Abstract
Randomly amplified polymorphic DNA (RAPD) PCR was used to analyze the temporal and spatial intraspecific diversity of 208 Vibrio vulnificus strains isolated from Galveston Bay water and oysters at five different sites between June 2000 and June 2001. V. vulnificus was not detected during the winter months (December through February). The densities of V. vulnificus in water and oysters were positively correlated with water temperature. Cluster analysis of RAPD PCR profiles of the 208 V. vulnificus isolates revealed a high level of intraspecific diversity among the strains. No correlation was found between the intraspecific diversity among the isolates and sampling site or source of isolation. After not being detected during the winter months, the genetic diversity of V. vulnificus strains first isolated in March was 0.9167. Beginning in April, a higher level of intraspecific diversity (0.9933) and a major shift in population structure were observed among V. vulnificus isolates. These results suggest that a great genetic diversity of V. vulnificus strains exists in Galveston Bay water and oysters and that the population structure of this species is linked to changes in environmental conditions, especially temperature.
Vibrio vulnificus is a marine and estuarine bacterium commonly found in water and shellfish of the Gulf of Mexico and other temperate environments (11, 16, 32, 35). It is capable of producing septicemia and severe wound infections in susceptible persons following consumption of raw oysters or exposure of open wounds to seawater. Individuals vulnerable to infection include those who have underlying chronic diseases or who are immunocompromised (8, 14).
The occurrence of the organism in Gulf Coast estuarine environments is favored by high water temperatures and relatively low salinities (11, 16, 28, 32). Most shellfish-associated V. vulnificus illnesses occur during the warm months, when V. vulnificus concentrations in Gulf Coast oysters are at their highest (8, 24). However, it has been suggested that human infections are caused by only a few strains among the heterogeneous populations present in the implicated oysters and that the infectious dose should be determined for the specific virulent strains instead of the total numbers of V. vulnificus bacteria in oysters (9).
Raw oysters are most often implicated as the source of V. vulnificus infections in the United States (5, 8, 12, 24). The seasonal ecology of V. vulnificus in Galveston Bay, the primary oyster-producing area in Texas, has been reported (32), but very little is known about the population structure and molecular evolution of this species in its native habitats. A study of intraspecific diversity of V. vulnificus in the natural environment will provide a better understanding of the ecology and even the epidemiology of V. vulnificus as a species of human concern.
Randomly amplified polymorphic DNA (RAPD) PCR, together with other techniques, such as ribotyping, pulsed-field gel electrophoresis, and amplified fragment length polymorphism, has been used for intraspecific differentiation of V. vulnificus (1, 2, 3, 4, 31, 33). The application of this technique in microbiology has been reviewed by Power (21).
Previous reports by Aznar et al. (3) and Arias et al. (2) have shown that RAPD PCR performed with the universal primers M13 and T3 can be used to differentiate V. vulnificus strains and that the correspondence between results obtained by ribotyping and RAPD PCR is better when primer M13 is used to generate RAPD PCR profiles. In this study, RAPD PCR with primer M13 was optimized and used to analyze the temporal and spatial intraspecific diversity of V. vulnificus in Galveston Bay water and oysters.
MATERIALS AND METHODS
Sample collection.
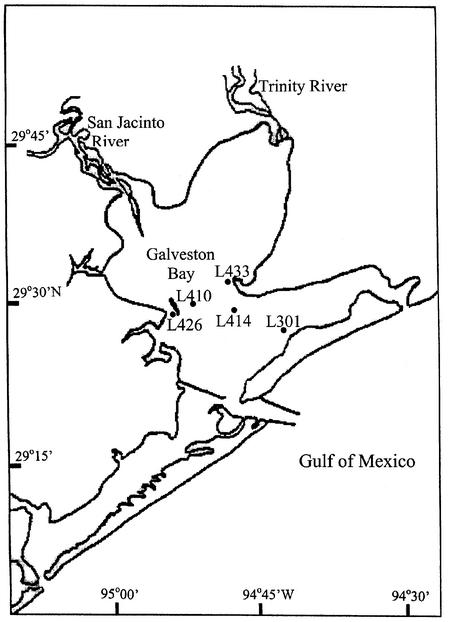
Water and oyster samples were collected at five Galveston Bay sites (private oyster leases) over a 13-month period from June 2000 to June 2001 (Fig. 1). Samples were generally collected biweekly during the spring and summer months and monthly during the autumn and winter months. Water temperature and salinity were determined in situ with a YSI model 30 salinity meter (YSI Inc., Yellow Springs, Ohio). A sterile 125-ml Nalgene bottle attached to an acid-caustic safety sampler (Fox Scientific Inc., Alvarado, Tex.) was used to collect water samples 0.5 m below the surface. Oyster samples, consisting of 15 individual shellstock oysters, were collected by dredging and immediately chilled by placing bagged ice on top of the oysters inside an insulated 54-quart cooler. Water samples were also cooled in the same manner. Bubble wrap was placed between the samples and the ice packs to prevent direct contact. The chilled oyster and water samples were immediately transported to the Seafood Safety Laboratory at Texas A&M University at Galveston for analysis. Upon receipt, the meat temperatures of three oysters from each sampling site were recorded to ensure that the temperature in the container had remained low enough to prevent the replication of V. vulnificus during the transport process. All samples were analyzed for V. vulnificus within 12 h of collection.
FIG. 1.
Sampling sites in Galveston Bay included in this study.
Isolation and enumeration of V. vulnificus.
Oysters were scrubbed, shucked, and diluted in equal amounts of phosphate-buffered saline (PBS), and the oyster meat and liquor were homogenized for 2 min in a Waring blender. The first 10−1 dilution was prepared by weighing 20 g of the homogenate into a sterile dilution bottle containing 80 g of PBS. For water samples, the first 10−1 dilution was prepared by transferring 10 ml of water to a sterile dilution bottle containing 90 ml of PBS. Subsequent serial 10-fold dilutions were prepared in PBS on a volume-per-volume basis. The three-tube most-probable-number (MPN) procedure described in the U.S. Food and Drug Administration Bacteriological Analytical Manual (6) was used for enumeration of V. vulnificus bacteria. This included overnight enrichment in alkaline peptone water, isolation on modified colistin-polymyxin B-cellobiose agar, and confirmation of suspect colonies by enzyme immunoassay (29).
RAPD PCR.
Two or three V. vulnificus strains were randomly selected from each sample that was positive for V. vulnificus, and a total of 208 V. vulnificus isolates were included in this study. Genomic DNA of each isolate was extracted from overnight V. vulnificus cultures grown in heart infusion broth (Difco Laboratories, Detroit, Mich.) with the QIAamp DNA Mini Kit (Qiagen Inc., Valencia, Calif.) in accordance with the manufacture's instructions. DNA concentration and quality were determined by UV light absorption at wavelengths of 260 and 280 nm with an MBA 2000 spectrophotometer (Perkin-Elmer Corp., Norwalk, Conn.). Primer M13 (5′GAAACAGCTATGACCATG3′; Sigma-Genosys, The Woodlands, Tex.) was used in the RAPD PCR assay. Each 50-μl RAPD reaction mixture contained 0.8 to 1.0 μg of genomic DNA, 5.0 μl of GeneAmp 10× PCR buffer II (100 mM Tris-HCl [pH 8.3], 500 mM KCl; Applied Biosystems, Foster City, Calif.), 29.5 μl of diethyl pyrocarbonate-treated H2O, 5 μl of 25 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (Applied Biosystems), 4 μM primer M13 (Sigma-Genosys), 2.5 μl of dimethyl sulfoxide (Stratagene, La Jolla, Calif.), and 5.0 U of AmpliTaq Gold DNA polymerase (Applied Biosystems). Amplifications were performed in a 9700 thermal cycler (Perkin-Elmer). The reaction mixtures were subjected to initial denaturation at 95°C for 10 min, followed by 35 cycles of 94°C for 20 s, 44°C for 30 s, and 72°C for 70 s and a final extension step of 72°C for 10 min. The amplification products were electrophoresed on 12% polyacrylamide gels (ISC BioExpress, Kaysville, Utah) at 105 V in 0.5× Tris-borate-EDTA buffer (Sigma Chemical Co., St. Louis, Mo.). The gel was then stained with SYBR Gold nucleic acid gel stain (Molecular Probes, Inc., Eugene, Oreg.) and photographed under UV light. A 100-bp DNA ladder (New England Biolabs, Inc., Beverly, Mass.) was used as a molecular weight marker. All strains were subjected to RAPD PCR analysis three times, yielding reproducible results.
Computer analysis of RAPD PCR profiles.
RAPD PCR gel photographs were scanned with an HP Scanjet 6300Cxi scanner (Hewlett-Packard, Inc.). The images were calibrated and analyzed with the Rflpscan program included in the Gene Profiler software package (version 4.03; Scanalytics, Inc., Fairfax, Va.). For band matching, the match tolerance was set at 2.0% of the molecular weight of each band. The Treecon program (version 1.3b) included in the Gene Profiler software package (Scanalytics) was used on the Rflpscan output to estimate genetic distances by the method of Link et al. (15) and to create a dissimilarity matrix. Cluster analysis with the unweighted pair group method using arithmetic averages (UPGMA) (25) was performed to infer a dendrogram from the dissimilarity matrix. The gene diversity among each month's isolates was calculated by the method of Nei (17) with Arlequin software (version 2.000; available at http://lgb.unige.ch/arlequin).
Statistical methods.
The impact of water temperature and salinity on the abundances of V. vulnificus in water and oyster samples was evaluated by regression analysis. Differences among the five sampling sites with respect to temperature, salinity, and V. vulnificus densities in water and oyster samples were estimated by analysis of variance. MPN counts were log10 transformed before being subjected to analysis. MPNs that were indeterminate (<3.0) were assigned a value equal to one-half the limit of detection (i.e., 1.5). Regression analysis and analysis of variance were performed with the Statistical Analysis System (SAS Institute Inc., Cary, N.C.).
RESULTS
Environmental parameters.
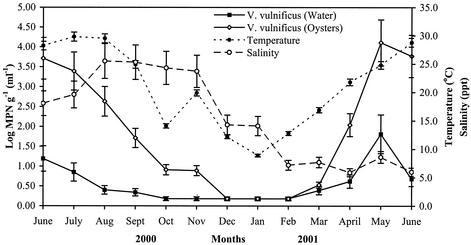
Water temperature and salinity varied widely over the 13-month study period. The mean monthly water temperature ranged from a low of 8.9°C in January 2000 to a maximum of 29.8°C in July 2000, while the mean monthly salinity ranged from a low of 5.9 ppt in April 2001 to a high of 25.5 ppt in August 2000 (Fig. 2).
FIG. 2.
Effects of water temperature and salinity on V. vulnificus concentrations in Galveston Bay water (per milliliter) and oysters (per gram). Nondetectable levels were assigned an MPN of 1.5 per g or ml (one-half the limit of detection) when log10 transformed. The values plotted are the mean monthly values for the five sampling sites studied. Error bars represent standard deviations.
Occurrence and distribution of V. vulnificus in Galveston Bay water and oysters.
Thirty-nine (45%) of 86 water samples and 64 (74%) of 86 oyster samples were found to be positive for V. vulnificus during the study period. V. vulnificus was not detected in any water samples collected between October 2000 and March 2001 or in oyster samples collected between December 2000 and February 2001 at any of the five sampling sites.
The seasonal distribution of V. vulnificus is also shown in Fig. 2. A significant correlation (P = 0.0001) was observed between water temperature and V. vulnificus densities in both water and oysters samples, whereas salinity did not influence V. vulnificus counts in oyster samples. However, there was a slight but significant (P = 0.045) negative correlation (r = −0.22) between salinities and V. vulnificus concentrations in water samples.
RAPD PCR analysis of V. vulnificus isolates.
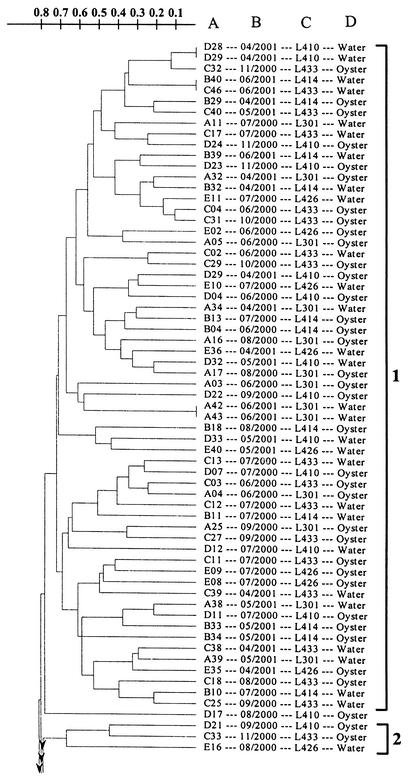
A total of 208 V. vulnificus strains, isolated from Galveston Bay water and oysters between June 2000 and June 2001, were subjected to RAPD PCR analysis and yielded reproducible profiles. Examples of RAPD PCR profiles are shown in Fig. 3. The results of the cluster analysis of the RAPD PCR profiles are shown in Fig. 4. Eleven clusters were defined at the 23% similarity level, and two strains (one August 2000 isolate and one June 2000 isolate) remained ungrouped. Isolates from different sampling sites and sources of isolation were distributed throughout the dendrogram. Table 1 lists, by sampling month, the numbers of V. vulnificus isolates, the numbers of different RAPD PCR profiles obtained, gene diversity, and the distribution of the isolates in RAPD clusters. A high level of intraspecific diversity (0.9167 to 1.0000) was observed within each month's isolates. March isolates were found to be more closely grouped than any other month's isolates, with eight of the nine isolates being grouped in cluster 8 (Fig. 4 and Table 1), and were therefore relatively more homogeneous. Cluster 1 isolates were found consistently in all of the months when V. vulnificus was detected, except March, while cluster 8 isolates were only found in March (Table 1).
FIG. 3.
Representative RAPD PCR profiles of V. vulnificus isolates. Lanes: 1 and 10, DNA size standards (values are given in base pairs on the left and right); 2, B14; 3, C14; 4, D10; 5, D11; 6, E12; 7, E13; 8, E14; 9; D12. The tracks show the processed band patterns after calibration and adjustment of the background with the Rflpscan program.
FIG. 4.
UPGMA cluster analysis of RAPD PCR profiles of 208 V. vulnificus strains isolated from Galveston Bay water and oysters during June 2000 through June 2001. The scale indicates dissimilarity. The information next to the dendrogram includes the strain designation (column A), the isolation month and year (column B), the sampling site (column C), and the sampling source (column D). The 11 clusters are shown on the far right.
TABLE 1.
Numbers of isolates subjected to RAPD analysis, numbers of different RAPD profiles obtained, gene diversity, and distribution of RAPD clusters for V. vulnificus isolates from Galveston Bay by month
| Sampling date (mo/yr) | No. of isolates subjected to RAPD analysis | No. of different RAPD profiles obtained | Gene diversityb (SD)c | Distribution of V. vulnificus isolates in RAPD clustersd (no. of strains in the cluster) |
|---|---|---|---|---|
| 06/2000 | 32 | 32 | 1.0000 (0.0078) | 1 (9), 3 (1), 5 (3), 6 (9), 7 (2), 9 (3), 11 (4) |
| 07/2000 | 44 | 44 | 1.0000 (0.0048) | 1 (15), 2 (1), 3 (6), 4 (1), 5 (6), 6 (3), 7 (4), 9 (3), 10 (4), 11 (1) |
| 08/2000 | 22 | 21 | 0.9957 (0.0153) | 1 (4), 3 (4), 7 (2), 9 (5), 10 (3), 11 (3) |
| 09/2000 | 25 | 25 | 1.0000 (0.0120) | 1 (4), 2 (1), 3 (9), 5 (3), 7 (2), 9 (3), 11 (3) |
| 10/2000 | 6 | 6 | 1.0000 (0.0962) | 1 (2), 5 (4) |
| 11/2000 | 9 | 9 | 1.0000 (0.0524) | 1 (3), 2 (1), 3 (1), 5 (2), 6 (2) |
| 12/2000 | —a | |||
| 01/2001 | — | |||
| 02/2001 | — | |||
| 03/2001 | 9 | 7 | 0.9167 (0.0920) | 8 (8), 9 (1) |
| 04/2001 | 25 | 23 | 0.9933 (0.0134) | 1 (11), 3 (2), 6 (3), 9 (9) |
| 05/2001 | 20 | 20 | 1.0000 (0.0158) | 1 (8), 3 (3), 6 (1), 9 (8) |
| 06/2001 | 16 | 14 | 0.9833 (0.0278) | 1 (5), 3 (5), 4 (2), 6 (1), 7 (1), 9 (2) |
DISCUSSION
Previous studies of the intraspecific diversity of V. vulnificus have mainly concentrated on sporadic environmental and/or clinical V. vulnificus strains obtained from different geographic origins and sources or on strains isolated from a very limited number of samples (1, 2, 3, 4, 23, 31). Little effort has been put forward to extensively analyze the temporal and spatial genetic diversity of V. vulnificus in a marine or estuarine environment. We believe the present study is the first systematic investigation of the intraspecific diversity of V. vulnificus from an estuarine environment, i.e., Galveston Bay, Tex.
V. vulnificus was present in oysters at concentrations comparable to those reported from other Gulf of Mexico regions (16). The bacterium was found more frequently and at higher concentrations in oysters than in water in synoptic water and oyster samples, probably because of the persistence and replication of the organism in oyster tissues (7, 30). The inability to isolate V. vulnificus from both water and oyster samples during the winter months (December to February) is thought to be due to entrance of the organism into a viable but nonculturable state, a survival strategy used by V. vulnificus in response to low-temperature stress (18, 19, 34).
The densities of V. vulnificus in Galveston Bay water and oysters were positively correlated with water temperature, as has been reported by other researchers (11, 16, 20). The lack of an obvious correlation between salinity and V. vulnificus densities in oysters may be explained by the fact that mean monthly salinity levels fluctuated between 5 and 25 ppt. Salinities within this range do not limit the growth of V. vulnificus and play little role in controlling V. vulnificus concentrations in water or oysters (10, 16).
The results of this study indicate that Galveston Bay contains a very dynamic and diverse population of V. vulnificus strains. Water temperature increases beginning in March correlated with subsequent increases in intraspecific diversity and shifts in the population structure of V. vulnificus in Galveston Bay. V. vulnificus strains first detected and isolated in March after winter's absence were relatively homogeneous (Table 1). A higher level of gene diversity was observed among V. vulnificus strains isolated from April to November (Table 1), the latest month of the year during the study period that V. vulnificus was recovered from Galveston Bay oysters. It was interesting that most of the March isolates belong to cluster 8 and this cluster's isolates were only found in March and not in any other month. On the other hand, cluster 1 isolates first appeared in April and were present in all subsequent months when V. vulnificus was detected. The relatively homogeneous March strains were then replaced by genetically very heterogeneous V. vulnificus strains during the warmer months (Table 1 and Fig. 4); i.e., almost every isolate analyzed yielded a unique RAPD PCR profile (Fig. 4).
The intraspecific diversity of V. vulnificus was not correlated with the sampling site or source of isolation. V. vulnificus isolates, regardless of the sampling site or source of isolation, appeared to be randomly distributed throughout the dendrogram, and strains with identical RAPD PCR profiles were isolated from different sampling sites. Probable explanations for this finding may be (i) a common response of the bacteria to similar environmental conditions at the five sampling sites, (ii) movement of bacteria among the different sites driven by water currents, and (iii) exchange of bacteria between water and oysters through the filter-feeding activities of oysters. Furthermore, no relationship between the genetic diversity of the strains and the month of isolation was observed, except that March isolates were relatively homogeneous and isolates collected during the rest of the months were highly heterogeneous.
RAPD PCR has been shown in recent years to be a useful technique for analysis of the intraspecific diversity of V. vulnificus and other bacterial species (1, 2, 13, 22, 26, 27) Application of the RAPD PCR technique in this study demonstrated the dynamic nature of the population structure and the high level of intraspecific diversity of V. vulnificus strains in Galveston Bay water and oysters. It remains to be determined how different factors promote such diversity and the possible relationship between such diversity and human infections.
Acknowledgments
This work was supported in part by grants (010298-0012b-1997 and 010298-0002-1999) from the State of Texas THECB Advanced Technology Program.
We thank the Texas Department of Health for assistance in sample collection and shipping. We also appreciate the technical support of Mona Hochman, Stephen Burkett, Karen Juntunen, and Justin Weems.
REFERENCES
- 1.Arias, C. R., L. Verdonck, J. Swings, E. Garay, and R. Aznar. 1997. Intraspecific differentiation of Vibrio vulnificus biotypes by amplified fragment length polymorphism and ribotyping. Appl. Environ. Microbiol. 63:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, C. R., M. J. Pujalte, E. Garay, and R. Aznar. 1998. Genetic relatedness among environmental, clinical, and diseased-eel Vibrio vulnificus isolates from different geographic regions by ribotyping and randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 64:3403-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aznar, R., W. Ludwig, and K.-H. Schleifer. 1993. Ribotyping and randomly amplified polymorphic DNA analysis of Vibrio vulnificus biotypes. Syst. Appl. Microbiol. 16:303-309. [Google Scholar]
- 4.Buchrieser, C., V. V. Gangar, R. L. Murphree, M. L. Tamplin, and C. W. Kaspar. 1995. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogeneous electric field gel electrophoresis. Appl. Environ. Microbiol. 61:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook, D. W., P. O'Leary, J. C. Hunsucker, E. M. Sloan, J. C. Bowers, R. J. Blodgett, and A. DePaola. 2002. Vibrio vulnificus and Vibrio parahaemolyticus in US retail shell oysters: a national survey from June 1998 to July 1999. J. Food Prot. 65:79-87. [DOI] [PubMed] [Google Scholar]
- 6.Elliot, E. L., C. A. Kaysner, L. Jackson, and M. L. Tamplin. 1995. Vibrio cholerae, V. parahaemolyticus, V. vulnificus, and other Vibrio spp., p. 9.01-9.27. In U.S. Food and Drug Administration bacteriological and analytical manual, 8th ed. Association of Official Analytical Chemists, Gaithersburg, Md.
- 7.Harris-Young, L., M. L. Tamplin, W. S. Fisher, and J. W. Mason. 1993. Effects of physicochemical factors and bacterial colony morphotype on association of Vibrio vulnificus with hemocytes of Crassostrea virginica. Appl. Environ. Microbiol. 59:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hlady, W. G. 1997. Vibrio infections associated with raw oyster consumption in Florida, 1981-1994. J. Food. Prot. 60:353-357. [DOI] [PubMed] [Google Scholar]
- 9.Jackson, J. K., R. L. Murphree, and M. L. Tamplin. 1997. Evidence that mortality from Vibrio vulnificus infection results from single strains among heterogeneous populations in shellfish. J. Clin. Microbiol. 35:2098-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaspar, C. W., and M. L. Tamplin. 1993. Effect of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl. Environ. Microbiol. 59:2425-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly, M. T. 1982. Effect of temperature and salinity on Vibrio (Beneckkea) vulnificus occurrence in a Gulf Coast environment. Appl. Environ. Microbiol. 44:820-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klontz, K. C., L. Williams, L. M. Baldy, and M. Campos. 1993. Raw oyster-associated Vibrio infections: linking epidemiologic data with laboratory testing of oysters obtained from a retail outlet. J. Food Prot. 56:977-979. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence, L. M., J. Harvey, and A. Gilmour. 1993. Development of a random amplification of polymorphic DNA typing method for Listeria monocytogenes. Appl. Environ. Microbiol. 59:3117-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine, W. C., and P. M. Griffin. 1993. Vibrio infections on the Gulf Coast: results of first year of regional surveillance. J. Infect. Dis. 167:479-493. [DOI] [PubMed] [Google Scholar]
- 15.Link, W., C. Dixkens, M. Singh, M. Schwall, and A. E. Melchinger. 1995. Genetic diversity in European and Mediterranean fava bean germ plasm revealed by RAPD markers. Theor. Appl. Genet. 90:27-32. [DOI] [PubMed] [Google Scholar]
- 16.Motes, M. L., A. Depaola, D. W. Cook, J. E. Veazey, J. C. Hunsucker, W. E. Garthright, R. J. Blodgett, and S. J. Chirtel. 1998. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nei, M. 1987. Molecular evolutionary genetics, p. 180. Columbia University Press, New York, N.Y.
- 18.Oliver, J. D. 1995. The viable but non-culturable state in the human pathogen Vibrio vulnificus. FEMS Microbiol. Lett. 133:203-208. [DOI] [PubMed] [Google Scholar]
- 19.Oliver, J. D., F. Hite, D. McDougald, N. L. Andon, and L. M. Simpson. 1995. Entry into, and resuscitation from, the viable and nonculturable state by Vibrio vulnificus in an estuarine environment. Appl. Environ. Microbiol. 61:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Neill, K. R., S. H. Jones, and D. J. Grimes. 1992. Seasonal incidence of Vibrio vulnificus in the Great Bay estuary of New Hampshire and Maine. Appl. Environ. Microbiol. 58:3257-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Power, E. G. M. 1996. RAPD typing in microbiology—a technical review. J. Hosp. Infect. 34:247-265. [DOI] [PubMed] [Google Scholar]
- 22.Quiberoni, A., P. Tailliez, P. Quénée, V. Suárez, and J. Reinheimer. 1998. Genetic (RAPD-PCR) and technological diversities among wild Lactobacillus helveticus strains. J. Appl. Microbiol. 85:591-596. [Google Scholar]
- 23.Ryang, D. W., S. W. Cho, M. G. Shin, J. H. Shin, and S. P. Suh. 1997. Molecular typing of Vibrio vulnificus isolates by random amplified polymorphic DNA (RAPD) analysis. Jpn. J. Med. Sci. Biol. 50:113-121. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro, R. L., S. Altekruse, L. Hutwagner, R. Bishop, R. Hammond, S. Wilson, B. Ray, S. Thompson, R. V. Tauxe, and P. M. Griffin. 1998. The role of Gulf Coast oysters harvested in warmer months in Vibrio vulnificus infections in the United States, 1988-1996. J. Infect. Dis. 178:752-759. [DOI] [PubMed] [Google Scholar]
- 25.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification. W. H. Freeman & Co., San Francisco, Calif.
- 26.Stephan, R., H. Schraft, and F. Untermann. 1994. Characterization of Bacillus licheniformis with the RAPD (randomly amplified polymorphic DNA). Lett. Appl. Microbiol. 18:260-263. [DOI] [PubMed] [Google Scholar]
- 27.Tailliez, P., J. Tremblay, S. D. Ehrlich, and A. Chopin. 1998. Molecular diversity and relationship within Lactococcus lactis, as revealed by randomly amplified polymorphic DNA (RAPD). Syst. Appl. Microbiol. 21:530-538. [DOI] [PubMed] [Google Scholar]
- 28.Tamplin, M. L., G. E. Rodrick, N. J. Blake, and T. Cuba. 1982. Isolation and characterization of Vibrio vulnificus from two Florida estuaries. Appl. Environ. Microbiol. 44:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamplin, M. L., A. L. Martin, A. D. Ruple, D. W. Cook, and C. W. Kaspar. 1991. Enzyme immunoassay for identification of Vibrio vulnificus in seawater, sediment, and oysters. Appl. Environ. Microbiol. 57:1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamplin, M. L., and G. M. Capers. 1992. Persistence of Vibrio vulnificus in tissues of Gulf Coast oysters, Crassostrea virginica, exposed to seawater disinfected with UV light. Appl. Environ. Microbiol. 58:1506-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamplin, M. L., J. K. Jackson, C. Buchrieser, R. L. Murphree, K. M. Portier, V. Gangar, L. G. Miller, and C. W. Kaspar. 1996. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl. Environ. Microbiol. 62:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanoy, R. W., M. L. Tamplin, and J. R. Schwarz. 1992. Ecology of Vibrio vulnificus in Galveston Bay oysters, suspended particular matter, sediment and seawater: detection by monoclonal antibody-immunoassay-most probable number procedure. J. Ind. Microbiol. 9:219-223. [Google Scholar]
- 33.Warner, J. M., and J. D. Oliver. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other Vibrio species. Appl. Environ. Microbiol. 65:1141-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf, P. W., and J. D. Oliver. 1992. Temperature effects on the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol. Ecol. 101:33-39. [Google Scholar]
- 35.Wright, A. C., R. T. Hill, J. A. Johnson, M.-C. Roghman, R. R. Colwell, and J. G. Morris, Jr. 1996. Distribution of Vibrio vulnificus in Chesapeake Bay. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]