Abstract
A so-called R-gene renders the yellow-striped flea beetle Phyllotreta nemorum L. (Coleoptera: Chrysomelidae: Alticinae) resistant to the defenses of the yellow rocket Barbarea vulgaris R.Br. (Brassicacea) and enables it to use it as a host plant in Denmark. In this study, genetic markers for an autosomal R-gene, inherited as a single, dominant locus in flea beetles from the Danish locality “Kværkeby” are described, and a genetic linkage map around this particular R-gene is constructed, using the technique of AFLP (Amplified Fragment Length Polymorphism).
Keywords: resistance allele, genetic linkage map, genetic marker
Introduction
A key goal in evolutionary biology is to understand the process of adaptation in populations encountering (sudden) adverse conditions or progressive environmental change. Much information has arisen from studies on resistance to toxic compounds (e.g. organophosphate insecticides or plant secondary compounds) in various commercially interesting insects (i.e. pests) such as the sheep blowfly Lucilia cuprina (Clarke and McKenzie, 1987; McKenzie and Clarke, 1988; Freebairn et al., 1996; Clarke et al., 2000), the mosquito Culex pipiens (Guillemaud et al., 1998; Raymond et al., 2001), and Phyllotreta flea beetles (Nielsen, 1977, 1997a; Palaniswamy and Lamb, 1998; Palaniswamy et al., 1998; De Jong and Nielsen, 1999).
In the coevolution of plants and herbivores, plants have evolved the ability to produce (sometimes toxic) secondary plant compounds to counteract herbivory (Thompson, 1994; Pilson, 2000). Plants can respond to herbivory either by defense (a decrease in susceptibility to herbivore damage) or tolerance (a decrease in the per unit effect of herbivory on plant fitness). Plant defense mechanisms include, amongst others, feeding barriers (e.g. wax-layers on leaves) and the production of secondary compounds that act as feeding deterrents or as toxins. It is not uncommon that plants use various strategies simultaneously to minimize herbivory (Nielsen, 1977; Palaniswamy et al., 1998; Pilson, 2000; Shinoda et al., 2002; Agerbirk et al., 2003a). If herbivores feeding on these plants have no alternative host they need to adapt to these defenses. Their counter defenses represent natural adaptations that have evolved over a long time, in contrast, for example, to the resistance to pesticides that represents accelerated evolution (Wilson, 2001)
The mechanisms of detoxification in insects are relatively well studied in relation to pesticides but there are few detailed examinations of the mechanisms of the insect detoxification system of plant defenses in natural situations (Francis et al., 2005). It will be important to determine whether similar mechanisms are involved, and to understand how herbivore resistance will evolve in a population. Model systems at the interface of the biochemistry of detoxification of natural compounds and co-evolution are crucial to addressing these goals. These model systems are very attractive as they provide a wealth of information on evolutionary processes in the field as well. The findings are furthermore of great interest when commercially interesting plant species and their herbivores are involved, like for example flea beetles and crucifers (Palaniswamy and Lamb, 1998; Palaniswamy et al., 1998).
Many phytophagous insects have become specialized in utilizing a rather limited number of plant species. This is considered to reflect evolution of particular adaptations to shared defensive chemistry across the host plants (Nielsen, 1977; Thompson, 1994). The ability to utilize crucifers (Capparales: Brassicaceae) as host plants, for example, has evolved several times within the Chrysomelidae (Coleoptera) (Nielsen, 1988). Many crucifers are crop plants of commercial value. Examples include yellow rocket Barbarea vulgaris, rapeseed Brassica napus, cabbage and cauliflower Brassica oleracea. Flea beetles feeding on these crucifer species are considered pests. The fact that some (wild) crucifers are unsuitable as host plants has attracted considerable interest. The B. vulgaris defense mechanism has proved useful in dead end trap cropping (Shelton and Nault, 2004) and some of the crucifers are regarded as potential sources of defense genes that might be transferred to the species used as crops to create genetically-engineered, herbivory-free cultivars (Palaniswamy and Lamb, 1998; Tattersall et al., 2001). Thus rather than using insecticides, the spread and numbers of the flea beetles can be controlled in a more environmentally friendly way by using a manipulation of plant secondary compounds (Gavoski et al., 2000). However, just as insects can evolve resistance to insecticides (cf. L. cuprina and C. pipiens), they might also become resistant to plant secondary compounds, in a parallel manner to their ability to use novel crucifers. The yellow-striped flea beetle Phyllotreta nemorum in Denmark serves as a good example to explore these processes in a natural insect-plant system.
The model system used in this study is that of the yellow-striped flea beetle, Phyllotreta nemorum L. (Coleoptera: Chrysomelidae: Alticinae), and its atypical host plant in Denmark, the yellow rocket, Barbarea vulgaris. Two chemically and morphologically distinguishable types of Barbarea vulgaris ssp. arcuata have been identified (Nielsen, 1997b), a flea-beetle resistant G-type (Glabrous) and a non-resistant P-type (Pubescent). Several flea beetle populations in Denmark have been found to be polymorphic in their ability to use Barbarea vulgaris ssp. arcuata G-type as a host plant. The so-called resistant beetles can feed throughout the seasons when these plants are available, while the majority of P. nemorum beetles (so-called susceptible beetles) can only do so in autumn when the plants are not resistant (Nielsen, 1997b; De Jong and Nielsen, 1999). Besides the morphological difference between the P- and the G-type (pubescence), there are numerous chemical differences (Agerbirk et al., 2003a). Interestingly, the G-type but not the P-type contains a triterpenoid saponin (Agerbirk et al., 2003b). A similar saponin has been reported to be responsible for the resistance of B. vulgaris to the diamondback moth, Plutella xylostella (Shinoda et al., 2002). So far, the chemical basis of flea beetle resistance in the G-type is unknown, but one or more saponins may potentially be responsible (Shinoda et al., 2002; Agerbirk et al., 2003b).
The ability to survive on G-type plants depends on the presence of at least one dominant allele of a so-called resistant (R) gene. Although several populations in Denmark have been identified with resistant beetles, it appears that each population may have a different R-gene (and sometimes more than one), as they can be present on the X- and Y-chromosome as well as on the autosome(s) (Nielsen, 1997a, 1999; De Jong et al., 2000). Whether these genes in different populations are similar or even identical, is at present unknown. It is not unlikely that different R-genes have a similar effect (Raymond et al., 2001). The main objective of the present study is to find genetic markers for the presence of the dominant resistant allele of the single autosomal gene in the P. nemorum flea beetles from the Danish region of Kværkeby and to construct a genetic linkage map around this particular R-gene. We use the technique of AFLP (amplified fragment length polymorphism) to amplify a random set of restriction fragments. This technique is considered a reliable and easy tool to study genetic variability (Vos et al., 1995).
Genetic mapping and identifying an AFLP marker for the R-gene will help to provide a basis for future genetic research in P. nemorum, including identifying the nature of the R-gene(s), the molecular mechanism of the resistance, and eventually the biochemical pathway involved in detoxifying the saponin. The results will be of great value in studies investigating the commercially important crucifers and the flea beetles feeding on them, especially when one wants to create genetically modified crops resistant to herbivory by flea beetles. Furthermore, molecular markers will allow straightforward genotyping of beetles in ecological and evolutionary experiments without the need of laborious bioassays. The presence of the R-gene has severe negative effects on survival, which is presumably one of the reasons why resistance is not widespread in Denmark (De Jong and Nielsen, 2000).
Materials and Methods
Animals
The AFLP technique was used to construct a genetic linkage map of AFLP markers around the R-gene in P. nemorum flea beetles. This technique amplifies a random set of restriction fragments (Vos et al., 1995). The number of DNA fragments detected depends not only on the primer sets used, but also on the genetic variability of the DNA. Therefore, in order to maximize the possibility of detecting a genetic marker for the R-gene the following crossing schedule was chosen.
Flea beetles used in this study originated from two source populations in Denmark, about 32 km apart, at Taastrup (ST) and Kværkeby (Kv). Details on the origin and maintenance of the ST line can be found in De Jong and Nielsen (2000). The ST beetles do not survive on G-type plants and have the rr genotype (i.e. they are “susceptible”; ‘ST’ = susceptible line from Taastrup). ST beetles used in the present study have been kept under laboratory conditions for more than 15 generations. Kv beetles were collected in a B. vulgaris ssp. arcuata G-type field in Kværkeby (Denmark) in May 2002. Kv beetles have been shown in previous studies and surveys (involving crossing and subsequent genotyping through bioassays) to be homozygous resistant at one, single, major autosomal locus (De Jong et al., 2000). The collected beetles were reared in the laboratory and various crosses and genotyping through bioassays confirmed the results of the previous studies (unpublished data). The Kv beetles, therefore, have the RR, resistant, genotype. They were reared for three generations in the laboratory prior to the present study.
A virgin Kv female was mated with a virgin ST male (= P). The (virgin) male and female offspring (F1) were separated one to three days after emergence, a period during which no matings occur. The F1 females were kept virgin for up to three weeks in a climate cabinet at 24 ± 1° C and a LD 16:8 light-dark cycle in plastic vials containing a moist gypsum-charcoal bottom layer (De Jong et al., 2000). To obtain a segregating population for the R gene, ten randomly chosen virgin F1 females were then each mated individually with a virgin ST male. In this study the results of the single mating that yielded the largest number of viable BC1 (BackCrossed) beetles (> 140) will be presented. The unfed and virgin male and female offspring (BC1) were separated one to three days after emergence. The animals were then fed ad libitum with fresh radish cotyledons for 3 consecutive days. They were then kept for one day without food, after which leaf discs (diameter of 19 mm) of B. vulgaris ssp. arcuata G-type leaves (see below for details on these plants) were presented to each of them individually, to perform a bioassay. After three days the total amount of plant material eaten was determined, from which the presence or absence of the resistance allele could be determined, and thus the genotype of the BC1 beetle.
G-type plants
Clones of B. vulgaris ssp. arcuata G-type were used for bioassays (Breuker et al., unpublished). The plants were grown on a mixture of soil and vermiculite in climate chambers under such conditions (20 ± 2° C, L18:D6 photoperiod, under 400W HPI/T-lamps supplying 160-200 mol quanta m-2 s-1 on the level of the leaf surface) that all plants were fully toxic (as tested in bioassays using ST-larvae). Plants were 4–8 weeks old and still in the vegetative state when used for bioassays (cf. De Jong and Nielsen, 2000). Because cloned G-type plants were used, variation in feeding can be assumed to be due to the presence or absence of R-genes, rather than to any variation in toxicity among individual plants.
DNA extraction and AFLP markers
The DNA was extracted from 117 BC1 and 2 F1 (the parents: male and female) beetles using a commercially available protocol (Dneasy Tissue Kit, formerly known as the QIAamp Tissue Kit, QIAGEN, www.qiagen.com). Frozen beetles (−80° C) were homogenized by individually grinding them in liquid nitrogen with a pestle, before thawing in buffer ATL, and addition of proteinase K. DNA was eluted into 400 µl of buffer AE, and subsequently stored at −20° C.
The eluted DNA was then used to construct samples for AFLP analysis (full details of the protocol can be found at http://bio.leidenuniv.nl/mollab/webAFLPprotocolhtm.html). The protocol consists of the restriction of the DNA, ligation of the adapters and a two-step amplification (preamplification and selective amplification) (Vos et al., 1995).
Restriction and ligation were performed at the same time. The flea beetle DNA (125 ng per sample) was restricted with 0.12 µl MseI (10 U/µl) and 0.0625 µl EcoRI (20 U/µl), ligated with 0.6 Weiss units T4 ligase, 2.5 µl 10 × T4 ligase buffer, 1.7 µl of a mix of EcoRI adapter (5′-CTCGTAGACTGCGTACC-3′ + 3′- CATCTGACGCATGGTTAA-5′) and MseI adaptor (5′- GACGATGAGTCCTGAG-3′ + 3′- TACTCAGGACTCAT-5′) (the adaptor solution contains a 1 to 10 ratio of Eco to Mse adaptors) in a final volume of 20 µl for 2 hours at 37° C. The ligase activity was inactivated by incubating for 10 minutes at 65° C.
The first step in the amplification process is a preamplification, in which only one selective basepair is added, followed by a selective amplification in which a further two base pairs are added. The two-step procedure has less background noise and avoids the occurrence of nonspecific bands (Vos et al., 1995). In the preamplification, 2 µl of the restriction-ligation mix per sample was added to 13 µl of a PCR mix consisting of: 11.25 µl AFLP core mix (Applied Biosystems, www.appliedbiosystems.com), 0.15 µl EcoRI – A primer (20 µM, 3 pmol, 5′- GACTGCGTACCAATTCA-3′), 1.15 µl MseI – C primer (20 µM, 23 pmol, 5′- GATGAGTCCTGAGTAAC-3′), and 0.45 µl ultrapure water. The mixture was heated in a thermocycler at 72° C for 2 minutes, 94° C for 2 minutes, followed by 20 cycles of 5 seconds at 94° C, 30 seconds at 56° C, and 2 minutes at 72° C.
In the selective amplification, per sample, 1 µl of the preamp-mix was diluted 10 times in a selective amplification reaction mix consisting of: 7.50 µl AFLP core mix (Applied Biosystems), 0.50 µl EcoRI + AXX primer (1 µM, 1 pmol, 5′- GACTGCGTACCAATTCAXX-3′), 0.125 µl MseI + CX primer (20 µM, 2.5 pmol, 5′- GATGAGTCCTGAGTAACX-3′), and 0.875 µl ultrapure water. We have used the following MseI + CX primers: × = A, G, C. The EcoRI- AXX primers were fluorescently labeled with either 5-FAM (ACA), 6-FAM (AGC), NED (AAC) or JOE(AGG) . The following 7 combinations were used: 5-FAM-CA, 5-FAM-CC, 5-FAM-CG, 6-FAM-CG, JOE-CA, JOE-CG, and NED-CA. A PCR in a thermocycler was used with 9 cycles of 5 seconds at 94° C, 30 seconds at 65° C, and 2 minutes at 72° C, whereby the annealing temperature was lowered by 1°C each cycle. These 9 cycles were then followed by 25 cycles of 5 seconds at 94° C, 30 seconds at 56° C, and 2 minutes at 72° C, then 35 minutes at 72° C.
To visualize the amplified fragments, the (selectively) amplified products were analyzed by gel electrophoresis. Per sample, a mixture of 1.2 µl AFLP loading buffer (GeneScan 500 Rox) and 0.5 µl of 5-FAM or 6-FAM labeled products, or 0.9 µl of JOE or 0.8 µl of Ned labeled products, was loaded on a 5% denaturing Long Ranger polyacrylamide gel (length 36 cm, 0.20 mm thick), and run on an automated sequencing machine (Applied Biosystems, ABI Prism 377) for 3 hours. Prior to running the electrophoresis, the samples were denatured, for 3 minutes at 94° C. Data were processed and initial analyses carried out in ABI Genescan Analysis 3.1 (Applied Biosystems). The presence or absence of fragments in the range of 80 bp and 500 bp was assessed in Genographer, using the thumbnail option (freeware, http://hordeum.msu.montana.edu/genographer).
Markers were named according to the fluorescent label used on the EcoRI side, the selective base pairs on the MseI side and the size in basepairs of the fragment.
Data analysis
Joinmap® version 3.0 (Van Ooijen and Voorrips, 2001) was used to construct a genetic linkage map of the obtained AFLP markers using the CP procedure (i.e. using a calculation procedure based on a so-called Cross Pollination population structure: a population resulting from a cross between two heterogeneously heterozygous and homozygous diploid parents, linkage phases originally (possibly) unknown). Besides the AFLP markers, an additional “marker” was added to the dataset. Individuals were either resistant or not (1/0). This allowed mapping of AFLP markers around the resistance gene. Markers were grouped using a minimum LOD score of 3 (maximum recombination fraction thresholds at 0.40), and recombination fractions were translated into map distances (cM) with Kosambi's mapping function (see also Pannebakker et al., 1998). Separate maps were generated for markers segregating in the male and female parent.
Results
A total of 84 segregating AFLP markers were found, of which 14 (16.67%) showed distorted segregation. Six of these markers showed highly significant segregation distortion (chi-square testing for deviation from Mendelian segregation ratios (depending on the genotype of the parents for a certain trait either 1:1 or 1:2:1, p < 0.001) (cf. Pannebakker et al., 1998), and were excluded from the analyses. The R gene segregated in the F1 parent.
Segregation in the R gene did not significantly deviate from Mendelian inheritance (resp. µ2= 0.2, Df=1, ns). Of the 78 AFLP markers used in the analyses, 42 segregated in the F1 parent, 26 in the ST parent, and 10 in both parents.
Results from Joinmap allowed the inference from the AFLP data that 11 linkage groups were segregating in the F1 parent, and 4 linkage groups in the ST parent. Linkage group 4 in the ST parent was identical to linkage group 11 in the F1 parent, while linkage group 2 in both parents had markers in common. A maximum of 14 linkage groups was thus found with a minimum of 11 groups. Karyotype analysis shows that P. nemorum has 15 chromosomes, including the sex chromosome (Marek, personal communication).
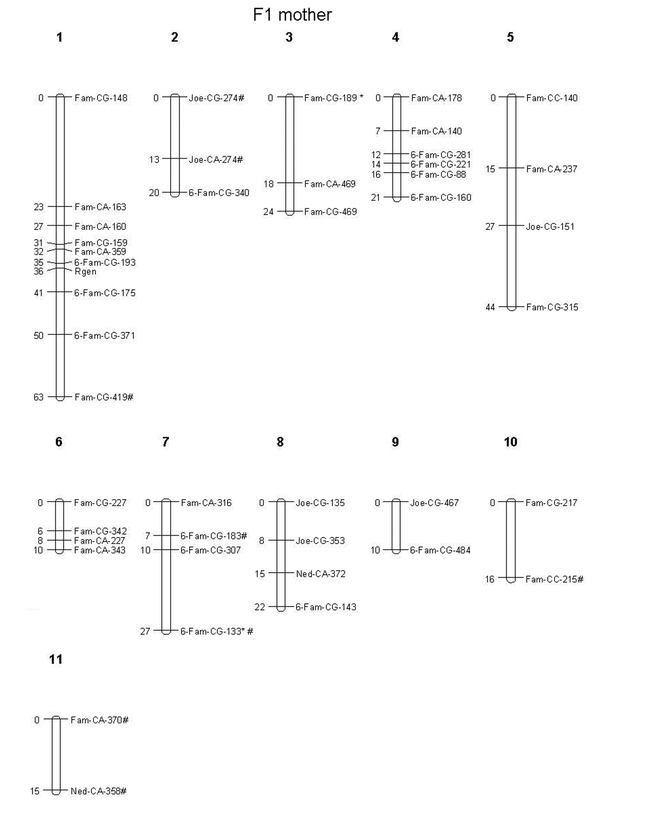
The resistance gene was mapped on the genetic linkage map for the female parent alongside 43 AFLP markers, of which two showed a distorted segregation (and these were not located near the R-gene). A total of 9 AFLP markers remained unlinked. The male map consists of 22 AFLP markers, of which two showed distorted segregation, but were not in the same linkage group as the R-gene. Fourteen markers remained unlinked. The male map covers 163 cM with a mean (± SD) intermarker distance of 9.06 cM (± 6.50). The female map covers 272 cM with a mean (± SD) intermarker distance of 8.24 cM (± 5.82). 4 AFLP markers were found within a 5 cM range from the R-allele (Fig. 1), with 1 AFLP-marker (6-FAM-CG-193) only 1 cM away from the R-allele. The presence of the latter restriction fragment correlates in a highly significant manner with the presence of the R-allele (as do 6-FAM-CG-175 and 5-FAM-CG-159). The presence of 5-FAM-CA-359 indicates the presence of the r-allele. Further investigation (e.g. sequencing) of this region of the genome may reveal DNA sequences of candidate R-gene(s), and eventually elucidate the nature of the R-gene.
Fig. 1A.
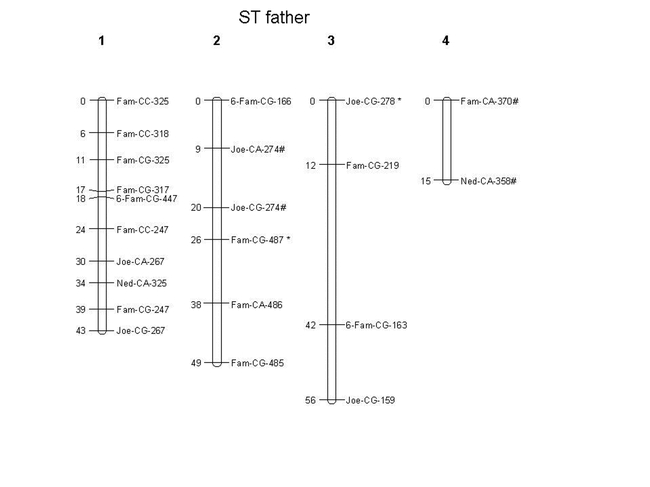
Genetic linkage map created with Joinmap® of the flea beetle Phyllotreta nemorum genome. AFLP markers are named as follows: (Eco-fluorescent label)-(Mse-CXX)-(number of bp). AFLP markers with distorted segregation ratios (chi-square analyses, p < 0.05) are indicated with an asterisk (*), and are ordered at LOD ≥ 3.0. The R-gene is positioned on the map as well. Genetic distances (in cM) are indicated on the left in each linkage group. A) Linkage groups segregating in the ST father, B) linkage groups segregating in the F1 mother. # indicates markers segregating in both parents.
Fig. 1B.
Discussion
The ability to use B. vulgaris ssp. arcuata G-type plants in P. nemorum flea beetles originating from Kværkeby depends on the occurrence of only one dominant allele at a single major autosomal locus, unlike for some other sampled Danish populations where at least two sex-linked genes seem to be involved - on the X- and Y-chromosome - as well as one or more autosomal genes (Nielsen, 1997a; De Jong and Nielsen, 1999; Nielsen, 1999; De Jong et al., 2000; De Jong and Nielsen, 2002). We have concentrated on the Kværkeby R-allele, as the mode of inheritance is straightforward and we have descriptions of the pleiotropic effects of this particular R-allele on life history traits and developmental stability (Breuker et al., unpublished).
Further work using the marker established in this study may identify the Kv R-allele, and hence potentially elucidate the mechanism of B. vulgaris resistance by revealing the biochemical activity associated with the gene. It seems possible that the R-gene is responsible for detoxification of the resistance factor in the plant, which may be one or more saponins, or encodes a variant of a target protein that is insensitive to the resistance factor. Because the nature of the R-allele is unknown at present, it is unclear how fitness traits are affected by the presence of the R-allele and precisely how the high mortality comes about in the early stages of development (Breuker et al., unpublished). The discovery of several AFLP markers associated with the R-gene (three indicating the presence of the R-allele, and one indicating the presence of the r-allele) will: 1) make accurate genotyping of beetles possible, 2) enable positional cloning and thereby 3) offer the possibility of sequencing the part of the genome around the R-gene.
It will also be possible to map and identify candidate genes, which may aid the rapid isolation of R-genes and provide evidence for their involvement in the mechanisms of resistance. An additional advantage of the AFLP markers is that it is possible to develop a specific primer for the presence of the R-gene, and thus avoid laborious bioassays. Furthermore, this primer of the R-gene can also be used in genetic surveys of insects collected from other regions where other P. nemorum populations are able to feed on B. vulgaris ssp. arcuata G-type plants. This will provide more insight into whether the same genes or molecular mechanisms are involved in conferring resistance. These advances should enable fundamental insights about the process of evolution of the ability of an herbivorous insect to feed on a novel host plant.
Acknowledgments
We are very grateful to J.K. Nielsen for useful suggestions at an early stage of the research, and for his hospitality and help in collecting flea beetles in Denmark. We would also like to thank F. Marek for sharing unpublished data with us, B. Slieker and M. van de Kamp for collecting some of the AFLP data, H. Nel and N. Wurzer for their technical support, and M. Gibbs and an anonymous reviewer for useful comments on the manuscript. The work was funded by the Dutch Organization for Scientific Research, on a NWO grant (810.34.007).
References
- Agerbirk N, Orgaard M, Nielsen JK. Glucosinolates, flea beetle resistance, and leaf pubescence as taxonomic characters in the genus Barbarea (Brassicaceae) Phytochemistry. 2003a;63:69–80. doi: 10.1016/s0031-9422(02)00750-1. [DOI] [PubMed] [Google Scholar]
- Agerbirk N, Olsen CE, Bibby BM, Frandsen HO, Brown LD, Nielsen JK, Renwick AA. A saponin correlated with variable resistance of Barbarea vulgaris to the diamondback moth Plutella xylostella. Journal of Chemical Ecology. 2003b;29:1417–1433. doi: 10.1023/a:1024217504445. [DOI] [PubMed] [Google Scholar]
- Clarke GM, McKenzie JA. Developmental stability of insecticide resistant phenotypes in blowfly; a result of canalizing selection. Nature. 1987;325:345–346. [Google Scholar]
- Clarke GM, Yen JL, McKenzie JA. Wings and bristles:character specificity of the asymmetry phenotype in insecticide-resistant strains of Lucilia cuprina. Proceedings of the Royal Society of London Series B Biological Sciences. 2000;267:1815–1818. doi: 10.1098/rspb.2000.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong PW, Nielsen JK. Polymorphism in a flea beetle for the ability to use an atypical host plant. Proceedings of the Royal Society of London, Series B, Biological Sciences. 1999;266:103–111. [Google Scholar]
- De Jong PW, Nielsen JK. Reduction in fitness of flea beetles which are homozygous for an autosomal gene conferring resistance to defenses in Barbarea vulgaris. Heredity. 2000;84:20–28. doi: 10.1046/j.1365-2540.2000.00613.x. [DOI] [PubMed] [Google Scholar]
- De Jong PW, Nielsen JK. The possible role of coadapted gene complexes in host plant use in phytophagous insects. Entomologia Experimentalis et Applicata. 2002;104:207–215. [Google Scholar]
- De Jong PW, Frandsen HO, Rasmussen L, Nielsen JK. Genetics of resistance against defenses of the host plant Barbarea vulgaris in a Danish flea beetle population. Proceedings of the Royal Society of London, Series B, Biological Sciences. 2000;267:1663–1670. doi: 10.1098/rspb.2000.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis F, Vanhaelen N, Haubruge E. Glutathione S-transferases in the adaptation to plant secondary metabolites in the Myzus persicae aphid. Archives of Insect Biochemistry and Physiology. 2005;58:166–174. doi: 10.1002/arch.20049. [DOI] [PubMed] [Google Scholar]
- Freebairn K, Yen JL, McKenzie JA. Environmental and genetic effects on the asymmetry phenotype:Diazinon resistance in the Australian sheep blowfly, Lucilia cuprina. Genetics. 1996;144:229–239. doi: 10.1093/genetics/144.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavoski JE, Ekuere U, Keddie A, Dosdall L, Kott L, Good AG. Identification and evaluation of flea beetle (Phyllotreta cruciferae) resistance within Brassicaceae. Canadian Journal of Plant Science. 2000;80:881–887. [Google Scholar]
- Guillemaud T, Lenormand T, Bourguet D, Chevillon C, Pastuer N, Raymond M. Evolution of resistance in Culex pipiens: allele replacement and changing environment. Evolution. 1998;52:443–453. doi: 10.1111/j.1558-5646.1998.tb01644.x. [DOI] [PubMed] [Google Scholar]
- McKenzie JA, Clarke GM. Diazinon resistance, fluctuating asymmetry and fitness in the Australian sheep blowfly, Lucilia cuprina. Genetics. 1988;120:213–220. doi: 10.1093/genetics/120.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JK. Host plant relationships of Phyllotreta nemorum L. (Coleoptera: Chrysomelidae) Zeitschrift für Angewandte Entomologie. 1977;84:396–407. [Google Scholar]
- Nielsen JK. 1988 Crucifer-feeding Chrysomelidae: mechanisms of host plant finding and acceptance. In: Jolivet P, Petitpierre E, Hsiao TH, editors. Crucifer-feeding Chrysomelidae: mechanisms of host plant finding and acceptance. 25–40.Kluwer Academic Publishers, Dordrecht. [Google Scholar]
- Nielsen JK. Genetics of the ability of Phyllotreta nemorum larvae to survive in an atypical host plant, Barbarea vulgaris ssp. arcuata. Entomologia Experimentalis et Applicata. 1997a;82:37–44. [Google Scholar]
- Nielsen JK. Variation in defenses of the plant Barbarea vulgaris and in counteradaptations by the flea beetle Phyllotreta nemorum. Entomologia Experimentalis et Applicata. 1997b;82:25–35. [Google Scholar]
- Nielsen JK. Specificity of a Y-linked gene in the flea beetle Phyllotreta nemorum for defenses in Barbarea vulgaris. Entomologia Experimentalis et Applicata. 1999;91:359–368. [Google Scholar]
- Palaniswamy P, Lamb RJ. Feeding preferences of a flea beetle, Phyllotreta cruciferae (Coleoptera: Chrysomelidae), among wild crucifers. Canadian Entomologist. 1998;130:241–242. [Google Scholar]
- Palaniswamy P, Lamb RJ, Bodnaryk RP. Resistance to the flea beetle Phyllotreta cruciferae (Coleoptera: Chrysomellidae) in false flax, Camelina sativa (Brassicaceae) Canadian Entomologist. 1998;130:235–240. [Google Scholar]
- Pannebakker BA, Beukeboom LW, Van Alphern JJM, Brakefield PM, Zwaan BJ. The genetic basis of male fertility in relation to haplodiploid reproduction in Leptopilina clavipes. Genetics. 2004;168:341–349. doi: 10.1534/genetics.104.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilson D. The evolution of plant response to herbivory:simultaneously considering resistance and tolerance in Brassica rapa. Evolutionary Ecology. 2000;14:457–489. [Google Scholar]
- Raymond M, Berticat C, Weill M, Pasteur N, Chevillon C. Insecticide resistance in the mosquito Culex pipiens: what have we learned about adaptation? Genetica. 2001;112–113:287–296. [PubMed] [Google Scholar]
- Shelton AM, Nault BA. Dead-end trap cropping:a technique to improve management of the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) Crop Protection. 2004;23:497–503. [Google Scholar]
- Shinoda T, Nagao T, Nakayama H, Serizawa H, Koshioka M, Okabe H, Kawai A. Identification of a triterpenoid saponin from a crucifer, Barbarea vulgaris, as a feeding deterrent to the diamondback moth, Plutella xylostella. Journal of Chemical Ecology. 2002;28:587–599. doi: 10.1023/a:1014500330510. [DOI] [PubMed] [Google Scholar]
- Tattersall DB, Bak S, Jones PR, Olsen CE, Nielsen JK, Hansen ML, Hoj PB, Moller BL. Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science. 2001;293:1826–1828. doi: 10.1126/science.1062249. [DOI] [PubMed] [Google Scholar]
- Thompson JN. 1994 The Coevolutionary process. Chicago & London: The University of Chicago Press. [Google Scholar]
- Van Ooijen JW, Voorrips RE. 2001 Joinmap® 3.0, Software for the calculation of genetic linkage maps. Plant Research International, Wageningen, Netherlands. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP:a new technique for DNA fingerprinting. Nucleic Acid Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TG. Resistance of Drosophila to toxins. Annual Review of Entomology. 2001;46:545–571. doi: 10.1146/annurev.ento.46.1.545. [DOI] [PubMed] [Google Scholar]


