Abstract
The use of antibiotic resistance analysis (ARA) for microbial source tracking requires the generation of a library of isolates collected from known sources in the watershed. The size and composition of the library are critical in determining if it represents the diversity of patterns found in the watershed. This study was performed to determine the size that an ARA library needs to be to be representative of the watersheds for which it will be used and to determine if libraries from different watersheds can be merged to create multiwatershed libraries. Fecal samples from known human, domesticated, and wild animal sources were collected from six Virginia watersheds. From these samples, enterococci were isolated and tested by ARA. Based on cross-validation discriminant analysis, only the largest of the libraries (2,931 isolates) were found to be able to classify nonlibrary isolates as well as library isolates (i.e., were representative). Small libraries tended to have higher average rates of correct classification, but were much less able to correctly classify nonlibrary isolates. A merged multiwatershed library (6,587 isolates) was created and was found to be large enough to be representative of the isolates from the contributing watersheds. When isolates that were collected from the contributing watersheds approximately 1 year later were analyzed with the multiwatershed library, they were classified as well as the isolates in the library, suggesting that the resistance patterns are temporally stable for at least 1 year. The ability to obtain a representative, temporally stable library demonstrates that ARA can be used to identify sources of fecal pollution in natural waters.
The contamination of streams, rivers, and estuaries with untreated fecal material from nonpoint sources continues to be a major environmental problem. Nonpoint source fecal pollution can contain high levels of nitrogen and phosphorus, which contribute to eutrophication. Pollution from fecal sources can also contain pathogenic bacteria, protozoa, and viruses, which can contribute to an increased public health risk. Although methods of quantifying the amount of fecal pollution in natural waters are well established, there is a critical need for the development of methods for the identification of the sources of the pollution to facilitate the remediation of the polluted waters.
Several methods of source tracking methods have been proposed, and the strengths and weaknesses of these methods have been recently reviewed (27, 30). Some methods detect source-specific indicators such as viruses and bacteriophages (3, 4, 18, 21, 26), Bacteroides (1, 2), bifidobacteria (1, 25), endospore-formers (9), and Enterococcus (32). Other methods rely on detecting differences among populations of Escherichia coli and enterococci found in different sources. These methods, which require the generation of a “library” of patterns from bacteria from known sources, include genotypic methods such as ribotyping (6, 8, 16, 24), pulsed-field gel electrophoresis (22, 28, 29, 31), amplified fragment length polymorphism (13, 31), 16S rRNA sequencing (13), and PCR-based methods, such as randomly amplified polymorphic DNA (RAPD) (31) and rep-PCR (7). Phenotypic library-based methods have also been developed, such as those that measure differences in carbon-source utilization patterns (15) and resistance to antibiotics. Differences in antibiotic resistance among bacteria from various potential fecal sources have been measured by the multiple antibiotic resistance method, which uses single concentrations of each drug (13, 19, 23), and by antibiotic resistance analysis (ARA), which uses multiple concentrations of each antibiotic (5, 12, 14, 17, 20, 33-35).
For all library-based methods, the size and composition of the library are critical for accurate and reliable source determination. In order for a library to reliably identify fecal sources, it needs to be representative of the sources that are present in the watersheds (i.e., it should contain examples of all of the patterns found in the bacteria from each of the source types that are found in the watershed). Additionally, an ideal library should be representative enough to be able to classify fecal isolates from other geographic areas and should be stable over time so that new libraries do not need to be continually created. Our laboratory has focused on using ARA for source identification and has created libraries of enterococci from six Virginia watersheds. The purpose of this paper is to examine the relationship between the size of ARA libraries and their representativeness and to determine if libraries from different watersheds can be merged to create multiwatershed libraries that are geographically representative and temporally stable.
MATERIALS AND METHODS
Watersheds and library composition.
Fecal samples from known sources were collected from six Virginia watersheds (Table 1). Land use of these watersheds varied, including urban (high proportions of paved surface, with a high density of homes connected to a sewer system), rural (primarily animal or crop agricultural land use), and wooded (undeveloped land predominately covered by trees). Fecal samples were collected from three types of sources: human (septic tanks and sewage influent), domesticated animal (beef, chicken, dairy, dog, goat, horse, sheep, and turkey), and wild animal (deer, goose, duck, bird, groundhog, and raccoon). When possible, feces from three to five animals were combined into one sample to maximize the diversity of the library. Human samples were obtained from septic tank pumpouts and from the primary influent to sewage treatment plants. Domesticated animal samples were obtained from farms that raised the animals or (in the case of dogs) from city parks. Wild animal scat samples were collected in the field or from trapped animals and were identified by wildlife biologists. The number of samples of each source type varied among the libraries and ranged from 9 to 191 samples per type per library (Table 2). Fecal samples were collected from as many unique locations within the watershed as time and budgets allowed.
TABLE 1.
Watershed characteristics
| Watershed | Duration of sampling (mo/yr) | Land use | Location (Va county) | Drainage (river) | Size (acres) |
|---|---|---|---|---|---|
| Thumb Run | 7/01-9/01 | Rural, wooded | Fauquier | Rappahannock | 23,123 |
| Goose Creek | 12/01-5/02 | Rural, wooded | Loudon and Fauquier | Potomac | 247,921 |
| Blacks Run | 6/99-12/00 | Urban, rural | Rockingham | North | 12,253 |
| Moores Creek | 6/00-3/01 | Urban, rural | Albemarle | Rivanna | 22,348 |
| Holmans Creek | 7/99-5/01 | Rural | Shenandoah | Shenandoah | 11,990 |
| Long Glade Creek | 5/01-12/01 | Rural | Augusta | North | 11,871 |
TABLE 2.
Numbers of samples collected over the duration of the study from each of the sources in each of the libraries and numbers of unique isolatesa obtained from those samples
| Library | Type of sample
|
Total
|
||||||
|---|---|---|---|---|---|---|---|---|
| Human
|
Domesticated
|
Wild
|
||||||
| No. of samples | No. of unique isolates | No. of samples | No. of unique isolates | No. of samples | No. of unique isolates | No. of samples | No. of unique isolates | |
| Thumb Run | 11 | 100 | 17 | 129 | 20 | 165 | 48 | 394 |
| Goose Creek | 12 | 91 | 35 | 224 | 21 | 153 | 68 | 468 |
| Blacks Run | 11 | 172 | 12 | 157 | 27 | 356 | 50 | 685 |
| Moores Creek | 9 | 121 | 59 | 606 | 19 | 150 | 87 | 877 |
| Holmans Creek | 42 | 518 | 45 | 625 | 9 | 89 | 96 | 1,232 |
| Long Glade Creek | 111 | 968 | 191 | 1,604 | 47 | 359 | 349 | 2,931 |
| Merged (all 6) | 196 | 1,970 | 359 | 3,345 | 143 | 1,272 | 698 | 6,587 |
Isolates from a given sample with unique antibiotic resistance patterns.
In addition, libraries were created either by merging or by removing isolates from the six individual libraries. A multiwatershed merged library was created by combining the isolates from each of the six individual libraries. For testing the similarity between libraries, combinations of five libraries were used to classify isolates from the remaining sixth library into the three source types: human, domesticated, and wild. For determining the effect of library size on classification success and representativeness, sublibraries of various sizes (5, 10, 20, 40, 60, or 80% of the full library) were randomly generated (along with their corresponding groups of held-out isolates) from the Long Glade library by using the SURVEYSELECT procedure of SAS (SAS version 8.1; SAS Institute, Inc.) To generate the sublibraries, 5, 10, 20, 40, 60, or 80% of the isolates in each of the 349 samples in the Long Glade library were randomly selected. Three sublibraries were generated for each sublibrary size.
ARA.
Enterococci were isolated from the fecal samples by previously described methods (35). Generally, 10 to 12 strains were isolated from approximately 0.5 g of each sample. The total numbers of isolates that were obtained from each watershed were as follows: Thumb Run, 551; Goose Creek, 735; Blacks Run, 959; Moores Creek, 1,248; Holmans Creek, 1,777; and Long Glade Creek, 3,973. ARA was performed as previously described (35) with the following drugs and concentrations: bacitracin, 10, 25, 50, and 100 μg/ml; cephalothin (sodium salt), 10, 15, and 50 μg/ml; chlortetracycline hydrochloride, 20, 60, and 80 μg/ml; erythromycin, 10, 30, and 50 μg/ml; gentamicin, 5, 10, and 20 μg/ml; kanamycin monosulfate, 10, 15, 30, and 50 μg/ml; neomycin, 10, 15, 30, and 50 μg/ml; oxytetracycline hydrochloride, 20, 40, and 80 μg/ml; streptomycin sulfate, 20, 40, 60, and 80 μg/ml; tetracycline, 10, 30, and 50 μg/ml; and vancomycin, 5, 10, and 30 μg/ml.
Statistical analysis.
For each of the six individual libraries and for the multiwatershed merged library, the data for each of the known isolates' ability to grow in the presence of each concentration of each antibiotic were classified by discriminant analysis by the DISCRIM procedure of SAS (prior probabilities, equal; covariance matrix, pooled). Within each sample, only isolates with unique resistance patterns were analyzed. (Isolates with identical patterns were discarded by using the NODUPRECS option of the SORT procedure.) The percentage of unique isolates ranged from 64% in Goose Creek to 76% in Blacks Run, and the average number of unique isolates per sample ranged from 7 (Goose Creek) to 14 (Blacks Run).
The average rate of correct classification (ARCC) of the various libraries, sublibraries, and sets of held-out isolates was computed by averaging the percentages of correctly classified isolates for each source (35). Additionally, the rates of misclassification were calculated. To determine the extent of misclassification of isolates from other sources into a given source type, the percentages of each of the other source types that were misclassified as that type were averaged. This value was termed the “expected frequency of misclassification” by Harwood et al. (17). As a way to determine the lower limit for considering a source to be a significant contributor to a watershed, Whitlock et al. (33) averaged the expected frequencies of misclassification for all sources and then added four times the value of the standard deviation to the average. We propose calling this value the “minimum detectable percentage” (MDP). Thus, if a source is found at levels above the MDP, it can be reasonably assumed that this is not the result of misclassification of other sources and therefore is present in the watershed.
Representativeness testing.
The representativeness of a library can be estimated by how well the library can classify nonlibrary isolates (isolates from the same watershed that are not included in the library). If the nonlibrary isolates are classified (on average) as well as the library isolates, then the library contains enough representation of the patterns to provide confidence in the classification of unknown isolates from the watershed. In the standard type of discriminant analysis (called “resubstitution analysis”), each isolate is classified based on the patterns in the entire library, including its own pattern. As a result, the ARCCs from resubstitution analysis may overestimate representativeness, because each isolate is classified against itself (i.e., it is classified by using a library that contains its resistance pattern). Therefore, in addition to resubstitution analyses, cross-validation analyses (also called “jackknife” or “leave-one-out” analyses) were performed to determine the representativeness of the libraries. For the cross-validation analyses, an individual isolate (or all of the isolates from the same sample) was removed from the library one at a time. Then, the removed isolate or isolates were classified based on the library comprised of the remaining isolates, and the ARCC for these removed isolates was calculated. Two types of cross-validation analyses were performed: pulled-isolate analysis and pulled-sample analysis. For the pulled-isolate analyses, each isolate in the library was removed separately. However, isolates from the same sample might have similar patterns, which would make the library seem more representative than it actually is. To eliminate this possibility, pulled-sample analyses were performed in which all of the isolates from a common sample were removed at the same time.
Temporal and geographical testing.
To determine the temporal stability of the libraries, fecal samples were collected from the Moores Creek, Holmans Creek, and Long Glade Creek watersheds approximately 1 year after the original library samples were collected. To measure the geographic range of the libraries, samples were collected from watersheds in southwestern Virginia and in southern Florida. The isolates obtained from these samples will be referred to as “temporal validation isolates” and “geographical validation isolates,” respectively.
RESULTS
Classification of the known isolates was performed for each library. When resubstitution analysis was used, the ARCCs ranged from 65 to 81% (Table 3). The ARCC of the merged library (57%) was lower than those of any of the individual libraries. Human isolates were generally classified the best, with domesticated and wild isolates classified approximately equally well. Cross-validation of these libraries by the pulled-isolate method resulted in slightly lower ARCCs (Table 4), with the differences between the two methods ranging from 2% for the Moores Creek and Long Glade libraries up to 8% for the Goose Creek library. The ARCC of the merged library did not change when analyzed by the pulled-isolate method. When pulled-sample cross-validation analyses were performed, there was a more marked reduction in ARCCs (Table 5). The differences between this method and the resubstitution analyses were large for most of the libraries (ranging from 8 to 24%), but were much smaller for the largest library (Long Glade Creek) and for the merged library (4 and 3%, respectively).
TABLE 3.
Resubstitution discriminant analysis of six Virginia watersheds
| Watershed | % of isolates correctly classified as:
|
ARCC | ||
|---|---|---|---|---|
| Human | Domesticated | Wild | ||
| Thumb Run | 80 | 69 | 75 | 75 |
| Goose Creek | 67 | 59 | 67 | 65 |
| Blacks Run | 85 | 76 | 82 | 81 |
| Moores Creek | 70 | 76 | 69 | 72 |
| Holmans Creek | 67 | 64 | 80 | 70 |
| Long Glade Creek | 67 | 65 | 62 | 65 |
| Merged (all 6) | 63 | 54 | 55 | 57 |
TABLE 4.
Pulled-isolate discriminant analysis of six Virginia watersheds
| Watershed | % of isolates correctly classified as:
|
ARCC | ||
|---|---|---|---|---|
| Human | Domesticated | Wild | ||
| Thumb Run | 77 | 63 | 72 | 70 |
| Goose Creek | 54 | 54 | 65 | 57 |
| Blacks Run | 82 | 69 | 78 | 77 |
| Moores Creek | 68 | 75 | 67 | 70 |
| Holmans Creek | 64 | 62 | 73 | 66 |
| Long Glade Creek | 66 | 64 | 60 | 63 |
| Merged (all 6) | 62 | 54 | 55 | 57 |
TABLE 5.
Pulled-sample discriminant analysis of six Virginia watersheds
| Watershed | % of isolates correctly classified as:
|
ARCC | ||
|---|---|---|---|---|
| Human | Domesticated | Wild | ||
| Thumb Run | 73 | 49 | 58 | 60 |
| Goose Creek | 45 | 44 | 58 | 49 |
| Blacks Run | 74 | 55 | 66 | 65 |
| Moores Creek | 61 | 71 | 59 | 64 |
| Holmans Creek | 54 | 55 | 29 | 46 |
| Long Glade Creek | 65 | 61 | 58 | 61 |
| Merged (all 6) | 61 | 52 | 50 | 54 |
One way to estimate the representativeness of a library is to determine how well isolates obtained from new samples from the watershed are classified compared to how well library isolates from that source are classified. If isolates from new samples (nonlibrary isolates) from a given source are classified (on average) as well as the isolates from that source that are in the library, then the library can be considered representative for that source. The testing of new isolates can be simulated by cross-validation analyses (which remove isolates from the library and then classify them). If just one isolate per sample had been used to construct the library, the pulled-isolate method of cross-validation (which classifies each isolate based on a library from which it has been removed) would be a good method to use. However, because the libraries in this study contain multiple isolates from the same sample, the pulled-isolate method may not give a true measure of representativeness, because the similarity of isolates within samples may be greater than the similarity between samples. Therefore, the pulled-isolate method is inappropriate because it may overestimate representativeness. The pulled-sample cross-validation analysis avoids this problem by removing all of the isolates from each sample and then classifying them against the remaining isolates, thus simulating the testing of a set of multiple isolates from a new sample. Thus, by comparing the difference between the ARCC of the resubstitution analysis (which classifies each isolate based on a library containing all of the isolates) and the ARCC of the pulled-sample analysis (which classifies each isolate based on a library from which the isolates from its own sample have been removed), the representativeness of a library can be estimated. If the difference is small (less than 5%), then the library can be considered representative (i.e., new isolates are classified almost as well as isolates in the library). Based on this criterion, only the large libraries (Long Glade and the merged library) can be considered representative of the sources of enterococci.
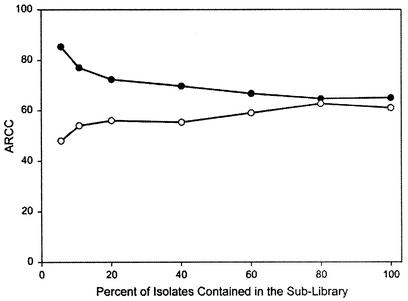
Another way to estimate the representativeness of a library is to hold out a portion of the isolates from an existing library and then classify them by using the sublibrary that contains the remaining isolates. As with the cross-validation analyses, if these held-out isolates are classified (on average) as well as the isolates in the sublibrary, then the entire library can be considered representative. When various sizes of randomly selected sublibraries of the Long Glade library were created, the smallest libraries had the highest ARCCs, but were the poorest in classifying the held-out isolates (Fig. 1). For example, the smallest Long Glade sublibrary (5% of the full library) had an ARCC of 85%, but when the held-out isolates (the remaining 95% of the full library) were classified based on this small library, they were correctly classified at an average rate of just 48%. As the size of the sublibraries increased, the difference decreased. The held-out isolates were correctly classified about as well as the sublibrary isolates at sizes of about 80% of the full library. Thus, approximately 2,300 isolates were required to produce a representative library.
FIG. 1.
Comparison of the correct classification of randomly chosen sublibraries and the corresponding held-out isolates from the Long Glade Creek library. Solid circles, isolates included in the sublibraries; open circles, held-out isolates that were classified by using the sublibraries. Each data point is the mean of three random selections.
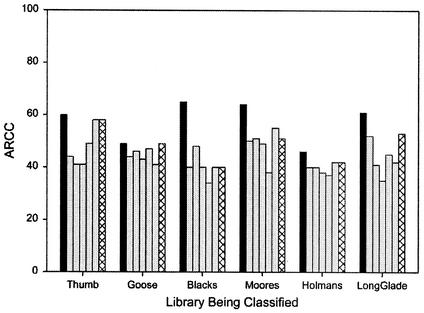
Because of the large number of isolates that are required for representativeness, it may be impractical to collect enough isolates from a single watershed. However, if the resistance patterns of isolates from different watersheds are similar, several smaller libraries could be combined to produce a larger, more representative multiwatershed library. To determine the similarity of the six libraries to each other, the isolates from each of the libraries were classified by using each of the other five libraries. Generally, the isolates were better classified against isolates from the same library (based on pulled-sample analysis) than they were against any of the other libraries (Fig. 2). The average of the differences between the pulled-sample ARCC and the ARCC of each library classified against each of the other five libraries was 14%.
FIG. 2.
Classification of each of the libraries against itself (pulled-sample method) (black bars), each of the other five individual libraries (gray bars), or against a library comprised of all of the remaining five libraries (cross-hatched bars).
To determine how well the isolates from each library would be classified by a larger, more representative library, each of the libraries was classified by using a library comprising the other five libraries combined (i.e., the merged library with the individual library removed). Several of the individual libraries were classified nearly as well against the other five libraries combined as they were against themselves (by the pulled-sample method) (Fig. 2). The average of these differences was 9%. The difference probably results from the lack of representativeness of several of the smaller libraries, as well as some geographical differences between the watersheds. However, the previously demonstrated representativeness of the merged six-watershed library suggests that the resistance patterns do not vary greatly over the geographical range of these watersheds (northern and central Virginia).
As a further test of the geographic range of the merged library, samples from known sources from south Florida and from southwestern Virginia were tested in our laboratory and then classified by the merged library. In general, classification of the isolates from these geographical validation samples was lower than the ARCC of the merged library, with correct classification of just 45% of Florida human isolates and 37% of Florida domesticated isolates (Table 6). The classification of southwest Virginia isolates was varied, with human isolates being classified better than the isolates in the library (71% compared to 61%), but with domesticated isolates classified much worse (18% compared to 52%).
TABLE 6.
Classification of known geographic validation isolates collected from different watersheds and of known temporal validation isolates collected from the same watershed approximately 1 year after the library had been completed
| Type of isolate by watershed/region | Source type | No. of isolates (samples) | % Correct by:
|
|
|---|---|---|---|---|
| Individual library | Merged library | |||
| Temporal validation | ||||
| Moores Creek | Human | 11 (1) | 27 | 64 |
| Domesticated | 24 (3) | 75 | 79 | |
| Wild | 8 (1) | 38 | 38 | |
| Average | 47 | 60 | ||
| Holmans Creek | Human | 25 (4) | 60 | 52 |
| Domesticated | 64 (2) | 45 | 64 | |
| Wild | 5 (1) | 40 | 80 | |
| Average | 48 | 65 | ||
| Long Glade Creek | Domesticated | 49 (6) | 84 | 84 |
| Geographic validation | ||||
| South Florida (Hillsborough County) | Human | 69 (5) | 45 | |
| Domesticated | 62 (13) | 37 | ||
| Average | 41 | |||
| Southwest Virginia (Franklin and | ||||
| Montgomery Counties) | Human | 41 (4) | 71 | |
| Domesticated | 153 (8) | 18 | ||
| Wild | 43 (3) | 53 | ||
| Average | 47 | |||
If the merged library is to be most useful, the resistance patterns should be stable over time. To determine how stable the resistance patterns are, additional samples from three of the watersheds were collected approximately 1 year after the original library was collected. When compared to the original Moores Creek library, the isolates from these temporal validation samples from Moores Creek were classified worse than the isolates in the library (with an average correct classification rate of just 47%) (Table 6). However, when the merged library was used, they were classified at a higher rate (average = 60%). Similarly, the temporal validation isolates from Holmans Creek were classified better by using the merged library (average =65%) than by using just the Holmans library (average = 48%). Long Glade Creek temporal validation isolates were classified better than the isolates in the library (84%) by using both the Long Glade library and the merged library. On average, all three sets of temporal validation isolates were classified better than the isolates from the merged library, demonstrating that the patterns are temporally stable over a period of 1 year.
Given that the merged library is representative and is temporally stable, this would be the library of choice for the identification of unknown isolates. Although the ARCC of the library (Table 7) is 57%, it can still provide useful information, because the classification success rate is well above the rate that would be produced by random classification (33%). For this analysis, the expected frequencies of misclassification for human, domesticated, and wild sources were 20, 22, and 22%, respectively. The average rate at which the entire library was misclassified was 21% ± 1% (mean ± standard deviation), which resulted in an MDP of 25%. Thus, if an average of more than 25% of a particular source type is detected in a watershed, it can be reasonably assumed that it is a true source and not just a result of misclassification of the other source types.
TABLE 7.
Classification of 6,587 isolates of enterococci from known human, domesticated, and wild sources in six Virginia watershedsa
| Source (n) | No. (%) of isolates classified as:
|
||
|---|---|---|---|
| Human | Domesticated | Wild | |
| Human (1,970) | 1,228 (63) | 381 (19) | 361 (18) |
| Domesticated (3,345) | 659 (20) | 1,813 (54) | 873 (26) |
| Wild (1,272) | 262 (21) | 305 (24) | 705 (55) |
The ARCC for this analysis is 57%, and the MDP is 25%.
DISCUSSION
The representativeness of the libraries was tested by several techniques. If multiple isolates are included from each sample, the pulled-isolate method does not give a true measure of representativeness, because the similarity of isolates within samples is greater than the similarity between samples. Therefore, the pulled-sample method, even though it is harder to perform, is a better measure of representativeness. By using the pulled-sample method, it is clear that larger libraries are more representative. Both the Long Glade Creek library and the 6,500-isolate merged library show approximately equal classification success when isolates from a nonlibrary sample (one that has been pulled out) are classified and thus are representative. At least 2,300 of the isolates in the Long Glade library were required for it to be representative, which suggests that this is the magnitude of a minimum size for a representative library. Unfortunately, these larger libraries also have the lowest ARCCs. This lowered classification success of the larger libraries probably results from variability in the resistance patterns of the isolates within each source type in the watersheds. In other words, the more isolates of each source type that are contained in the library (i.e., the more representative it is), the greater the chance they will vary in their resistance patterns, which would thus result in lower classification success. If there is variability in the patterns in a watershed, then the ARCC of a small library could be misleading, because it would be unable to classify the large number of unknown isolates that have a pattern that is not included in it. Thus, it is unwise to rely only on the ARCC of a library without also knowing its representativeness.
The levels of classification of the temporal validation isolates are encouraging for maintaining a merged library over the period of 1 year. Although the long-term usefulness of ARA libraries has yet to be demonstrated, many source tracking studies are conducted over the course of a single year, so the temporal stability that we observed for this library would be sufficient for these studies. Because the creation of a representative library requires considerable effort, it would be desirable to have long-term temporal stability. To determine this, we are continuing to monitor the resistance patterns in the Holmans Creek and Long Glade Creek watersheds.
There were some differences in classification success from watershed to watershed. This may be indicative of real differences between watersheds (feeding practices, antibiotic usage, types of animals, etc.), or it may be a result of nonrepresentativeness of the individual libraries. When the libraries were merged, the differences decreased, suggesting that the lack of representativeness was causing some of the differences and that geographically close watersheds share some patterns. Unfortunately, the lower classification rates of the geographical validation isolates suggest that libraries will have to be comprised of isolates from a specific region for them to be useful. Further research will be needed to determine the size of these regions.
The ARCCs for the six libraries, especially those of the large Long Glade Creek library and the merged library, are generally lower than the values obtained in previous studies. However, for the watersheds presented here, only unique isolates have been included in the analyses. The presence of duplicate isolates can artificially raise the ARCC by the inclusion of more similar patterns of each source type. Thus, the ARCCs in this paper are more truly indicative of the actual classification abilities of ARA. Also, the Long Glade and merged libraries are larger than previously published libraries, and larger libraries have lower ARCCs. Because ARA is rapid and inexpensive, however, large numbers of unknown isolates can be tested, which can overcome the lower ARCC. Even with an ARCC of 57% and an MDP of 25%, the merged library can be useful for source tracking studies. Often, only the major source(s) of fecal pollution will need to be identified for these studies, and this library can reliably identify sources that are present at average levels above 25%.
The ability of ARA to differentiate among the different source types demonstrates the validity of our working hypothesis that differential antibiotic use selects for bacterial populations with different resistance patterns in animals of different sources. This is an advantage over other source tracking methods that do not rely on detecting a selectable difference. Many library-based methods look for overall genetic differences among the strains, but not for any specific difference. However, overall genetic differences may be hard to demonstrate because of the diverse (10; G. A. Dykes, Letter, Appl. Environ. Microbiol. 68:4698, 2002) and changeable (11) nature of the gut microflora. One specific selectable difference between strains that is commonly proposed is that of adhesion or attachment to the intestinal wall. However, recent evidence shows that although strains of Lactobacillus that adhere to the mucosa of the gut wall are host specific, they are very different from the strains present in the feces (36). By relying on a selectable difference that is exogenously applied to the guts of the source animals, ARA can somewhat reduce the effects of the variation in gut composition.
In conclusion, jackknife analysis of multiple ARA libraries has shown that large libraries are required for representativeness, that libraries from several watersheds can be merged to produce a library that is large enough to be representative, and that the resistance patterns within watersheds are stable over the span of 1 year. The ability to obtain a representative, temporally stable library demonstrates that ARA can be used to identify sources of fecal pollution in natural waters.
Acknowledgments
We thank J. Herrick for critical reading of the manuscript and C. Hagedorn and J. Harwood for discussion and helpful comments.
This work was supported by the James Madison University Department of Biology and by grants from the United States Department of Agriculture (no. 2001-35102-10790), the Virginia Department of Conservation and Recreation, the Thomas Jefferson Planning District Commission, the Lord Fairfax Planning District Commission, the Interstate Commission on the Potomac River Basin, and the Rappahannock-Rapidan Regional Commission.
REFERENCES
- 1.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes coding for 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brion, G. M., and S. Lingireddy. 1999. A neural network approach to identifying non-point sources of microbial contamination. Water Res. 33:3099-3106. [Google Scholar]
- 4.Brion, G. M., J. S. Meschke, and M. D. Sobsey. 2002. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res. 36:2419-2425. [DOI] [PubMed] [Google Scholar]
- 5.Butaye, P., L. A. Devriese, and F. Haesebrouck. 2001. Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animals. Antimicrob. Agents Chemother. 45:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson, A. C., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farag, A. M., J. N. Goldstein, D. F. Woodward, and M. Samadpour. 2001. Water quality in three creeks in the backcountry of Grand Teton National Park, USA. J. Freshw. Ecol. 16:135-143. [Google Scholar]
- 9.Fujioka, R. S. 2001. Monitoring coastal marine waters for spore-forming bacteria of faecal and soil origin to determine point from non-point source pollution. Water Sci. Technol. 44:181-188. [PubMed] [Google Scholar]
- 10.Geornaras, I., J. W. Hastings, and A. von Holy. 2001. Genotypic analysis of Escherichia coli strains from poultry carcasses and their susceptibilities to antimicrobial agents. Appl. Environ. Microbiol. 67:1940-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 12.Graves, A. K., C. Hagedorn, A. Teetor, M. Mahal, A. M. Booth, and R. B. Reneau, Jr. 2002. Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Qual. 31:1300-1308. [DOI] [PubMed] [Google Scholar]
- 13.Guan, S., R. Xu, S. Chen, J. Odumeru, and C. Gyles. 2002. Development of a procedure for discriminating among Escherichia coli isolates from animal and human sources. Appl. Environ. Microbiol. 68:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagedorn, C., J. B. Crozier, K. A. Mentz, A. M. Booth, A. K. Graves, N. J. Nelson, and R. B. Reneau, Jr. 2003. Carbon source utilization profiles as a method to identify sources of fecal pollution in water. J. Appl. Microbiol. 94:792-799. [DOI] [PubMed]
- 16.Hartel, P. G., J. D. Summer, J. L. Hill, J. V. Collins, J. A. Entry, and W. I. Segars. 2002. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed] [Google Scholar]
- 17.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, S., R. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaspar, C. W., J. L. Burgess, I. T. Knight, and R. R. Colwell. 1990. Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can. J. Microbiol. 36:891-894. [DOI] [PubMed] [Google Scholar]
- 20.Kern, J., B. Petrauskas, P. McClellan, V. O. Shanholtz, and C. Hagedorn. 2002. Bacterial source tracking: a tool for total maximum daily load development, p. 125-142. In T. Younos (ed.), Advances in water monitoring research. Water Resource Publications, LLC, Highlands Ranch, Colo.
- 21.Ley, V., J. Higgins, and R. Fayer. 2002. Bovine enteroviruses as indicators of fecal contamination. Appl. Environ. Microbiol. 68:3455-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parveen, S., N. C. Hodge, R. E. Stall, S. R. Farrah, and M. L. Tamplin. 2001. Genotypic and phenotypic characterization of human and nonhuman Escherichia coli. Water Res. 35:379-386. [DOI] [PubMed] [Google Scholar]
- 23.Parveen, S., R. L. Murphree, L. Edmiston, C. W. Kaspar, K. M. Portier, and M. L. Tamplin. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes, M. W., and H. Kator. 1999. Sorbitol-fermenting bifidobacteria as indicators of diffuse human faecal pollution in estuarine watersheds. J. Appl. Microbiol. 87:528-535. [DOI] [PubMed] [Google Scholar]
- 26.Schaper, M., and J. Jofre. 2000. Comparison of methods for detecting genotypes of F-specific RNA bacteriophages and fingerprinting the origin of faecal pollution in water samples. J. Virol. Methods 89:1-10. [DOI] [PubMed] [Google Scholar]
- 27.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons, G. M., Jr., S. A. Herbein, and C. M. James. 1995. Managing nonpoint fecal coliform sources to tidal inlets. Water Resour. Update 100:64-74. [Google Scholar]
- 29.Simmons, G. M., Jr., D. F. Waye, S. Herbein, S. Myers, and E. Walker. 2002. Estimating nonpoint source fecal coliform sources using DNA profile analysis, p. 143-168. In T. Younos (ed.), Advances in water monitoring research. Water Resources Publications, LLC, Denver, Colo..
- 30.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 31.Vancanneyt, M., A. Lombardi, C. Andrighetto, E. Knijff, S. Torriani, K. J. Björkroth, C. M. A. P. Franz, M. R. Foulquié Moreno, H. Revets, L. De Vuyst, J. Swings, K. Kersters, F. Dellaglio, and W. H. Holzapfel. 2002. Intraspecies genomic groups in Enterococcus faecium and their correlation with origin and pathogenicity. Appl. Environ. Microbiol. 68:1381-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheeler, A. L., P. G. Hartel, D. G. Godfrey, J. L. Hill, and W. I. Segars. 2002. Potential of Enterococcus faecalis as a human fecal indicator for microbial source tracking. J. Environ. Qual. 31:1286-1293. [DOI] [PubMed] [Google Scholar]
- 33.Whitlock, J. E., D. T. Jones, and V. J. Harwood. 2002. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Res. 36:4273-4282. [DOI] [PubMed] [Google Scholar]
- 34.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiggins, B. A., R. W. Andrews, R. A. Conway, C. L. Corr, E. J. Dobratz, D. P. Dougherty, J. R. Eppard, S. R. Knupp, M. C. Limjoco, J. M. Mettenburg, J. M. Rinehardt, J. Sonsino, R. L. Torrijos, and M. E. Zimmerman. 1999. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 65:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. L. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]