Abstract
Quinones can function as redox mediators in the unspecific anaerobic reduction of azo compounds by various bacterial species. These quinones are enzymatically reduced by the bacteria and the resulting hydroquinones then reduce in a purely chemical redox reaction the azo compounds outside of the cells. Recently, it has been demonstrated that the addition of lawsone (2-hydroxy-1,4-naphthoquinone) to anaerobically incubated cells of Escherichia coli resulted in a pronounced increase in the reduction rates of different sulfonated and polymeric azo compounds. In the present study it was attempted to identify the enzyme system(s) responsible for the reduction of lawsone by E. coli and thus for the lawsone-dependent anaerobic azo reductase activity. An NADH-dependent lawsone reductase activity was found in the cytosolic fraction of the cells. The enzyme was purified by column chromatography and the amino-terminal amino acid sequence of the protein was determined. The sequence obtained was identical to the sequence of an oxygen-insensitive nitroreductase (NfsB) described earlier from this organism. Subsequent biochemical tests with the purified lawsone reductase activity confirmed that the lawsone reductase activity detected was identical with NfsB. In addition it was proven that also a second oxygen-insensitive nitroreductase of E. coli (NfsA) is able to reduce lawsone and thus to function under adequate conditions as quinone-dependent azo reductase.
Azo dyes are widely used as dyes for textiles, food, cosmetics, and various other purposes (51). They are usually very stable under aerobic conditions and therefore resist biodegradation in conventional aerobic sewage treatment systems. In contrast, under anaerobic conditions various bacterial strains are able to reduce azo dyes to amines and thus decolorize the dyes (10, 39, 43). These anaerobic reactions are usually extremely unspecific with regard to the organisms involved and the dyes converted. In these processes, low-molecular-weight redox mediators (e.g., quinones) are often involved which are enzymatically reduced by the bacteria. The reduced mediator compounds (e.g., hydroquinones) can then reduce the azo compounds outside of the cells in a purely chemically and therefore extremely unspecific redox reaction (17, 23).
It was recently shown that among different quinones tested, especially anthraquinone-2-sulfonate (AQS) and lawsone (2-hydroxy-1,4-naphthoquinone) considerably increased the anaerobic reduction rates of azo dyes by Sphingomonas xenophaga BN6, Escherichia coli, and various other bacterial strains from different bacterial groups. For S. xenophaga BN6, AQS was identified as the most effective redox mediator from all quinones investigated and it was suggested that the membrane bound NADH:ubiquinone oxidoreductase of the respiratory chain was responsible for this activity (23, 35). In contrast to S. xenophaga BN6, cells of E. coli demonstrated much higher reduction rates with lawsone compared to AQS (35). These results suggested that the reduction of lawsone by E. coli (and also other bacterial strains) was catalyzed by a different enzyme system than the reduction of AQS by S. xenophaga BN6. It was recently demonstrated that the presence of lawsone also enabled whole cells of E. coli to reduce anaerobically polymeric azo compounds which may be used for the colon-specific delivery of drugs (36). In the present study it was therefore attempted to identify the enzymatic activity which is responsible for the ability of E. coli (and presumably other bacterial strains) to perform a lawsone-dependent azo reductase reaction.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli K-12 was used as the standard strain for most experiments. In addition, different mutant strains of E. coli were used (Table 1). The strains were routinely cultured aerobically at 37°C in nutrient broth (NB) (plus ampicillin [100 μg/ml], if appropriate).
TABLE 1.
Mutant strains of E. coli used in this study
| E. coli strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| NFR 402 | NfsA− | 9 |
| NFR 502 | NfsA− NfsB− | 9 |
| SIL 41 | NfsB− | 9 |
| JVQ1 | NfsA− | 46 |
| JVQ2 | NfsA− NfsB− | 46 |
| AB1157 | NfsA+ NfsB+, parent strain for JVQ1 + JVQ2 | 46 |
| JM83 | NfsA+ NfsB+, parent strain for JM83(pNR3) and JM83(pAJ13NH) | 28 |
| JM83(pNR3) | NfsB overproducing | 49 |
| JM83(pAJ13NH) | NfsA overproducing | S. Zenno |
Conversion of lawsone or nitrofurazone by resting cells of E. coli.
The cultures (100 ml) were grown aerobically under the conditions indicated above until they reached the late exponential growth phase. Cells were harvested by centrifugation (8,000 × g), washed, and resuspended in Na/K-phosphate buffer (50 mM, pH 7.7) to an optical density at 546 nm of about 5. These cell suspensions were transferred to serum bottles, and oxygen was removed by repeated evacuation and flushing with nitrogen gas. The serum bottles were transferred to an anaerobic incubation chamber (Toepfer Lab System, Göppingen, Germany) and aliquots (usually 20 μl) transferred under strictly anaerobic conditions to the wells of a 96-wells microtiter plate. The wells of the microtiter plates contained under the standard test conditions in a total volume of 200 μl 50 mM Na/K-phosphate buffer (pH 7.7), 10 mM glucose, 0.1 mM of lawsone or nitrofurazone and cells (OD at 546 nm = 0.5). The microtiter plates were transferred to a microtiter plate reader (Powerwave 340-I; Biotek Kontron, Neufahrn, Germany), which was located inside the anaerobic chamber and the decrease in absorbance determined at 400 or 450 nm for 30 min at 30°C (using 1-min measuring intervals).
Preparation of cell extracts.
The cells were grown in NB medium until they reached the late exponential growth phase, harvested by centrifugation (8,000 × g, 10 min), washed and finally resuspended in Na/K-phosphate buffer (50 mM, pH 7.7). An aliquot of DNase I was added and the cells disrupted using a French Press (Aminco, Silver Spring, Md.) at 80 MPa. Cell debris was removed by centrifugation at 80,000 × g and 4°C for 35 min. Protein was determined by the method of Bradford (5) with bovine serum albumin being used as a standard.
Preparation of cell membranes.
For the preparation of cell membranes, the cells were washed and broken using a French press as described above. These extracts were centrifuged for 10 min at 8,000 × g to remove whole cells and larger particles. The supernatant was then centrifuged for 2 h at 120,000 × g. This resulted in the formation of a transparent pellet which was resuspended in Na/K-phosphate buffer (50 mM, pH 7.7) and used in the enzyme assays as the membrane fraction (27). The supernatant from the ultracentrifugation was used in certain experiments as the “cytosolic fraction.”
Standard assay for the determination of the lawsone reductase activity with cell extracts or purified enzyme preparations.
The lawsone reductase activity was routinely assayed in 96-well microtiter plates using a microtiter plate reader, which was located inside the anaerobic incubation chamber (see above). The wells of the microtiter plates contained in a total volume of 200 μl 50 mM Na/K-phosphate buffer (pH 7.7), 0.25 mM lawsone, and different amounts of protein. (All solutions were freed of oxygen prior to use by repeated flushing with nitrogen gas.). The reactions were started by the addition of NADH (0.1 mM) and the decrease in absorbance determined for 30 min at 450 nm at 37°C (using 1-min measuring intervals).
Determination of the substrate specificity of the lawsone reductase.
The enzyme assays were performed with the enzyme preparations obtained after the final Mono-Q column chromatography (Table 2) under anaerobic conditions at 37°C in microtiter plates. The reaction mixtures contained in 200 μl Tris-HCl buffer (50 mM, pH 7.7), 0.1 mM NADH, 0.5 mg l−1 protein, and 100 μM of the appropriate substrate. The reactions were started by the addition of the NADH solution. The microtiter plates were shaken for 5 s (17 Hz), and the reaction rates were determined spectrophotometrically using a microtiter plate reader.
TABLE 2.
Purification of lawsone reductase activity from E. coli K-12a
| Purification step | Vol (ml) | Total protein (mg) | Sp. act (U/mg) | Total activity (U) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Crude extract | 23 | 308 | 0.058 | 18.0 | 100 | 1 |
| Q-Sepharose | 5 | 8.4 | 1.75 | 14.6 | 81 | 30 |
| Mono-P | 6 | 0.6 | 9.9 | 6.25 | 35 | 170 |
| Mono-Q | 1 | 0.2 | 29 | 5.8 | 32 | 500 |
Experimental details are given in Materials and Methods.
Enzyme purification.
Protein was purified at room temperature under anaerobic conditions in an anaerobic incubation chamber (Toepfer Lab System, Göppingen, Germany) by use of a fast-performance liquid chromatography system consisting of an LCC 500 controller, a pump 500, a UV-1 monitor, a REC-482 recorder, and a FRAC autosampler from Pharmacia (Uppsala, Sweden).
The crude extract was applied to a Q-Sepharose-FF column (HR 16/10; Pharmacia). Protein was eluted with 180 ml of a linear gradient of Tris-HCl (50 mM, pH 7.7) into Tris-HCl (50 mM, pH 7.7) plus 1 M NaCl at a flow rate of 2 ml/min. Fractions (5 ml) were collected, and the lawsone reductase activity was determined at a λ of 450 nm under anaerobic conditions using a microtiter plate reader. The lawsone reductase activity was eluted as a single peak at a concentration of about 0.18 M NaCl. The active fraction was dialysed against 25 mM Bis-Tris buffer (pH 6.3) and then loaded on a Mono-P column (HR 5/20). The proteins were eluted with 40 ml of Polybuffer 74 (Pharmacia) at a flow rate of 1 ml/min using a linear pH gradient from pH 6.3 to 3.9. Fractions (2 ml each) were collected, and the pH of the fractions was determined by using a pH electrode. The active fraction eluted at pH 5.3. The active fractions were pooled, dialysed against Tris-HCl buffer (50 mM, pH 7.7), and applied to a Mono-Q column (HR5/5). Protein was eluted with 50 ml of a linear gradient of Tris-HCl (50 mM, pH 7.7) into Tris-HCl (50 mM, pH 7.7) plus 1 M NaCl at a flow rate of 1 ml/min. The active fractions eluted at about 0.1 M NaCl.
PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (24). Gels were silver stained (3) or stained with Coomassie blue by the method of Weber and Osborn (45).
The subunit size of the lawsone reductase activity was determined by SDS-PAGE using the standard proteins of a low molecular weight electrophoresis calibration kit (Amersham, Pharmacia) as reference.
In certain experiments nondenaturing gel electrophoresis was used for the visualization and detection of quinone or azo reductase activities in cell extracts (33, 34). The basic composition of the gel was the same as that used for the SDS-PAGE but under omission of SDS. The gel electrophoresis was performed under aerobic conditions. The gels were transferred after finishing the electrophoresis to the anaerobic incubation chamber and washed with 5 ml of anaerobic Na/K-phosphate buffer (50 mM, pH 7.7). The buffer solution was carefully removed and the gel covered with 2 ml of an anaerobic solution of amaranth (1 mM in 50 mM Na/K-phosphate buffer). After 2 min of incubation, 2 ml of lawsone or AQS (1 mM in 50 mM Na/K-phosphate buffer) and 2 ml of NAD(P)H (1 mM in water) were added. The active protein bands became usually visible as decolorized zones in the gels after 15 to 30 min of incubation.
Western blot.
The proteins were electrophoretically transferred to an Immobilon-P transfer membrane (Millipore, Bedford, Mass.) according to the standard procedures described by Ausubel et al. (2).
Determination of amino acid sequences.
The NH2-terminal amino acid sequence of the protein was determined by automated Edman degradation using an Applied Biosystems (Foster City, Calif.) model 491 sequencer.
Chemicals.
Nitrofurazone was supplied by ICN (Aurora, Ohio). The sources of all other chemicals have been described before (35).
RESULTS
Localization of the lawsone reducing activity in the cytosolic fraction of E. coli.
It was previously demonstrated that in S. xenophaga BN6 the major AQS-reducing activity is present in the cell membranes (23). In order to identify the localization of the lawsone reductase activity in E. coli, cells were broken and the cell constituents separated in a membrane and a soluble fraction. Thus, it was clearly demonstrated that the main lawsone reducing activity was present in the cytosolic fraction (37 U/g of protein) and that only a very low activity was present in the membrane fraction (1.4 U/g of protein). (With whole cells a lawsone reductase activity of 32 U/g of protein was determined.) This strongly indicated that the lawsone reductase activity was distinct from the previously studied membrane-bound AQS reducing system.
Visualization of a lawsone-dependent azo reductase activity in cell extracts by nondenaturing gel electrophoresis.
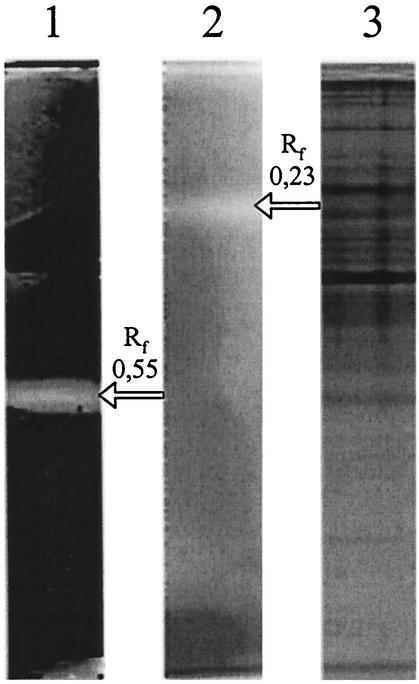
The presence of a distinct cytosolic lawsone-dependent azo reductase activity was demonstrated by activity staining. Cell extracts from E. coli were fractionated in native polyacrylamide gels and subsequently incubated under anaerobic conditions with the azo dye amaranth, NADH, and lawsone; or amaranth, NADH, and AQS. This experiment demonstrated that probably only one major enzyme activity was responsible for the NADH- and lawsone-dependent azo reductase activity and that this activity was different from a second enzyme that was able to catalyze an AQS-dependent decolorization of amaranth (Fig. 1).
FIG. 1.
Detection of different quinone-dependent azo reductase activities in cell extracts of E. coli K-12 by nondenaturing PAGE. E. coli K-12 was grown under aerobic conditions in NB medium, cell extracts were prepared, and 18 μg of protein was transferred to each lane of a nondenaturing polyacrylamide gel (10% polyacrylamide). After the end of the electrophoresis, the respective lanes were separated and two lanes were transferred to an anaerobic chamber. The proteins in the lanes were treated with amaranth and NADH (1 mM each). Then the proteins in lane 1 were additionally treated with a 1 mM lawsone and those in lane 2 with 1 mM AQS. Protein bands with quinone-dependent azo reductase activities were detected by the decolorization of the amaranth stained gels. The different background colors of lanes 1 and 2 were caused by the different colors of the quinones used. In lane 3 all soluble proteins were stained under aerobic conditions with Coomassie blue.
Purification of the lawsone reductase activity.
The lawsone reductase activity was purified by column chromatography under anaerobic conditions using the procedure shown in Table 2. The enzyme was purified about 500-fold, giving a specific activity of 29 U/mg of protein. The overall yield was 32% of the activity present in cell extracts. The pooled fractions of the active protein obtained after the last chromatographic step demonstrated a slight yellow color. Therefore, a sample was concentrated and a UV-visible spectrum determined. This spectrum showed an absorption maximum at a λ of 456 nm, which indicated the presence of a flavin chromophor in the active protein fraction. In order to obtain a completely homogenous preparation, the sample was further purified on a native polyacrylamide gel, the lawsone reductase activity identified by activity staining and finally removed from the gel either by cutting out the relevant part of the PAGE gel (for a subsequent analysis of protein purity by SDS-PAGE) or by Western blotting (for the determination of the amino-terminal amino acid sequence). The SDS-PAGE demonstrated in combination with a silver staining that only a single peptide with a molecular mass of approximately 24 kDa was present in the preparation.
The NH2-terminal amino acid sequence of the purified enzyme was determined by automated Edman degradation as DIISVALKRHSTKAFDA.
Identity of the lawsone reductase activity with the oxygen-insensitive NfsB of E. coli.
The amino acid sequence obtained was used to screen the NCBI database and it was found that only a single protein with the same amino acid sequence is present in the complete genome of E. coli. This protein has previously been described as oxygen-insensitive nitroreductase B (NfsB) (GenBank accession number P38489). It had been previously shown that this enzyme is able to reduce a variety of nitroaromatics by the simultaneous transfer of two electrons from NAD(P)H via a flavin cofactor to the substrates. Furthermore, it was known that the enzyme also reduced the quinones menaquinone and 1,4-benzoquinone, but lawsone had not been previously described as substrate of the enzyme (29, 31, 49).
The identity of the lawsone reductase activity with NfsB was also suggested by the observed similarities in the subunit size and the presence of a flavin cofactor in both proteins. In order to obtain some more biochemical evidence for the identity of the lawsone reductase activity with NfsB, the conversion of different known substrates of NfsB was tested with the purified lawsone reductase activity (Table 3). These experiments demonstrated that the substrate specificity of the lawsone reductase was indeed very similar to the substrate specificity of NfsB previously described by Zenno et al. (49). Furthermore, the lawsone reductase activity was also inhibited by dicoumarol and could also use NADPH as source of reduction equivalents (Table 4) as previously demonstrated by Zenno and coworkers for NfsB. It was therefore concluded that the lawsone reductase activity was indeed identical with NfsB.
TABLE 3.
Activity of the purified lawsone reductase with different substrates and comparison of the results with values described earlier for NfsBe
| Substrated | λ (nm) | ɛ (mM−1 cm−1)f | Activity (U mg of protein−1) of:
|
|
|---|---|---|---|---|
| Lawsone reductasea | Nitroreductase Bb | |||
| Flavin mononucleotide | 450 | 13 | 0.26 | 1 |
| Flavin adenine dinucleotide | 450 | 9.2 | 0.32 | 1 |
| Riboflavin | 450 | 10.7 | 0.27 | 2 |
| 1,4-Benzoquinonec | 340 | 6.72g | 41.7 | 251 |
| Menaquinone | 358 | 6.7g | 16.7 | 60 |
| Nitrofurazone | 400 | 13 | 20.6 | 13 |
| Nitrofurantoin | 400 | 14.2 | 25.3 | 21 |
| 4-Nitrobenzoate | 358 | 4.7g | 0.10 | 1 |
| 4-Nitrotoluene | 358 | 4.7g | 0.04 | 0.4 |
| 4-Nitrophenol | 400 | 14 | 0.75 | 0.1 |
| 4-Nitroaniline | 400 | 14 | 0.02 | 0.1 |
| K3[Fe(CN)6] | 420 | 1 | 360 | 387 |
| 2,6-Dichloroindophenol | 605 | 20 | 0.36 | 2 |
| Methylenebluec | 605 | 30 | 0 | 2 |
Experimental details are given in Materials and Methods.
Nitroreductase B activities given by Zenno et al. (49). The authors determined the activity under aerobic conditions in a spectrophotometrical cuvette test at 23°C in Tris-HCl buffer (pH 7) using a 100 μM concentration of each of the respective substrates.
1,4-Benzoquinone and methyleneblue were reduced under the anaerobic test conditions spontaneously by NADH. The activities indicated were corrected for these spontaneous reactions.
The activities for 1,4-benzoquinone, menaquinone, and the nitrocompounds were measured with 5% methanol in the test system to ensure proper dissolution of the substrates.
The experiment was performed twice and the mean values are given.
The reaction rates were determined at the indicated wavelength.
Decrease of NADH was measured at the indicated wavelengths.
TABLE 4.
Activity of purified lawsone reductase activity with nitrofurazone and effect of known NfsB inhibitors on enzyme activitya
| Inhibitor | Reductase activity (U mg of protein−1) with:
|
||
|---|---|---|---|
| Nitrofurazone (+ NADH) | Lawsone
|
||
| + NADH | + NADPH | ||
| None | 20.6 | 33.7 | 17 |
| Diphenyliodonium (50 μM) | 8.6 | 6.3 | 4.7 |
| Dicoumarol (50 μM) | 0.1 | 0.1 | 0.1 |
The reactions were performed under anaerobic conditions in a microtiter plate. The reaction mixtures contained in a total volume of 200 μl 50 mM Tris-HCl-buffer (pH 7.7), lawsone, or nitrofurazone (100 μM each) and the purified enzyme (0.5 mg of protein per ml). The reactions were started by the addition of NADH or NADPH (100 μM each), and the decrease in the absorbance was measured at 400 or 450 nm in a microtiter plate photometer.
Quinone reductase activity of NfsB.
Zenno and coworkers only determined the reduction of two quinones (menaquinone and 1,4-benzoquinone) by NfsB. Our results demonstrated that NfsB was also able to reduce lawsone and was therefore responsible for the lawsone-dependent azo reductase activity of E. coli. It was previously shown that aside of lawsone also other quinones with standard redox potentials (E0′) < −50 mV could function as redox mediators in the reduction of azo dyes (35). Therefore, a series of different quinones was tested as potential substrates for NfsB. It was shown that NfsB also reduced some other quinones but that quinones with a more negative redox potential than lawsone were not converted or were only converted with very low reaction rates (Table 5).
TABLE 5.
Activity of purified lawsone reductase (NfsB) with different quinonesa
| Substrate | λ (nm) | ɛ (mM−1 cm−1) | Redox potential E0′ (mV) | Activity (U mg of protein−1) |
|---|---|---|---|---|
| 1,4-Benzoquinone | 340 | 6.72 | 270 | 41.7 |
| 2,3-Dimethoxy-5-methyl-1,4-benzoquinone | 407 | 0.72 | 174 | 47.3 |
| 1,4-Naphthoquinone | 358 | 6.8 | 40 | 35.5 |
| Menaquinone | 358 | 6.7 | −10 | 16.7 |
| Plumbagine | 407 | 3.4 | −29 | 1.3 |
| Lawsone | 450 | 2.8 | −137 | 33.7 |
| Lapachol | 450 | 2.0 | −156 | 0.2 |
| Anthraquinone-2,6-disulfonate | 358 | 3.9 | −184 | 0.4 |
| Anthraquinone-2-sulfonate | 358 | 4.0 | −225 | 0.9 |
The reaction rates were determined at the indicated wavelength under the same conditions as described in Table 3.
NfsA as lawsone reductase.
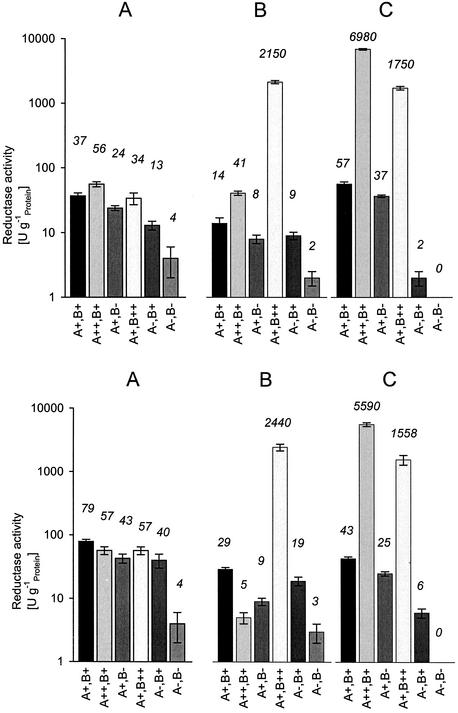
NfsB has been described as “minor” oxygen-insensitive nitroreductase in E. coli, which is synthesized only in a small proportion in relationship to the “major” oxygen-insensitive nitroreductase (NfsA) which requires NADPH and shows only very low activities with NADH (9, 20, 47). NfsA reduces a similar substrate range as NfsB, but shows only a very low degree of sequence identity with NfsB (7%) (20). In order to test if also NfsA was able to act as lawsone reductase, recombinant E. coli strains overexpressing NfsA or NfsB were compared with the wild-type organism and mutants defective in one or both nitroreductases for their nitro- and lawsone reductase activities (Fig. 2). The control experiments with nitrofurazone as substrate clearly demonstrated that cell extracts with high activities of NfsB reduced the model nitro compound nitrofurazone in the presence of NADH with high specific activities (Fig. 2, upper part). If NADPH was used as cofactor, cell extracts from NfsA or NfsB overexpressing strains showed an increased activity. These results were in accordance with the known cosubstrate requirements of NfsA (almost exclusively NADPH) and NfsB (NADH and NADPH) (20, 47, 49). When nitrofurazone was replaced by lawsone as substrate to be reduced (Fig. 2, lower part), basically the same results were obtained, which gave strong evidence that NfsA and NfsB could function (at least in vitro) as lawsone reductases. In an additional experiment (data not shown) it was demonstrated by nondenaturing PAGE and subsequent activity staining with amaranth, lawsone, and NADH or NADPH that after the addition of NADPH indeed two different lawsone-dependent azo reductase activities were present in the cell extracts which were clearly correlated with the levels of expression of the oxygen-insensitive nitroreductases.
FIG. 2.
Nitroreductase (upper part) and lawsone reductase (lower part) activities of whole cells (A) and cell extracts (B and C) of various E. coli (mutant) strains which differed in their activities of the oxygen-insensitive nitroreductases NfsA and NfsB. The following E. coli strains were used (the relevant nitroreductase activities are indicated in brackets): JM83 [NfsA+ NfsB+]; JM83(pAJ13NH) [NfsA++ NfsB+]; SIL41 [NfsA+ NfsB−]; JM83(pNR3) [NfsA+ NfsB++]; JVQ1 [NfsA− NfsB+]; JVQ2 [NfsA− NfsB−]. The strains were grown under aerobic conditions (with ampicillin if appropriate) until they reached the late exponential growth phase. The cells were harvested by centrifugation, washed, and resuspended in Na/K-phosphate buffer (50 mM, pH 7.7). One part of these cell suspensions was used directly for anaerobic resting cell experiments in microtiter plates (A). The wells of the microtiter plates contained in 200 μl Na/K-phosphate buffer (50 mM, pH 7.7), glucose (10 mM), and nitrofurazone (0.1 mM) (upper part of the figure) or lawsone (lower part of the figure) and cells (≈0.3 to 0.6 g of protein liter−1). The respective second part of the cell suspensions were broken by using a French press and cell extracts prepared by ultracentrifugation. The cytosolic nitroreductase activities were determined under anaerobic conditions in microtiter plates. The reaction mixtures contained in 200 μl Na/K-phosphate buffer (50 mM, pH 7.7), and nitrofurazone (0.1 mM) (upper part of the figure) or lawsone (0.1 mM) (lower part of the figure) and 3 to 200 mg l−1 protein. The reactions were started by the addition of 0.1 mM NADH (part B) or NADPH (part C). The decrease in absorbance due to nitrofurazone (upper part) or lawsone (lower part) were determined spectrophotometrically in the microtiter plate reader at 400 or 450 nm, respectively. The protein concentrations in all cell suspensions and cell extracts were determined using the Biuret reagent. The experiment was performed three times, and the relevant standard deviations (error bars) are indicated.
The whole-cell experiments with the wild-type strain and the mutant strain devoid of NfsA and NfsB (Fig. 2A) showed that the mutant strain possessed only about 5% of the lawsone reductase activity observed with the wild-type strain. This clearly demonstrated that also in vivo NfsA and NfsB were the main enzymes which catalyzed the reduction of lawsone.
The comparison of the reduction rates observed with whole cells of the wild type strain (NfsA+ NfsB+) and those strains which either overexpressed NfsA or NfsB demonstrated no significant differences in the reduction rates for nitrofurazone or lawsone. This suggested that in the mutant strains which demonstrated in vitro significantly increased enzyme activities, additional factors limit the reactions [e.g., membrane permeability or the supply of NAD(P)H].
DISCUSSION
It was recently suggested that quinoide redox mediators with standard redox potentials (E0′) between approximately −320 mV and −50 mV could in general function as effective redox mediators in the microbial reduction of sulfonated azo dyes. These limits are set on the one hand by the redox potentials of the cellular redox cofactors NAD(P)H and on the other hand by the redox potentials of the azo compounds, which approximately vary between −180 mV and −430 mV (11, 35). It is expected that for the functioning of a quinone as an effective mediator the proper redox potential is not sufficient, but that also the transport of the reduction equivalents from the cellular system to the azo compounds is important, because most relevant azo compounds are either too polar (e.g., polysulfonated azo dyes) and/or too large (e.g., polymeric azo compounds) to passively diffuse through the cell membranes. We have previously suggested for the system consisting of S. xenophaga BN6, AQS, and azo dyes that this problem can be circumvented by the reduction of the redox mediator AQS by a membrane bound part of the respiratory chain. It was suggested that in this system AQS has not to enter the cells and is reduced at the outward-facing part of the cell membrane (23).
Lawsone was originally detected in a comparative survey using different bacterial strains and quinones as especially effective redox mediator and it was found that whole cells of E. coli demonstrated in the presence of lawsone much higher azo reductase activities than in the presence of AQS. In contrast, for many other bacterial strains (e.g., S. xenophaga BN6) AQS was much more effective than lawsone (35). These differences in the specific activities with different quinones gave the first indications that the lawsone reductase was different from the previously described AQS-reducing activity. We could only find in the scientific literature one reference which described the metabolism of lawsone by E. coli (which involved an l-aspartate oxidase [40]), and there is even one publication that states that whole cells of E. coli do not reduce lawsone (25). In contrast, there are several reports which describe the reduction of other quinones either by the membrane-bound respiratory chain or some uncharacterized cytosolic activities (14, 15, 41, 42).
It was a rather surprising observation that the lawsone reducing activity observed with whole cells of E. coli was caused by a soluble cytoplasmic enzyme, but the experimental evidence for this assumption is rather strong: First, it could be demonstrated in the cell-fractioning experiments that the lawsone reductase activity was almost exclusively present in the cytoplasmic fraction and that only about 5% of the specific activity found in the cytoplasmic fraction was present in the membrane fraction. Furthermore, it could be demonstrated that the specific activity recovered in the cell extracts was about the same as demonstrated by the whole cells. The second strong indication that indeed the cytoplasmic oxygen-insensitive nitroreductases are responsible for the lawsone reductase activity were obtained by the investigations with the different mutant strains. These experiments clearly demonstrated that the mutant which was defective in NfsA and NfsB only showed about 5% of the lawsone reductase activity observed with the wild-type strain.
The so-called oxygen-insensitive nitroreductases have been found and studied in various enterobacteria (e.g., E. coli, Enterobacter cloacae, or Salmonella enterica serovar Typhimurium) but also in other bacterial strains, such as Bacillus subtilis or Helicobacter pylori (9, 16, 21, 44, 50). They have been described as oxygen-insensitive because they are able to reduce nitro compounds in the presence of oxygen and are therefore distinct from a second group of nitroreductases (the oxygen-sensitive nitroreductases) which are inhibited in the presence of oxygen (32). The oxygen-insensitive nitroreductases usually contain a flavin cofactor and transfer two-electrons in the course of the reaction to the nitro groups. These enzymes have originally been studied because they are responsible for the reductive activation of certain nitro groups containing antibiotics (e.g., nitrofurazone). Therefore, mutants in this enzyme activity are resistant against the lethal and mutagenic effects of nitrofurans (7, 8). Because the nitroreductases perform a rather specific activation of nitro groups containing prodrugs to toxic compounds, there is also considerable interest in coupling them with antibodies in order to obtain a site-specific delivery to tumor tissues (antibody directed enzyme prodrug therapy, ADEPT) (1, 19). More recently, it has been found that oxygen-insensitive nitroreductases can also be used for the degradation of environmental pollutants such as 2,4,6-trinitrotoluene or hexahydo-1,3,5-trinitro-1,3,5-triazine (13, 18).
In the present study, conclusive evidence was obtained that indeed the intracellular reduction of lawsone by nitroreductases is responsible for the lawsone-dependent reduction of azo compounds. In order for the reaction to proceed the reactants have to come in close contact to each other. We have recently demonstrated for two recombinant proteins (a flavin reductase and a true azo reductase) which show either anaerobically or aerobically azo reductase activities that the cell membranes of E. coli severely restrict the uptake of sulfonated azo dyes into the bacterial cells (4, 37). It is therefore much more probable that lawsone and its hydroquinone 1,2,4-trihydroxynaphthalene diffuse through the bacterial membranes than the sulfonated azo dyes. Thus, these mediator compounds enable the transfer of reducing power from the cytoplasm to the outside of the cells. A facilitated diffusion of lawsone compared to amaranth through the bacterial membranes is also suggested by the octanol buffer partition coefficients which have been experimentally determined at the relevant pH values used in this study as logP < −3 for amaranth and logP ≈ −1.05 for lawsone (data not shown). Thus, it appears that lawsone is not only because of its redox potential but also because of its lipophilicity a suitable redox mediator for the transfer of intracellular reduction power to extracellular substrates. A further reason for the extraordinary applicability of lawsone compared to other quinones is also suggested by a very recent study which was performed with the nitroreductase from E. cloacae (30). This study suggested that lawsone and other 2-hydroxy-1,4-naphthoquinones were reduced by this enzyme with considerable higher reaction rates than expected from their reduction potentials. Therefore, it was suggested that these quinones demonstrated an enhanced reactivity because they were able to bind at or near to the NADH binding site in the nitroreductase whereas other quinones used an alternative binding site (30).
In the present work we could demonstrate that the oxygen-insensitive nitroreductases from E. coli are responsible for the ability of E. coli to reduce lawsone and thus to reduce azo dyes under anaerobic conditions in the presence of lawsone. It is expected that the described or similar systems are not specific for E. coli but are also functioning in other bacterial strains, because nitroreductases homologous to NfsA and NfsB are found in many enterobacteria. Furthermore, NfsA and NfsB are members of (different) enzyme families which are present in various enterobacteria but also in other bacterial groups. Thus, NfsA shows significant sequence homologies to a certain group of flavin reductases (FRP) (47) and NfsB to another group of bacterial flavin reductases (FRaseI) (22, 48), which are, e.g., involved in the luminescence reaction of marine photobacteria. This suggests that enzymes similar to NfsA and NfsB with the ability to reduce lawsone may be present in many other organisms and thus may be able to function under anaerobic conditions under the appropriate conditions as lawsone dependent azo reductases.
Recently, several examples have been described which demonstrate the utilization of quinones for the transfer of reducing power from bacterial cells to various natural or xenobiotic compounds. Thus, quinones and quinoide compounds have been described which stimulate the reductive biotransformation of various compounds such as ferric iron, nitroaromatic compounds or polyhalogenated pollutants (6, 12, 26, 38). It is therefore very probable that enzyme systems similar to those studied in the present manuscript will also be involved in the reductive degradation of other environmental pollutants.
REFERENCES
- 1.Anlezark, G. M., R. G. Melton, R. F. Sherwood, B. Coles, F. Friedlos, and R. J. Knox. 1992. The bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954). I. Purification and properties of a nitroreductase enzyme from Escherichia coli: a potential enzyme for antibody-directed enzyme prodrug therapy (ADEPT). Biochem. Pharmacol. 44:2289-2295. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. D. Seidman, J. A. Smith, and K. Struhl (ed.). 1996. Current protocols in molecular biology, vol 3. John Wiley & Sons, New York, N.Y.
- 3.Blum, H., H. Beier, and H. I. Gross. 1987. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 8:93-99. [Google Scholar]
- 4.Blümel, S., H.-J. Knackmuss, and A. Stolz. 2002. Molecular cloning and characterization of the gene coding for the aerobic azoreductase from Xenophilus azovorans KF46F. Appl. Environ. Microbiol. 68:3948-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, P. M., F. H. Chapelle, and D. R. Lovley. 1998. Humic acids as electron acceptors for anaerobic microbial oxidation of vinyl chloride and dichloroethene. Appl. Environ. Microbiol. 64:3102-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant, C., and M. DeLuca. 1991. Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J. Biol. Chem. 266:4119-4125. [PubMed] [Google Scholar]
- 8.Bryant, C., L. Hubbart, and W. D. McElroy. 1991. Cloning, nucleotide sequence and expression of the nitroreductase gene from Enterobacter cloacae. J. Biol. Chem. 266:4126-4130. [PubMed] [Google Scholar]
- 9.Bryant, D. W., D. R. McCalla, M. Leeksma, and P. Laneuville. 1981. Type I nitroreductases of Escherichia coli. Can. J. Microbiol. 27:81-86. [DOI] [PubMed] [Google Scholar]
- 10.Chung, K.-T., S. E. Stevens, Jr., and C. E. Cerniglia. 1992. The reduction of azo dyes by the intestinal microflora. Crit. Rev. Microbiol. 18:175-190. [DOI] [PubMed] [Google Scholar]
- 11.Dubin, P., and K. L. Wright. 1975. Reduction of azo food dyes in cultures of Proteus vulgaris. Xenobiotica 5:563-571. [DOI] [PubMed] [Google Scholar]
- 12.Field, J. A., F. J. Cervantes, F. P. van der Zee, and G. Lettinga. 2000. Role of quinones in the biodegradation of priority pollutants: a review. Water Sci. Technol. 42:215-222. [Google Scholar]
- 13.Hannink, N., S. J. Rosser, C. E. French, A. Basran, J. A. H. Murray, S. Nicklin, and N. C. Bruce. 2001. Phytodetoxification of TNT by transgenic plants expressing a bacterial nitroreductase. Nat. Biotechnol. 19:1168-1172. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, M., K. Hasegawa, Y. Oguni, and T. Unemoto. 1990. Characterization of FMN-dependent NADH-quinone reductase induced by menadione in Escherichia coli. Biochim. Biophys. Acta 1035:230-236. [DOI] [PubMed] [Google Scholar]
- 15.Imlay, J., and I. Fridovich. 1992. Exogenous quinones directly inhibit the respiratory NADH dehydrogenase in Escherichia coli. Arch. Biochem. Biophys. 296:337-346. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen, M. A., M. A. Trend, S. L. Hazell, and G. L. Mendz. 2001. Potential involvement of several nitroreductases in metronidazole resistance in Helicobacter pylori. Arch. Biochem. Biophys. 392:180-191. [DOI] [PubMed] [Google Scholar]
- 17.Keck, A., J. Klein, M. Kudlich, A. Stolz, H.-J. Knackmuss, and R. Mattes. 1997. Reduction of azo dyes by mediators originating in the naphthalenesulfonic acid degradation pathway of Sphingomonas sp. BN6. Appl. Environ. Microbiol. 63:3684-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitts, C. L., C. E. Green, R. A. Otley, M. A. Alvarez, and P. J. Unkefer. 2000. Type I nitroreductases in soil enterobacteria reduce TNT (2,4,6-trinitrotoluene) and RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). Can. J. Microbiol. 46:278-282. [DOI] [PubMed] [Google Scholar]
- 19.Knox, R. J., F. Friedlos, M. Jarman, L. C. Davies, P. Goddard, G. M. Anlezark, R. G. Melton, and R. F. Sherwood. 1995. Virtual cofactors for an Escherichia coli nitroreductase enzyme: relevance to reductively activated prodrugs in antibody directed prodrug therapy (ADEPT). Biochem. Pharmacol. 49:1641-1647. [DOI] [PubMed] [Google Scholar]
- 20.Kobori, T., H. Sasaki, W. C. Lee, S. Zenno, K. Saigo, M. E. P. Murphy, and M. Tanokura. 2001. Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds. J. Biol. Chem. 276:2816-2823. [DOI] [PubMed] [Google Scholar]
- 21.Koder, R. L., and A.-F. Miller. 1998. Steady-state kinetic mechanism, stereospecifity, substrate and inhibitor specifity of Enterobacter cloacae nitroreductase. Biochim. Biophys. Acta 1387:395-405. [DOI] [PubMed] [Google Scholar]
- 22.Koike, H., H. Sasaki, T. Kobori, S. Zenno, K. Saigo, M. E. P. Murphy, E. T. Adman, and M. Tanokura. 1998. 1.8 Å Crystal structure of the major NAD(P)H:FMN oxidoreductase of a bioluminiscent bacterium, Vibrio fischeri: overall structure, cofactor-analog binding, and comparison with related flavoproteins. J. Mol. Biol. 280:259-273. [DOI] [PubMed] [Google Scholar]
- 23.Kudlich, M., A. Keck, J. Klein, and A. Stolz. 1997. Localization of the enzyme system involved in the anaerobic reduction of azo dyes by Sphingomonas sp. BN6 and effect of artificial redox mediators on the rate of azo dye reduction. Appl. Environ. Microbiol. 63:3691-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Learoyd, S. A., R. G. Kroll, and C. F. Thurston. 1992. An investigation of dye reduction by food-borne bacteria. J. Appl. Bacteriol. 72:479-485. [DOI] [PubMed] [Google Scholar]
- 26.Lovley, D. R., J. D. Coates, E. L. Blunt-Harris, E. J. P. Phillips, and J. C. Woodward. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445-448. [Google Scholar]
- 27.Matsushita, K., T. Ohnishi, and H. R. Kaback. 1987. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26:7732-7737. [DOI] [PubMed] [Google Scholar]
- 28.Messing, J., and J. Vieira. 1982. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene 19:269-276. [DOI] [PubMed] [Google Scholar]
- 29.Michael, N. P., J. K. Brehm, G. M. Anlezark, and N. P. Minton. 1994. Physical characterization of the Escherichia coli B gene encoding nitroreductase and its over-expression in Escherichia coli K12. FEMS Microbiol. Lett. 124:195-202. [DOI] [PubMed] [Google Scholar]
- 30.Nivinskas, H., S. Staskeviciene, J. Sarlauskas, R. L. Koder, A.-F. Miller, and N. Cenas. 2002. Two-electron reduction of quinones by Enterobacter cloacae NAD(P)H-nitroreductase: quantitative structure-activity relationships. Arch. Biochem. Biophys. 403:249-258. [DOI] [PubMed] [Google Scholar]
- 31.Parkinson, G. N., J. V. Skelly, and S. Neidle. 2000. Crystal structure of FMN-dependent nitroreductase from Escherichia coli B: a prodrug-activating enzyme. J. Med. Chem. 43:3624-3631. [DOI] [PubMed] [Google Scholar]
- 32.Peterson, F. J., R. P. Mason, J. Hovsepian, and J. L. Holtzman. 1979. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J. Biol. Chem. 254:4009-4014. [PubMed] [Google Scholar]
- 33.Rafii, F., and C. E. Cerniglia. 1990. An anaerobic nondenaturing gel assay for the detection of azoreductase from anaerobic bacteria. J. Microbiol. Methods 12:139-148. [Google Scholar]
- 34.Rafii, F., W. Franklin, and C. E. Cerniglia. 1990. Azoreductase activity of anaerobic bacteria isolated from the human intestinal microflora. Appl. Environ. Microbiol. 56:2146-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rau, J., H.-J. Knackmuss, and A. Stolz. 2002. Effects of different quinoide redox mediators on the anaerobic reduction of azo dyes by bacteria. Environ. Sci. Technol. 36:1497-1504. [DOI] [PubMed] [Google Scholar]
- 36.Rau, J., B. Maris, R. Kinget, C. Samyn, G. van den Mooter, and A. Stolz. 2002. Enhanced anaerobic degradation of polymeric azo compounds by Escherichia coli in the presence of low-molecular-weight redox mediators. J. Pharm. Pharmacol. 54:1471-1479. [DOI] [PubMed] [Google Scholar]
- 37.Russ, R., J. Rau, and A. Stolz. 2000. The function of cytoplasmatic flavin reductases in the bacterial reduction of azo dyes. Appl. Environ. Microbiol. 66:1429-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarzenbach, R. P., R. Stierli, K. Lanz, and J. Zeyer. 1990. Quinone and iron porphyrine mediated reduction of nitroaromatic compounds in homogeneous aqueous solution. Environ. Sci. Technol. 24:1566-1574. [Google Scholar]
- 39.Stolz, A. 2001. Basic and applied aspects in the microbial degradation of azo dyes. Appl. Microbiol. Biotechnol. 56:69-80. [DOI] [PubMed] [Google Scholar]
- 40.Tedeschi, G., L. Zetta, A. Negri, M. Mortarino, F. Ceciliani, and S. Ronchi. 1997. Redox potentials and quinone reductase activity of L-aspartate oxidase from Escherichia coli. Biochemistry 36:16221-16230. [DOI] [PubMed] [Google Scholar]
- 41.Thorn, J. M., J. Barton, N. E. Dixon, D. L. Ollis, and K. J. Edwards. 1995. Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH. J. Mol. Biol. 249:785-799. [DOI] [PubMed] [Google Scholar]
- 42.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 43.Walker, R. 1970. The metabolism of azo compounds: a review of the literature. Food. Cosmet. Toxicol. 8:659-676. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe, M., T. Nishino, K. Takio, T. Sofuni, and T. Nohmi. 1998. Purification and characterization of wild-type and mutant “classical” nitroreductases from Salmonella typhimurium. J. Biol. Chem. 273:23922-23928. [DOI] [PubMed] [Google Scholar]
- 45.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determination by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 224:4406-4412. [PubMed] [Google Scholar]
- 46.Whiteway, J., P. Koziarz, J. Veall, N. Sandhu, P. Kumar, B. Hoecher, and I. B. Lambert. 1998. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180:5529-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zenno, S., H. Koike, A. K. Kumar, R. Jayaraman, M. Tanokura, and K. Saigo. 1996. Biochemical characterization of NfsA, a Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi, flavin oxidoreductase. J. Bacteriol. 178:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zenno, S., H. Koike, M. Tanokura, and K. Saigo. 1996. Conversion of NfsB, a minor Escherichia coli nitroreductase, to a flavin reductase similar in biochemical properties to FRaseI, the major flavin reductase in Vibrio fischeri, by a single amino acid substitution. J. Bacteriol. 178:4731-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zenno, S., H. Koike, M. Tanokura, and K. Saigo. 1996. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRaseI, the major flavin reductase in Vibrio fischeri. J. Biochem. 120:736-744. [DOI] [PubMed] [Google Scholar]
- 50.Zenno, S., T. Kobori, M. Tanokura, and K. Saigo. 1998. Purification and characterization of NfrA1, a Bacillus subtilis nitro/flavin reductase capable interacting with the bacterial luciferase. Biosci. Biotechnol. Biochem. 62:1978-1987. [DOI] [PubMed] [Google Scholar]
- 51.Zollinger, H. 1991. Color chemistry, 2nd ed. Verlag Chemie, Weinheim, Germany.