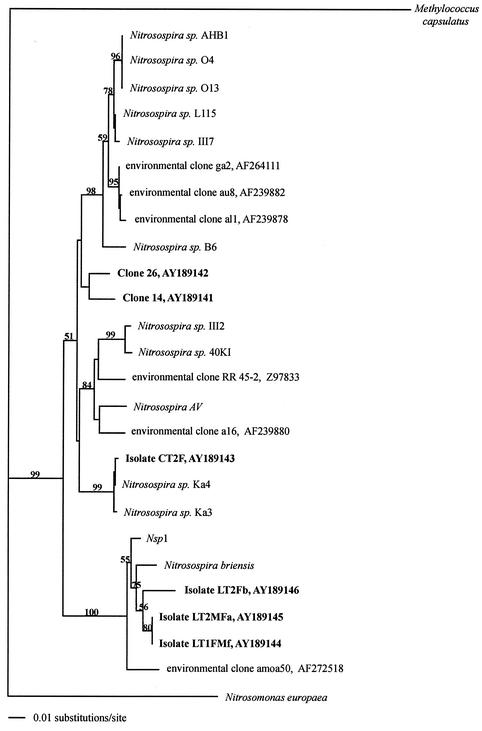
T-RFLP patterns of 14 of 15 isolates of AOB matched those obtained from the community fingerprints (Table 5). Isolate LT1F/Mf was the exception because the AluI 34-bp fragment could not be accurately detected in the Genescan analysis. The 491-bp fragment of amoA was sequenced from eight isolates representing the four different T-RFLP patterns and produced four unique sequences, which were phylogenetically placed in a neighbor-joining tree relative to other Nitrosospira spp. (Fig. 2). The amoA sequence of isolate CT2F, possessing the dominant soil T-RFLP fingerprint (TaqI283-AluI392-CfoI66), was found to be 99.8% similar to the amoA sequence of Nitrosospira sp. strain Ka4 of cluster 4. Isolates possessing the fragment sizes and combinations TaqI283-AluI491-CfoI66 and TaqI283-AluI491-CfoI66-68 clustered with Nitrosospira briensis and Nitrosospira sp. strain Nsp1 of cluster 3. Sequence analysis of 16S rDNA genes from two of the isolates (CT2F and LT1F/Md) confirmed their phylogenetic positions, with the CT2F sequence being identical to that of Nitrosospira sp. strain Ka4 and the sequence of LT1F/Md being 99% similar to N. briensis. Cloning and sequencing of soil DNA identified two additional amoA sequences (sequences 14 and 26) with fragment combinations that were also observed in the T-RFLP soil community profiles: sequence 26 (TaqI283-AluI491-CfoI135) and sequence 14 (TaqI283-AluI392-CfoI100). Although the relationship between the two cloned amoA sequences and the established Nitrosospira clusters remains equivocal, the clone 26 sequence was most similar (94%) to isolates associated with cluster 2 (O4, O13, L115, and III7).
TABLE 5.
T-RFLP patterns observed in NH3-oxidizer isolates and soil clones
| Isolatea | No. of isolates | Fragment length(s) (bp)
|
||
|---|---|---|---|---|
| CfoI | AluI | TaqI | ||
| CT1Ma, CT1M/Fb, CT1F, CT3Mb, CT3M/F CT2F*† | 6 | 66 | 392 | 283 |
| LT1F/Md*†, LT1Fd, LT2M/Fa* | 5 | 66 | 491 | 283 |
| LT1F/Mf* | 1 | 66 | 34 | 283 |
| LT1Ma, LT2Md, LT2Fb* | 3 | 66, 68 | 491 | 283 |
| Soil clone 26* | 100 | 392 | 283 | |
| Soil clone 14* | 135 | 491 | 283 | |
Asterisks indicate isolates and clones from which the amoA gene was sequenced; daggers indicate isolates from which the 16S rDNA gene was sequenced.
FIG. 2.
Phylogenetic analysis of amoA sequences performed in PAUP* by using a neighbor-joining method with a Kimura two-parameter distance measure. Bootstrap values were calculated from 100 replications with 397 characters. Isolates, clones, and accession numbers from this study are in bold.
DISCUSSION
Although extensive research has been carried out on the diversity of AOB in soils of Northern Europe and the United Kingdom, the present study is the first of its kind to describe AOB communities in a coniferous forest ecosystem of North America. Plant productivity of these vast ecosystems in the western United States is generally N limited; however, successions are usually initiated after fire or human disturbance with substantial inputs of N from pioneering actinorhizal plants and/or legume species. There is considerable interest in refining our understanding of how N cycling processes, and nitrification in particular, change as these ecosystems proceed through their different successional phases (9, 10, 15, 42, 52).
The recovery of isolates in possession of the amoA T-RFLP fingerprint found throughout meadow, transitional, and forest sites is an interesting finding for a variety of reasons. Although a number of 16S rDNA cloned sequences are available that phylogenetically delineate Nitrosospira cluster 4, few representatives of this cluster have been obtained into culture (1, 37). Kowalchuk et al. (27, 28) speculated that cluster 4 types might be disadvantaged in soils with high NH4+ or neutral pHs because these authors found that enrichments carried out at high NH4+ (5 mM) or at pH 7.5 contained a lower percentage of cluster 4 sequences than did enrichments carried out in lower NH4+and pH. In this context, we recovered Nitrosospira sp. strain Ka4-like isolates in traditional, slightly alkaline medium containing 20 mM NH4+ and subsequently found them to oxidize and grow in 20 mM NH4+ at a faster rate than in either 2 or 0.5 mM NH4+ (A. E. Taylor and P. J. Bottomley, unpublished data). Nonetheless, for unknown reasons, Ka4-like isolates were recovered only from the Carpenter site. If NH4+ sensitivity varies among Nitrosospira cluster 4, we might have needed lower levels of NH4+ in the growth medium to recover representatives of these types of isolates from the Lookout site.
Because the T-RFLP fingerprint of the Nitrosospira cluster 4 was found consistently throughout meadow and forest soil samples in which NPRs ranged >100-fold from 0.4 μg-N g of soil−1 h−1 to <LLD (<0.003 μg-N g of soil−1 h−1) and in which the pH ranged from 5.8 to 5.2, we speculate that the Ka4-like bacteria possess both competitive and survival abilities under conditions both advantageous and deleterious for nitrifying activity. Although physiological differences exist among strains of Nitrosospira (21), it was pointed out recently that our knowledge about the physiology of Nitrosospira spp. lags far behind that of Nitrosomonas spp. and that more studies are needed to better understand the ecophysiology of this genus (23). Unfortunately, our Ka4-like isolates are slow growing and seem particularly slow to recover from small inoculum back transfers into fresh medium. Work continues in our laboratory to optimize growth conditions suitable for the study and maintenance of these isolates.
Even though the Ka4-like T-RFLP fingerprint dominated the forest soil samples at both locations, it is possible that the amoA T-RFLP approach was too limited and/or insensitive to resolve all of the functional types of AOB in these soils. Furthermore, the contribution of the Ka4-like bacteria to nitrification in the forest soils remains unknown. For example, the mean NPRs (6 to 20 ng of N g of soil−1 h−1) of forest soil samples at Carpenter and Lookout, respectively, are similar to the gross rates of nitrification (∼40 ng of N g of soil−1 h−1) that have been measured by isotope dilution in intact coniferous forest soils of the western United States (15, 16, 45). A population density of between 4 × 104 and 1.5 × 105 cells g of soil−1 would be required to support these rates if we assume a maximum rate of NH3 oxidation by Nitrosospira sp. of ca. 10 × 10−15 mol cell−1 h−1 (5, 21). This population density is 40- to 150-fold greater than the average MPN values obtained from the forest soil samples at Carpenter (∼1,000 g of soil−1) and far greater than the estimates made at Lookout. Although such discrepancies indicate that heterotrophic nitrification might be a source of oxidized nitrogen in these coniferous forest soil environments (35), our finding that acetylene completely inhibited NPRs across all transect positions suggests this was likely to be autotrophic nitrification. In this context, others have shown that the size of populations of AOB determined by quantitative PCR can be 10 to 500 times greater than values determined by traditional MPN methods (28, 36). Furthermore, the greatest discrepancy between MPN and competitive PCR-derived population estimates occurred in soil samples where Nitrosospira clusters 2 and 4 made major contributions to the soil populations (28). Given the difficulties we have encountered in culturing and maintaining the Ka4-like bacteria from our soils, the MPN conditions might have been woefully inadequate to resuscitate and culture quantitatively AOB of this type.
In contrast to the Ka4-like bacteria, which were widespread across the transects, our data indicate that nitrosospirads of cluster 3 (TaqI283-AluI491-CfoI66 and TaqI283-AluI491-CfoI66/68), and an uncultured lineage associated with cluster 2 (TaqI283-AluI491-CfoI135) might be restricted primarily to meadow and transitional environments. These findings can be related to other studies from Europe and the United Kingdom on the influence of plant communities and land management on community composition of Nitrosospira and soil N cycling. For example, clusters 2 and 4 increased relative to cluster 3 in Dutch soils that had been taken out of agricultural production and unfertilized with N for 10 to 20 years (27, 28). Similarly, a greater diversity of AOB was found in unimproved United Kingdom hill soils relative to adjacent soils that had been fertilized annually with N for 30 years (55). In both the Dutch and the United Kingdom studies these population changes coincided with relatively modest changes (two- to fivefold) in the nitrification potentials and/or net N mineralization rates of the soils. The differences observed in net N mineralization and nitrification potentials across transect positions at both Carpenter and Lookout were at least of this magnitude and often much greater.
Although our work provides no clues about what physiological attributes might account for the widespread occurrence of cluster 4 nitrosospirads in these high-elevation, moderately acidic forest soils, further work will investigate the physiological characteristics of the isolates. In addition, we will also examine the composition and dynamics of the AOB community and the properties of nitrification in these coniferous forest ecosystems as they proceed through their fire- and human-induced vegetation successional changes.
Acknowledgments
Financial support for this work was provided by a grant from the National Science Foundation Microbial Observatory Program (MCB-9977933) and by the Oregon Agricultural Experiment Station.
We acknowledge the assistance of several past and present laboratory colleagues with field site layout, sampling, and soil processing and the staff of the Central Analytical Services Laboratory of the Center for Gene Research and Biotechnology for Genescan and nucleotide sequence analyses. We are especially grateful to Kevin Virgin and Stephanie Connan of the Department of Microbiology for assistance with aspects of molecular methodology and phylogenetic tree preparation.
Footnotes
Technical paper no. 11,947 of the Oregon Agricultural Experiment Station.
REFERENCES
- 1.Åakra, A., J. B. Utaker, I. F. Nes, and L. R. Bakken. 1999. An evaluated improvement of the extinction dilution method for the isolation of ammonia-oxidizing bacteria. J. Microbiol. Methods 39:23-31. [DOI] [PubMed] [Google Scholar]
- 2.Åakra, A., J. B. Utaker, and I. F. Nes. 2001. Comparative phylogeny of the ammonia monoxygenase subunit A and the 16S rRNA genes of ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 205:237-242. [DOI] [PubMed] [Google Scholar]
- 3.Åakra, A., J. B. Utaker, A. Pommerening-Roser, H. P. Koops, and I. F. Nes. 2001. Detailed phylogeny of ammonia-oxidizing bacteria determined by rDNA sequences and DNA homology values. Int. J. Syst. E vol. Microbiol. 51:2021-2030. [DOI] [PubMed] [Google Scholar]
- 4.Belser, L. W., and E. L. Schmidt. 1978. Diversity in the ammonia-oxidizing nitrifier population of a soil. Appl. Environ. Microbiol. 36:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belser, L. W., and E. L. Schmidt. 1980. Growth and oxidation of ammonia by three genera of ammonia oxidizers. FEMS Microbiol. Lett. 7:213-216. [Google Scholar]
- 6.Bollmann, A., M.-J. Bar-Gilissen, and H. J. Laanbroek. 2002. Growth at low ammonium concentration and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollmann, A., and H. J. Laanbroek. 2001. Continuous culture enrichments of ammonia-oxidizing bacteria at low ammonium concentration. FEMS Microb. Ecol. 37:211-221. [Google Scholar]
- 8.Bruns, M. A., J. R. Stephen, G. A. Kowalchuck, J. I. Prosser, and E. A. Paul. 1999. Comparative diversity of ammonia-oxidizer 16S rRNA gene sequences in native, tilled, and successional soils. Appl. Environ. Microbiol. 65:2994-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapin, F. S., L. R. Walker, C. L. Fastie, and L. C. Sharman. 1994. Mechanisms of primary succession following deglaciation at glacier bay, Alaska. Ecol. Monogr. 64:149-175. [Google Scholar]
- 10.Clein, J. S., and J. P. Schimel. 1995. Nitrogen turnover and availability during succession from alder to poplar in Alaskan taiga forests. Soil Biol. Biochem. 27:743-752. [Google Scholar]
- 11.Davidson, E. A., S. C. Hart, and M. K. Firestone. 1992. Internal cycling of nitrate in soils of a mature coniferous forest. Ecology 73:1148-1156. [Google Scholar]
- 12.De Boer, W., H. Duyts, and H. J. Laanbroek. 1989. Urea stimulated autotrophic nitrification in suspensions of fertilized, acid heath soil. Soil Biol. Biochem. 21:349-354. [Google Scholar]
- 13.De Boer, W., and G. A. Kowalchuk. 2001. Nitrification in acid soils: microorganisms and mechanisms. Soil Biol. Biochem. 33:853-866. [Google Scholar]
- 14.Hankinson, T. R., and E. L. Schmidt. 1984. Examination of an acid forest soil for ammonia- and nitrite-oxidizing autotrophic bacteria. Can. J. Microbiol. 30:1125-1132. [Google Scholar]
- 15.Hart, S. C., G. E. Nason, D. D. Myrold, and D. A. Perry. 1994. Dynamics of gross nitrogen transformations in an old-growth forest: the carbon connection. Ecology 75:880-891. [Google Scholar]
- 16.Hart, S., J. M. Stark, E. A. Davidson, and M. K. Firestone. 1994. Nitrogen mineralization, immobilization, and nitrification, p. 985-1018. In R. Weaver et al. (ed.), Methods of soil analysis. Part 2. Microbiological and biochemical properties. Soil Science Society of America, Inc., Madison, Wis.
- 17.Hastings, R. C., M. T. Ceccherini, M. Nerino, J. R. Saunders, M. Bazzica-lupo, and A. J. McCarthy. 1997. Direct molecular biological analysis of ammonia oxidizing bacteria populations in cultivated soil plots treated with swine manure. FEMS Microb. Ecol. 23:45-54. [Google Scholar]
- 18.Hiorns, W. D., R. C. Hastings, I. M. Head, A. J. McCarthy, and J. R. Saunders. 1995. Amplification of 16S ribosomal RNA genes of autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of Nitrosospiras in the environment. Microbiology 141:2793-2800. [DOI] [PubMed] [Google Scholar]
- 19.Horz, H. P., J. H. Rotthauwe, T. Lukow, and W. Liesack. 2000. Identification of major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J. Microbiol. Methods 39:197-204. [DOI] [PubMed] [Google Scholar]
- 20.Hyman, M. R., and P. M. Wood. 1985. Suicide inactivation and labeling of ammonia monooxygenase by acetylene. Biochem. J. 227:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, Q. Q., and L. R. Bakken. 1999. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30:171-186. [DOI] [PubMed] [Google Scholar]
- 22.Killham, K. S. 1990. Nitrification in coniferous forest soils. Plant Soil 128:31-44. [Google Scholar]
- 23.Koops, H. P., and A. Pommerening-Roser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 24.Kowalchuck, G. A., P. D. E. Bodelier, G. H. J. Helig, J. R. Stephen, and H. J. Laanbroek. 1998. Community analysis of ammonia-oxidizing bacteria, in relation to oxygen-availability in soils and root-oxygenated sediments, using PCR, DGGE, and oligonucleotide probe hybridisation. FEMS Microb. Ecol. 27:339-350. [Google Scholar]
- 25.Kowalchuck, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 26.Kowalchuck, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of β-proteobacteria ammonia-oxidizing bacteria in coastal sand dunes using denaturing gradient gel electrophoresis and sequencing of PCR amplified 16S rDNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowalchuck, G. A., A. W. Stienstra, G. H. J. Helilig, J. R. Stephen, and J. W. Woldendorp. 2000. Changes in the community structure of ammonia-oxidizing bacteria during secondary succession of calcareous grasslands. Environ. Microbiol. 2:99-110. [DOI] [PubMed] [Google Scholar]
- 28.Kowalchuck, G. A., A. W. Stienstra, G. H. J. Helilig, J. R. Stephen, and J. W. Woldendorp. 2000. Molecular analysis of ammonia-oxidizing bacteria in soil of successional grasslands of the Drentsche A (The Netherlands). FEMS Microbiol. Ecol. 31:207-215. [DOI] [PubMed] [Google Scholar]
- 29.Malhi, S. S., and W. B. McGill. 1982. Nitrification in three Alberta soils: effect of temperature, moisture, and substrate concentration. Soil Biol. Biochem. 14:393-399. [Google Scholar]
- 30.Martikainen, P. J., and E. L. Nurmiaho-Lassila. 1984. Nitrosospira, an important ammonium-oxidizing bacterium in fertilized coniferous forest soil. Can. J. Microbiol. 31:190-197. [Google Scholar]
- 31.McTavish, H., J. A. Fuchs, and A. B. Hooper. 1993. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 175:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oved, T., A. Shaviv, T. Goldrath, R. T. Mandelbaum, and D. Minz. 2001. Influence of effluent irrigation on community composition and function of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 67:3426-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson, D. 1994. Filamentous fungi, p. 330-350. In R. Weaver et al. (ed.), Methods of soil analysis. Part 2. Microbiological and biochemical properties. Soil Science Society of America, Inc., Madison, Wis.
- 35.Pederson, H., K. A. Dunkin, and M. K. Firestone. 1999. The relative importance of autotrophic and heterotrophic nitrification in a conifer forest soil as measured by 15N tracer and pool dilution techniques. Biogeochemistry 44:135-150. [Google Scholar]
- 36.Phillips, C. J., D. Harris, S. L. Dollhopf, K. L. Gross, J. I. Prosser, and E. A. Paul. 2000. Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl. Environ. Microbiol. 66:5410-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis; implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice, E. L., and S. K. Pancholy. 1972. Inhibition of nitrification by climax vegetation. Am. J. Bot. 59:1033-1040. [Google Scholar]
- 39.Robertson, G. P. 1982. Nitrification in forested ecosystems. Phil. Trans. Roy. Soc. Lond. 296:445-457. [Google Scholar]
- 40.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monoxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schimel, J. P., R. G. Cates, and R. Ruess. 1998. The role of Balsam poplar secondary chemicals in controlling soil nutrient dynamics through succession in the Alaskan taiga. Biogeochemistry 42:221-234. [Google Scholar]
- 43.Schmidt, E. L., and L. W. Belser. 1994. Autotrophic nitrifying bacteria, p. 159-177. In R. Weaver et al. (ed.), Methods of soil analysis. Part 2. Microbiological and biochemical properties. Soil Science Society of America, Inc., Madison, Wis.
- 44.Stark, J. M., and M. K. Firestone. 1996. Kinetic characteristics of ammonium-oxidizer communities in a California oak woodland-annual grassland. Soil Biol. Biochem. 28:1307-1317. [Google Scholar]
- 45.Stark, J. M., and S. C. Hart. 1997. High rates of nitrification and nitrate turnover in undisturbed coniferous forests. Nature 385:61-64. [Google Scholar]
- 46.Stehr, G., B. Bottcher, P. Dittberner, G. Rath, and H.-P. Koops. 1995. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol. Ecol. 17:177-186. [Google Scholar]
- 47.Stephen, J. R., G. A. Kowalchuck, M. A. Bruns, A. E. McCaig, C. J. Phillips, T. M. Embley, and J. I. Prosser. 1998. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl. Environ. Microbiol. 64:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stienstra, A. W., P. K. Gunnewiek, and H. J. Laanbroek. 1994. Repression of nitrification in soils under climax grassland vegetation. FEMS Microbiol. Ecol. 14:45-52. [Google Scholar]
- 50.Swofford, D. L. 2001. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0b6. Sinauer Associates, Sunderland, Mass.
- 51.Utaker, J. B., L. Bakken, Q. Q. Jiang, and I. F. Nes. 1995. Phylogenetic analysis of seven new isolates of ammonia-oxidizing bacteria based on 16S rRNA gene sequences. Syst. Appl. Microbiol. 18:549-559. [Google Scholar]
- 52.Van Cleve, K., J. Yarie, and R. Erickson. 1993. Nitrogen mineralization and nitrification in successional ecosystems on the Tanana river floodplain interior Alaska. Can. J. For. Res. 23:970-978. [Google Scholar]
- 53.Verchot, L. V., Z. Holmes, L. Mulon, P. M. Groffman, and G. M. Lovett. 2001. Gross versus net rates of N mineralization and nitrification as indicators of functional differences between forest types. Soil Biol. Biochem. 33:1889-1901. [Google Scholar]
- 54.Weber, D. F., and P. L. Gainey. 1962. Relative sensitivity of nitrifying organisms to hydrogen ions in soils and in solutions. Soil Sci. 94:138-145. [Google Scholar]
- 55.Webster, G., T. M. Embley, and J. I. Prosser. 2002. Grassland management regimes reduce small-scale heterogeneity and species diversity of β-proteobacterial ammonia oxidizer populations. Appl. Environ. Microbiol. 68:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woomer, P. 1994. Most probable number counts, p. 59-79. In R. Weaver et al. (ed.), Methods of soil analysis. Part 2. Microbiological and biochemical properties. Soil Science Society of America, Inc., Madison, Wis.