Abstract
The causes of cerebral accumulation of amyloid β-protein (Aβ) in most cases of Alzheimer’s disease (AD) remain unknown. We recently found that homozygous deletion of the insulin-degrading enzyme (IDE) gene in mice results in an early and marked elevation of cerebral Aβ. Both genetic linkage and allelic association in the IDE region of chromosome 10 have been reported in families with late-onset AD. For IDE to remain a valid candidate gene for late-onset AD on functional grounds, it must be shown that partial loss of function of IDE can still alter Aβ degradation, but without causing early, severe elevation of brain Aβ. Here, we show that naturally occurring IDE missense mutations in a well-characterized rat model of type 2 diabetes mellitus (DM2) result in decreased catalytic efficiency and a significant ∼15 to 30% deficit in the degradation of both insulin and Aβ. Endogenously secreted Aβ40 and Aβ42 are significantly elevated in primary neuronal cultures from animals with the IDE mutations, but there is no increase in steady-state levels of rodent Aβ in the brain up to age 14 months. We conclude that naturally occurring, partial loss-of-function mutations in IDE sufficient to cause DM2 also impair neuronal regulation of Aβ levels, but the brain can apparently compensate for the partial deficit during the life span of the rat. Our findings have relevance for the emerging genetic evidence suggesting that IDE may be a late-onset AD-risk gene, and for the epidemiological relationships among hyperinsulinemia, DM2, and AD.
Cerebral accumulation of amyloid β-protein (Aβ) is a pathological hallmark of Alzheimer’s disease (AD). Although an impressive number of studies support a central role for Aβ in the pathogenesis of this very common neurodegenerative disorder, the underlying causes for gradual accumulation of the peptide are unknown in the vast majority of cases. Overproduction of Aβ has been implicated in only a small portion of cases of early-onset familial AD, representing <2% of the entire AD population.1 Newly generated Aβ is rapidly cleared from the brain,2,3 suggesting that Aβ-degrading proteases could play a critical role in regulating cerebral levels of the peptide. Although much work has focused on the generation of Aβ, relatively little is known about Aβ proteolysis, which could be as or more important in the pathogenesis and potential prevention of AD.
Several proteases have been shown capable of degrading Aβ, including insulin-degrading enzyme (IDE, insulysin; EC 3.4.24.56),4–8 neprilysin (NEP),9,10 endothelin-converting enzyme,11 the uPA/tPA/plasminogen system,12,13 and angiotensin-converting enzyme.14 Mice with experimental deletions of the NEP,15 endothelin-converting enzyme,16 or IDE17 genes have elevated levels of endogenous cerebral Aβ of similar magnitude (∼60 to 100%). These animal models validate the role of proteolysis in the regulation of cerebral Aβ levels in vivo.
IDE, an ∼110-kd thiol zinc metalloendopeptidase located in cytosol, peroxisomes, endosomes, and on the cell surface,8,18–20 cleaves small proteins of diverse sequence, several of which share a propensity to form β-pleated sheet-rich amyloid fibrils, including insulin, Aβ, amylin, atrial natriuretic factor, and calcitonin.21,22 An unbiased screen of various cell lines for Aβ-degrading activity revealed that the major such activity was attributable to IDE.6,7 Direct comparison of IDE and NEP activity by transfecting their respective cDNAs into stable human APP-expressing cell lines showed that IDE markedly reduced the levels of soluble and insoluble intracellular Aβ, whereas NEP lowered the levels of only insoluble intracellular Aβ.23 It is not yet fully established which pool of Aβ is more pathogenic, although recent studies suggest that cognitive dysfunction in AD patients correlates more tightly with soluble than insoluble levels of cortical Aβ.24,25 It has been demonstrated that proteolysis of Aβ by IDE eliminates the neurotoxic effects of the peptide in rat primary neuronal cultures and prevents the deposition of Aβ onto synthetic amyloid aggregates.26 Recently, it has been reported that levels of IDE protein and transcripts are reduced in the hippocampi from AD patients with an apolipoprotein E-ε4 allele compared to either AD patients without this AD-risk allele or normal patients (with or without the allele).27
Consistent with a possible role for Aβ-degrading proteases in human disease, the IDE region of chromosome 10q has been shown to have genetic linkage to late-onset (conventional) AD28 and to age of disease onset in AD families.29 Some studies have also reported allelic association in or near the IDE gene with late-onset AD,28,30,31 whereas others have not.32,33 In separate reports, the occurrence of AD34 and elevated plasma Aβ42 levels35 were each linked to a region of chromosome 10q ∼40 Mb centromeric to the linkage peaks over the IDE region. It remains unclear whether these results signify the same underlying locus or two separate loci.
The IDE region of chromosome 10q has also been genetically linked to type 2 diabetes mellitus (DM2)36–38 and to fasting glucose levels (20-year means).38,39 In this regard, it was shown that transferring a ∼3.7-cM chromosomal region containing the IDE gene from an inbred rat model of DM2 [Goto-Kakizaki (GK) rat] to a normoglycemic rat recapitulated several features of the diabetic phenotype, including hyperinsulinemia and postprandial hyperglycemia.40 The GK allele of IDE in this chromosomal region was found to bear two missense mutations that, when transfected into COS-1 cells, resulted in 31% less insulin degradation by these cells than by cells transfected with the wild-type (WT) allele. Targeted homozygous deletion of the IDE gene in mice, in addition to elevating cerebral levels of Aβ, also produced hyperinsulinemia and glucose intolerance in the animals.17 Thus, although the gene(s) responsible for the linkage and association of AD and DM2 to chromosome 10q has not been identified, the IDE gene is a leading candidate in both diseases.37,41 In this regard, there is growing evidence that DM2 and, in particular, hyperinsulinemia are associated with an increased risk of developing AD (see Discussion).
Although it is now clear that experimental deletion of the IDE gene results in acute elevations of cerebral Aβ, it is not known whether naturally occurring, partial loss-of-function mutations in IDE can affect Aβ catabolism, and, if they do, whether the effect is subtle enough to be compatible with the genetic evidence that IDE may be a risk-conferring gene for late-onset AD. It is possible that, in the pertinent neuronal compartments, the protease is in sufficient excess relative to its Aβ substrate so that mild hypofunction of IDE will not alter Aβ turnover, or that there is a compensatory increase in the levels of the hypofunctional IDE or another Aβ-degrading protease such that there is no net effect on Aβ catabolism. For IDE to remain a valid candidate gene for late-onset AD risk, as suggested by the emerging genetic evidence, not only must it be shown that partial loss of IDE function is physiologically relevant, but also that it does not cause an early and severe elevation of Aβ levels. The magnitude of IDE hypofunction in humans would be expected to be significantly less severe than that of the IDE gene-deleted mice, with their complete absence of IDE function and >60% increases in cerebral Aβ by only 12 weeks of age.
In this study, we take advantage of the GK rat to ascertain whether, and to what extent, naturally occurring, partial loss-of-function mutations in IDE that are sufficient to cause features of the DM2 phenotype40 can also affect Aβ turnover and alter levels of Aβ in neuronal cultures and brain. Based on our findings, we discuss the potential relationship among mutations in IDE, DM2, and AD.
Materials and Methods
Sequencing of Rat IDE cDNA
GK and Wistar rats were purchased from Taconic M&B (Denmark) and Charles River Labs (Wilmington, MA), respectively. IDE cDNA was amplified from rat liver by reverse transcriptase-polymerase chain reaction with gene-specific primers, ligated into a pcDNA5/FRT (Invitrogen, Carlsbad, CA) vector, and sequenced using an ABI PRISM 377 sequencer.
Preparation and Immunoblotting of Brain Fractions
Fresh-frozen rat cerebral hemispheres were homogenized in 8 vol (w:v) of 250 mmol/L sucrose in 50 mmol/L Tris-HCl (pH 7.4) in a Potter-Elvehjem homogenizer. After pelleting nuclei and unbroken cells, the supernatant was spun again at 100,000 × g for 1 hour. This resulting supernatant was saved as the soluble fraction, and the membrane pellet was washed in 100 mmol/L Na2CO3 (pH 11.3) to linearize microsomes and strip loosely associated proteins.42 The membranes were pelleted again, resuspended in 50 mmol/L Tris-HCl (pH 7.4), and sonicated. Protein concentrations were determined using a bicinchoninic acid-based protein assay (Pierce, Rockford, IL). For immunoblotting, membranes were probed with antibodies raised to: IDE (IDE-1),8 NEP (56C6; Novacastra, Newcastle, UK); amyloid precursor protein and its proteolytic derivatives C83, C99 (antibody C8),43 and the secreted APP fragment (22C11; Chemicon, Temecula, CA); presenilin N-terminal (Ab14, gift from S. Gandy, Thomas Jefferson University) and C-terminal (4627)44 fragments; BACE (Ab-2; Oncogene), and ADAM10 (AB19026, Chemicon).
Primary Cell Cultures
For primary fibroblasts, tissue from the abdominal wall and penis of adult rats were minced and partially digested with 0.25% trypsin in Hanks’ balanced salt solution. The dissociated cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. For primary cortical neuronal cultures, neocortices and hippocampi from postnatal day 0 rat litters were partially digested in 0.025% trypsin in Hanks’ balanced salt solution, 10 mmol/L HEPES, 10 mmol/L glucose, and 10 mmol/L sucrose, mechanically dissociated, and plated at varying densities onto poly-l-lysine-coated dishes. Cells were maintained in a neuronal-selecting medium (Neurobasal medium with the growth supplement B27; Invitrogen).
Trichloroacetic Acid (TCA) Precipitation Degradation Assays
To measure proteolysis by brain fractions, 100 pmol/L synthetic 125I-Aβ1-40 (human or rodent) (Amersham, Arlington Heights, IL), or 100 pmol/L recombinant human 125I-insulin (Amersham) was incubated at 37°C with 100 μg/ml and 50 μg/ml of protein, respectively. To determine rates of proteolysis by intact primary fibroblast (22,000 cells/cm2) and neuronal cultures (80,000 and 1,600,000 cells/cm2), cells were washed with serum-free and supplement-free medium (Dulbecco’s modified Eagle’s medium and Neurobasal, respectively) containing 0.1% bovine serum albumin, then incubated in the same medium with 40 pmol/L of either 125I-insulin or 125I-Aβ1-40. At each time point, an aliquot of the sample was added to an equal volume of 15% TCA to allow the uncleaved peptides to precipitate. After centrifugation, gamma counts in the TCA-insoluble pellet (undegraded peptide) and TCA supernatant (degraded peptide fragments) were determined, and the percentage of radiolabeled substrate degraded was calculated.
Quantification of Endogenous Aβ
After primary neurons were allowed to condition Neurobasal media with B27 supplement, the media were removed, 1,10-phenanthroline and a protease inhibitor cocktail (Sigma, St. Louis, MO) were added, and the samples were centrifuged to remove any cellular material. Aβ was extracted from brain using either the previously described diethylamine protocol2,45 or Tris-buffered saline (TBS)/guanidine HCl. In brief, we homogenized cerebral hemispheres in either 9 vol (w/v) 0.2% diethylamine in 50 mmol/L NaCl or 5 vol (w/v) of TBS with the above protease inhibitors using a Potter-Elvehjem homogenizer. Homogenates were centrifuged at 100,000 × g for 1 hour. Supernatants from the diethylamine extractions were neutralized with 1/10 volume 500 mmol/L Tris-HCl (pH 6.8), and the pellets were discarded. Supernatants from the TBS extraction were saved as soluble Aβ, while the associated pellets were resuspended in a 5-mol/L guanidine HCl and 50-mmol/L Tris-HCl (pH 8.0) buffer and saved as insoluble Aβ. Aβ enzyme-linked immunosorbent assays (ELISAs) on neuronal media and DEA brain extracts were performed using the well-characterized BNT-77/BA-27 (Aβ X-40) and BNT-77/BC-05 (Aβ X-42) system.46 ELISAs on the guanidine HCl and TBS brain extracts and confirmatory ELISAs on the neuronal media were done with the 2G3/4G8 (Aβ X-40) and 21F12/4G8 ELISA system (Aβ X-42).47
Transfection of CHO Cells
Equal numbers CHO cells stably expressing human APP751 containing the V717F AD-causing missense mutation were transiently transfected with empty vector or vectors containing either the GK or WT IDE allele (as generated above) using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, the cells were washed and allowed to condition fresh media for 24 hours (OptiMEM, Invitrogen). The media was then treated identically to the neuronal conditioned media as above, except a different Aβ ELISA system was used.48,49 Thirty-six hours after transfection, cell lysates were made from duplicate dishes of the three transfected constructs, and IDE overexpression was confirmed by immunoblotting.
IDE Immunodepletion
Brain-soluble fractions were prepared as above without protease inhibitors. For each sample, 400 μg of brain-soluble protein were first precleared with protein G-agarose resin (Roche, Indianapolis, IN), then incubated with fresh resin and either no immunoglobulin (sham immunodepletion controls) or the anti-IDE monoclonal antibody 9B12 (titer of 1:100) (gift from R. Roth, Stanford University) at 4°C for 12 hours. The resin was then pelleted by centrifugation, and the IDE-immunodepleted supernatant was saved for Aβ degradation assays.
Results
We first confirmed that our GK rats, a strain generated by inbreeding Wistar rats having the highest glucose levels on a glucose tolerance test,50 carried the three single nucleotide polymorphisms in IDE previously reported40 and that our wild-type Wistar rats did not. We cloned and sequenced the complete coding regions of IDE cDNAs from two rats of each strain. As expected, the GK rats had three nucleotide differences from the Wistar controls: CAC→CGC at codon 18 (H18R), GCG→GTC at codon 890 (A890V), and GAT→GAC at codon 934 (silent). The WT Wistar sequence was identical to that in GenBank (accession no. NM 013159). Protein levels of IDE, neprilysin, APP, the APP proteolytic derivatives (C83, C99, secreted APP fragment, and AICD), presenilin NTF and CTF, BACE, and ADAM 10 were indistinguishable by immunoblotting in GK and control rat brain fractions and primary neuronal lysates (not shown).
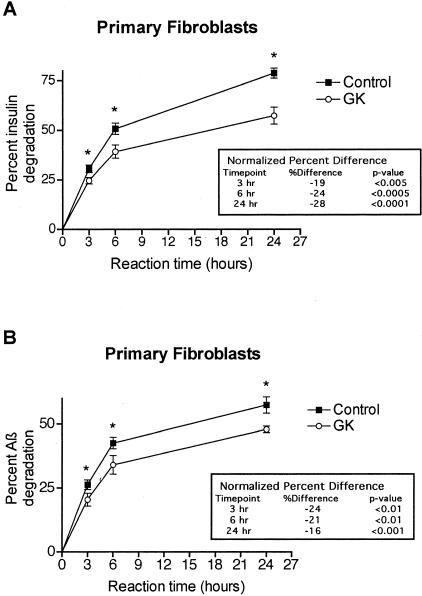
To initially assess degradative capacity for insulin and Aβ in intact, living cells of the GK rat, we incubated primary cultured fibroblasts of GK and WT rats with physiological levels (40 pmol/L) of radioiodinated human insulin or human Aβ1-40 in serum-free medium for 0, 3, 6, and 24 hours and then performed TCA precipitation assays at each time point as described in Materials and Methods. We found that the GK fibroblasts degraded insulin ∼24% less well than the controls (P < 0.005) (Figure 1A), in close agreement with the deficit in insulin degradation (∼21%) reported in isolated muscle fibers from rats with the GK IDE allele.40 The GK fibroblasts also degraded Aβ ∼20% less than controls (P < 0.01) (Figure 1B). Unlabeled insulin (10 μmol/L), a potent and relatively selective competitive inhibitor of IDE, blocked ∼90% of Aβ degradation in both the GK and control cultures, indicating that virtually all Aβ proteolysis by these intact cells is attributable to IDE.
Figure 1.
Insulin and Aβ proteolysis in primary fibroblast cultures of WT control and GK rats. Intact fibroblasts from two GK and two WT rats were incubated in serum-free medium with 40 pmol/L of either human 125I-insulin (A) or human 125I-Aβ1-40 (B). At the indicated times, degradation was quantified by TCA precipitation assays and reported as the percentage of radiolabeled substrate degraded. Graph points represent means, and error bars indicate SEM of 12 determinations (duplicate cultures from each of two rats analyzed in three independent assays; *, P < 0.01). The inset tables show the percent decrease in the GK proteolysis compared to the WT control at each time point after normalization to the mean WT percentage degradation in each assay and then combination of the data sets.
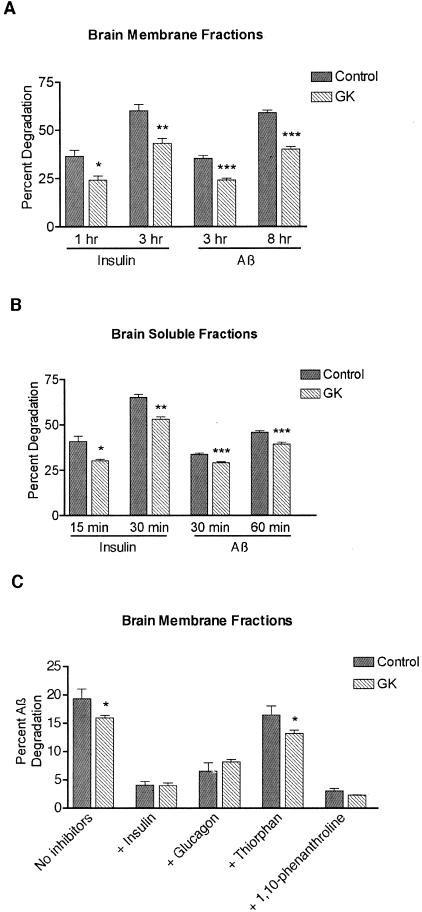
To extend this finding to the brain, the principal site of Aβ production and accumulation, we measured insulin and Aβ proteolysis in both brain-soluble and Na2CO3-washed membrane fractions from GK and WT rats. Brain membranes represent a physiological locus for IDE-mediated Aβ degradation, because Aβ, like IDE, is present at the cell surface and in endosomes.51 For insulin degradation assays, 50 μg/ml of protein was incubated with 100 pmol/L 125I-insulin for 0, 1, and 3 hours for membranes or 0, 15, and 30 minutes for the soluble fractions. For Aβ degradation assays, 100 μg/ml of protein was incubated with 100 pmol/L 125I-Aβ40 for 0, 3, and 8 hours for membranes or 0, 30, and 60 minutes for the soluble fractions. The GK brain membranes showed a highly significant ∼30% decrement in the degradation of both substrates at both time points. At 1 and 3 hours, insulin degradation in the WT rats was 37% and 60%, respectively, whereas in the GK rats, it was 24% and 43% (P < 0.01 at 1 hour, P < 0.001 at 3 hours) (Figure 2A). At 3 and 8 hours, the WT Aβ proteolysis was 35% and 59%, respectively, while in the GK it was 25% and 40% (P < 0.00001 for each time point). The GK brain-soluble fraction also showed degradation deficits. Insulin proteolysis was decreased 26% at 15 minutes (P < 0.01) and 19% at 30 minutes (P < 0.0001), whereas parallel Aβ assays showed a 14%, highly significant (P < 0.00001) decrease at both 30 and 60 minutes (Figure 2B).
Figure 2.
Insulin and Aβ proteolysis in brain fractions of WT control and GK rats. Washed brain membranes and soluble fractions were incubated with 100 pmol/L of 125I-insulin or 125I-Aβ1-40, and degradation was determined by the TCA precipitation assay. A: Insulin and Aβ degradation in brain membranes from three WT and four GK rats. Bars represent the mean ± SEM of 12 control or 16 GK determinations (duplicate samples from each rat analyzed in two independent assays; *, P < 0.005; **, P < 0.001; ***, P < 0.00001). B: Insulin and Aβ degradation in brain-soluble fractions from five WT and four GK rats. Bars represent the means ± SEM of 20 control or 16 GK determinations for insulin (duplicate samples from each rat analyzed in two independent assays), and 32 controls or 28 GK determinations for Aβ (duplicate samples from each rat analyzed in two to four independent assays; *, P < 0.005; **, P < 0.0001; ***, P < 0.00001). C: Inhibitor profile of Aβ proteolysis in brain membranes from two WT and three GK rats at 3 hours. The degradation assays were performed in the presence or absence of the indicated protease inhibitor at the concentrations mentioned in Results. Bars represent the means ± SEM of four to six determinations (duplicate samples from each rat analyzed in two to three independent assays; *, P < 0.05 for difference between WT control and GK degradation in a given condition).
To assess the relative roles of IDE and other proteases in Aβ degradation and acquire supportive evidence that IDE is responsible for the Aβ proteolytic dysfunction in GK rats, we examined the protease inhibitor profiles of the brain fractions (Figure 2C). In washed brain membranes, insulin inhibited 77% of control and 71% of GK-mediated degradation, resulting in identical residual proteolysis in the two genotypes, as would be expected if the GK deficit is because of IDE. Glucagon (10 μmol/L), a less avid competitive inhibitor of IDE than insulin, prevented ∼60% of the Aβ degradation. Thiorphan (1 mmol/L), a potent inhibitor of NEP and angiotensin-converting enzyme, and an inhibitor cocktail containing only serine and cysteine protease inhibitors (Roche) (not shown) each blocked only ∼15% of the degradation. In the presence of thiorphan and the serine and cysteine protease inhibitors, the GK membrane fraction continued to show a significant decrease in Aβ proteolysis, again supporting the attribution of this deficit to IDE. 1,10-Phenanthroline (2 mmol/L), a potent zinc metalloprotease inhibitor that can inhibit both IDE and NEP, prevented >80% of the Aβ proteolysis. In the brain-soluble fraction, Aβ degradation was inhibited >96% by either insulin or 1,10-phenanthroline, ∼75% by glucagon, <5% by thiorphan, and not at all by the serine and cysteine protease inhibitor cocktail (data not shown). Degradation assays done in brain membranes and soluble fractions using rodent 125I-Aβ40 iodinated at two different sites than the human peptide used above (ie, either His6 or His14 instead of Tyr10) yielded similar degradation deficits in the GK rats (not shown). These results indicate that the GK Aβ degradation deficit extends to the rodent peptide, with its three amino acid differences compared with human Aβ, and that the site of iodination does not affect the results of our Aβ degradation assays.
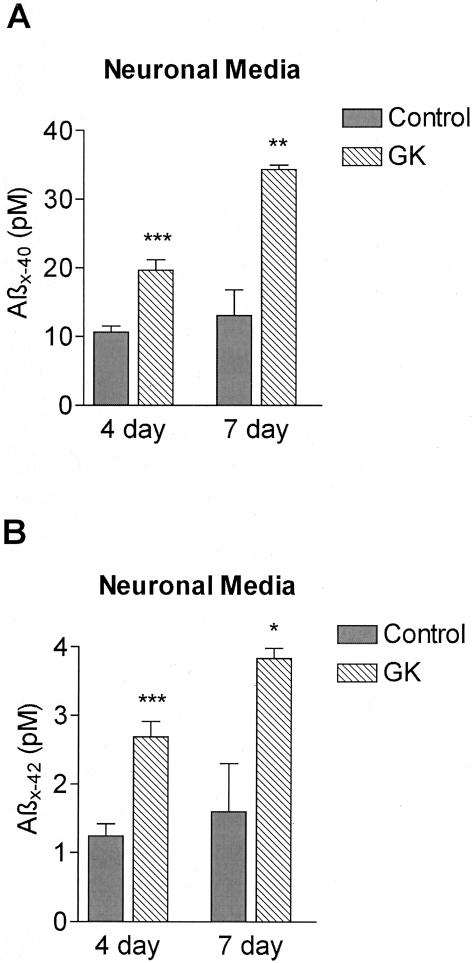
Next, we quantified insulin and Aβ proteolysis in intact primary cortical neurons. Neocortical cells from two GK and two WT litters were incubated with 40 pmol/L of 125I-insulin or 125I-Aβ40 in serum-free medium for 0, 3, 6, and 24 hours and TCA precipitation assays were performed. Overall, 125I-insulin proteolysis ranged from ∼45% at 3 hours to ∼85% at 24 hours, whereas 125I-Aβ degradation ranged from ∼10% at 3 hours to ∼65% at 24 hours. The GK neurons demonstrated less insulin and Aβ proteolysis than WT neurons at each time point, with the differences at 6 and 24 hours being statistically significant (6-hour values in Figure 3). Insulin degradation (Figure 3A) in the GK neurons at 3, 6, and 24 hours was 12% (P = 0.12), 15% (P = 0.005), and 4% (P = 0.03) less than WT, respectively. The Aβ proteolytic deficit (Figure 3B) was 8% (P = 0.07), 15% (P = 0.0008), and 7% (P = 0.0007) at 3, 6, and 24 hours, respectively. Unlabeled insulin prevented >90% of the neuronal Aβ degradation (P < 0.01), leaving identical amounts of residual activity in the GK and controls (Figure 3B), whereas glucagon blocked >50% of Aβ proteolysis (P < 0.05) (not shown). To determine whether the GK Aβ-degradation deficit documented in primary neurons occurs for the naturally produced, endogenous peptide, we measured Aβ levels in the neuronal conditioned media by ELISA. Neocortical cells from two GK and two WT postnatal day 0 litters plated at a density of 40,000 cells/cm2 were conditioned for 4 or 7 days. Compared to WT conditioned media, Aβ X-40 levels in the GK neuronal media were increased 84% after 4 days (P < 0.0005) and 162% after 7 days (P < 0.005) (Figure 4A). Aβ X-42 levels were also elevated in the GK neuronal media, increasing by 120% after 4 days (P < 0.0005), and 138% by 7 days (P < 0.05) (Figure 4B). In confirmatory experiments using a different ELISA system (see Materials and Methods), conditioned media were analyzed from primary neurons plated at 80,000 cells/cm2 (twice the density of the first set), and again, the GK media had highly significant elevations of both Aβ X-40 (60% after 4 days, P < 0.005; 57% after 7 days, P < 0.01) and Aβ X-42 (85% at 4 days, P < 0.001; 107% at 7 days (P < 0.05) (data not shown).
Figure 3.
Insulin and Aβ proteolysis in primary neurons of WT control and GK rats. Primary neurons from two GK and two WT postnatal day 0 litters (each litter containing seven to eight pups) were incubated with 40 pmol/L of radiolabeled substrate, and degradation was quantified by the TCA assay. Bars represent the mean degradation for the 6-hour time points. A: 125I-insulin degradation by neurons 17 to 19 days in vitro. Bars represent means ± SEM of seven to eight determinations (160,000 and 80,000 cells/cm2 cultures from each litter in two independent assays) after normalization to the mean WT percent degradation for each culture density, allowing combination of the two data sets. *, P = 0.005. B: 125I-Aβ1-40 degradation and inhibition by insulin by neurons 9 to 11 and 19 to 21 days in vitro. Degradation assays were performed in the presence and absence of 10 μmol/L insulin in the medium. Bars represent means ± SEM of 9 to 10 determinations (160,000 and 80,000 cells/cm2 cultures from each litter in two to three independent assays) after normalization to the mean WT percent degradation for each culture density, allowing combination of the data sets; **, P < 0.001for difference between WT and GK degradation).
Figure 4.
Endogenous Aβ levels in primary neuronal conditioned media of control and GK rats. Aβ X-40 (A) and Aβ X-42 (B) levels were quantified by ELISA in the media of primary neurons (40,000 cells/cm2) from two GK and two WT postnatal day 0 litters. The media were allowed to condition for 4 days (neurons, 6 to 10 days in vitro) or 7 days (neurons, 3 to 10 days in vitro). Bars represent means ± SEM of two to six determinations, and each determination is a mean of duplicate ELISA values. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Although the inbred GK line was originally derived from the Wistar rat, the two strains are genetically heterogeneous at more than just the IDE locus. The aforementioned evidence that IDE degrades Aβ, combined with the demonstrated insulin-degrading abnormality and the proteolytic inhibitor profile, all support the role of IDE in generating the GK Aβ catabolic deficit. To confirm that the GK allele of IDE is sufficient to explain the above Aβ-degrading deficit, we performed two types of experiments: overexpression and immundepletion of the GK and WT IDE isoforms. First, we transiently transfected either the GK or WT alleles of IDE into CHO cell lines stably expressing human APP, which additionally allowed us to determine whether endogenously secreted human Aβ also accumulates more as a result of the IDE mutations. The two IDE proteins were expressed at equal levels, as assessed by immunoblotting. The conditioned media from cells transfected with the GK and WT IDE alleles had 89.8% and 92.3% lower Aβ X-40 levels by ELISA than media from an equal number of cells transfected with empty vector (not shown). Compared to WT IDE transfected cells, those expressing the GK allele had 32% higher levels of secreted Aβ X-40 (P < 0.005) (Figure 5A). Aβ X-42 values were below the level of detection in the media of cells transfected with either IDE allele.
Figure 5.
Overexpression and immunodepletion of the GK and WT IDE alleles. A: Aβ in media of cells stably expressing human APP and transfected with the WT or GK IDE allele. Aβ levels were quantified in the media of CHO cells expressing human APP751 containing the V717F AD-causing missense mutation and transiently transfected with either the GK or WT IDE allele. Media were incubated on the cells for 24 hours and submitted for Aβ X-40 and Aβ X-42 ELISAs (Aβ X-42 levels were undetectable in the IDE-transfected cells). Bars represent means ± SEM of three determinations from two separate transfections (and each determination is a mean of quadruplicate ELISA values) after normalization to the WT Aβ value in each ELISA, allowing combination of the three data sets (*, P < 0.005). For reasons of scale, the normalized Aβ X-40 value of the vector alone (13.0) is not shown. B: Aβ degradation in brain-soluble fractions from two WT control and two GK rats before (pre-ID) and after (post-ID) IDE immunodepletion. TCA precipitation assays were performed after immunodepletion of IDE with the monoclonal antibody 9B12 (described in Materials and Methods) by incubating 100 pmol/L 125I-Aβ1-40 with 100 μg of protein/ml of the pre-ID and post-ID samples for 30 minutes at 37°C. Bars represent the mean degradation ± SEM of duplicate samples from each rat analyzed in two to three independent assays) (*, P < 0.001). Sham immunodepletion of the brain-soluble fractions (using protein G-agarose without immunoglobulin) had no affect on Aβ proteolysis (not shown).
In the second group of experiments, we immunodepleted IDE from the brain-soluble fractions of two GK and two WT rats using the IDE-specific monoclonal antibody 9B12, and performed 125I-Aβ degradation assays on the IDE-depleted supernates. Immunodepletion of IDE from the brain fractions removed ∼80% of the enzyme from each sample, as determined by immunoblotting (not shown) and removed ∼85% of the Aβ-degrading activity from the WT samples and ∼80% from the GK samples (Figure 5B). Before IDE immunodepletion (pre-ID), WT and GK Aβ degradation at 30 minutes was significantly different at 33% and 29%, respectively (P < 0.001), whereas after IDE removal (post-ID), residual Aβ degradation was identical at 5.2% (P = 0.49). Thus, the consistent difference observed in Aβ proteolysis between the GK and WT brain samples was eliminated by specific removal of IDE.
To determine whether the IDE-mediated deficit in Aβ degradation that elevates Aβ in neuronal cultures also increases steady-state levels of Aβ, in the brain, we quantified levels of the peptide in GK and WT brains by ELISA. Cerebral Aβ was extracted using two separate protocols, which yielded three distinct pools of Aβ. The diethylamine protocol, which extracts the aqueous soluble and a portion of the aqueous insoluble Aβ, was performed on one cerebral hemisphere from each of 34 rats divided into two age groups. The other hemisphere from a subset of these animals (19 rats) was first extracted in TBS to acquire the soluble Aβ, and then the TBS-insoluble material was extracted in guanidine HCl to obtain insoluble Aβ. There were no statistically significant differences between GK and WT brain Aβ X-40 and Aβ X-42 levels in any of the three types of extracts at either 5 to 7 months or 10 to 14 months of age (Table 1).
Table 1.
Aβ Levels in GK and WT Rat Brains (Means ± SEM, in pmol/g of Brain)
| Age | 5 to 7 months
|
10 to 14 months
|
7 months
|
|||
|---|---|---|---|---|---|---|
| Extraction | Diethylamine
|
Diethylamine
|
Guanidine HCl
|
|||
| Rat Strain | GK | WT | GK | WT | GK | WT |
| Number | 11 | 12 | 7 | 4 | 6 | 7 |
| Aβ X-40 | 0.55 ± 0.06 | 0.53 ± 0.05 | 0.54 ± 0.08 | 0.53 ± 0.12 | 0.91 ± 0.07 | 0.97 ± 0.08 |
| P = 0.39 | P = 0.47 | P = 0.31 | ||||
| Aβ X-42 | 0.41 ± 0.03 | 0.42 ± 0.02 | 0.41 ± 0.03 | 0.42 ± 0.06 | 0.42 ± 0.06 | 0.05 ± 0.00 |
| P = 0.34 | P = 0.43 | P = 0.13 | ||||
Table 1.
Continued
| 10 to 14 months
|
7 months
|
10 to 14 months
|
|||
|---|---|---|---|---|---|
| Guanidine HCl
|
TBS
|
TBS
|
|||
| GK | WT | GK | WT | GK | WT |
| 4 | 2 | 6 | 6 | 4 | 2 |
| 1.02 ± 0.09 | 1.07 ± 0.08 | 0.31 ± 0.04 | 0.37 ± 0.03 | 0.32 ± 0.04 | 0.36 ±0.01 |
| P = 0.35 | P = 0.17 | P = 0.24 | |||
| 0.05 ± 0.00 | 0.05 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.00 ± 0.00 |
| P = 0.48 | P = 0.39 | P = 0.08 | |||
To better understand the nature of the IDE proteolytic deficit conferred by the GK missense mutations, we performed quantitative kinetic analyses using a fluorescence polarization-based Aβ degradation assay.52 We used GK and WT brain-soluble fractions because, as shown above, >96% of the Aβ-degrading activity in these fractions is attributable to IDE. In each individual assay, Aβ degradation from 50 μg/ml of protein from one of three GK or three WT samples was analyzed. After completing eight assays per rat strain, the mean Vmax ± SEM and Km ± SEM were calculated for the GK and WT samples. The mean Vmax of Aβ degradation for the GK soluble fraction was significantly less relative to WT (30.0 ± 3.7 versus 47.6 ± 3.3 nmol/L/minute; P < 0.005), whereas the Km values did not differ significantly (1.5 ± 0.3 versus 2.1 ± 0.3 μmol/L, respectively). The catalytic efficiency (Vmax/Km) of the GK brain-soluble fraction was reduced by ∼15% relative to WT, which is in good agreement with the data presented above.
Discussion
Our findings demonstrate that naturally occurring, partial loss-of-function mutations in IDE, which are sufficient to cause a diabetic phenotype, result in significant impairment of Aβ degradation in brain membrane fractions and intact neurons, and this causes substantially elevated levels of neuronally secreted Aβ. Thus, there does not seem to be an increase in the levels of IDE or other Aβ-degrading proteases sufficient to compensate for the enzyme’s decreased catalytic efficiency in neurons. Furthermore, the relationship between IDE activity and available Aβ substrate is such that even mild IDE hypofunction alters turnover of the peptide and results in a rise in neuronally generated Aβ. The realization that IDE activity is rate-limiting in the neuronal proteolysis of Aβ recommends activation or disinhibition of IDE as a viable therapeutic target in patients with AD.
Although the relatively small (∼15 to 30%) decrement in Aβ proteolysis led to considerable increases in endogenous Aβ levels in neuronal cultures, we detected no quantifiable rise in steady-state levels of the peptide in brains from rats up to age 14 months. In the intact brain, microglia,6,7 astrocytes,53 and transport into the circulation could each play roles in Aβ clearance,54 and these may compensate for the relatively small decrease in IDE function. This is in sharp contrast to full loss of IDE function by homozygous deletion of the gene in mice, which was found to lead to a ∼65% elevation of endogenous cerebral Aβ40 levels by 3 months of age.17 Combining the results from these two different animal models strengthens the hypothesis, based on the recent human genetic data that IDE mutations could serve as a risk factor for late-onset AD. Whereas the gene-deleted mice provide proof-of-principle that IDE dysfunction can significantly elevate cerebral Aβ levels in vivo, the GK rats demonstrate that naturally occurring, subtle loss-of-function mutations in IDE are physiologically relevant (decreasing the catabolism of Aβ in intact neurons), but not so severe as to cause significant elevations of rodent Aβ in the brain during the 14-month life span of our rats. In late-onset AD patients, the imbalance between rates of Aβ production and clearance is such that the presymptomatic accumulation of cerebral Aβ occurs very slowly throughout decades, and the symptomatic phase of the disease is measured in years. If IDE turns out to be a genetic risk factor for late-onset AD, the magnitude of IDE hypofunction in humans would be expected to resemble that of the GK rat more than that of the IDE gene-deleted mice. It is possible that a modest decrease in neuronal Aβ clearance would only lead to detectably elevated cerebral Aβ levels after many years. We speculate that a subtle decrease in IDE function resulting in elevated levels of monomeric Aβ could lead to more accumulation of Aβ in humans than rodents, because the human peptide is far more prone to form stable aggregates that resist degradation by proteases than is the rodent form.
We have extended the finding that muscle fibers from rats with the GK allele degrade insulin less well than controls40 by showing that these diabetic rats also have a deficit in insulin proteolysis in primary fibroblasts, brain fractions, and intact neurons. The consequences of having chronic insulin-degrading deficits in the brain are unclear at present, but there is mounting evidence that insulin plays numerous roles in the central nervous system.55 When Fakhrai-Rad and colleagues40 transfected COS cells with the GK and IDE alleles, they were able to detect the GK insulin-degrading defect in intact cells, but not in lysates of the same cells. They hypothesized that the IDE defect is coupled to receptor-mediated internalization of insulin or other activities that require intact cell structures. In contrast, we clearly detected the GK IDE defect not only in untransfected intact fibroblasts and neurons, but also in tissue fractions, providing the simpler explanation that the IDE missense mutations directly decrease the ability of the protease to cleave insulin, independent of other processes. Furthermore, we have begun to characterize the kinetic effects of these insulin- and Aβ-elevating mutations, showing that they lower the Vmax without changing the Km of the protease, resulting in a modest but significant decrease in catalytic efficiency.
By demonstrating faulty Aβ degradation in a well-characterized animal model of DM2, our findings are directly relevant to emerging evidence that diabetes and, in particular, hyperinsulinemia, are associated with a heightened likelihood of developing AD. It is well known that DM2 is a risk factor for vascular diseases, including vascular dementia, but the relationship between the occurrence of DM2 and AD has only recently been addressed in large-scale epidemiological studies. Two of these studies in which other causes of dementia were carefully excluded showed that individuals with DM2 were approximately twice as likely to develop a clinical diagnosis of AD compared to normals, independent of vascular factors,56,57 whereas a third group found that diabetic patients had a 60% increased risk of developing either frank AD or mild cognitive impairment (a clinical state frequently progressing to AD).58 Another population-based study of Japanese-American men reported no association between AD and midlife diabetes,59 but a more recent report based on the same cohort concluded that diabetic patients’ relative risk of AD was 1.8 and that this increased risk was not related to vascular disease.60 The latter report hypothesized that the small number of diabetic patients included in first analysis limited its ability to find an association between DM2 and AD, but as the study population aged, the prevalence of DM2 more than doubled, giving the second study more power. Although it is not clear from the above epidemiological studies which particular aspect of the DM2 phenotype (eg, hyperglycemia, hyperinsulinemia) is most associated with an increased risk of AD, elevated plasma insulin levels in nondiabetic patients have been linked to dementia61 even after adjustment for vascular disease, and specifically AD.62,63 Supporting a role for IDE in these findings is evidence that the exaggerated insulin response to administered glucose seen in AD patients is secondary to an insulin clearance defect, rather than to overproduction of the hormone.64
Thus, although the mechanisms underlying the associations of DM2 and hyperinsulinemia with AD are not yet clear, the demonstration that naturally occurring partial loss-of-function mutations in IDE affect the catabolism of both insulin and Aβ, combined with the genetic linkage of both DM2 and AD to the IDE region of chromosome 10q, recommend IDE hypofunction as an intriguing explanation. Our results have attendant therapeutic implications in that compounds that subtly increase IDE activity, either directly or via blocking a natural IDE inhibitor, could chronically decrease Aβ levels in the human brain.
Acknowledgments
We thank Dominic Walsh for helpful discussions, Franziska Baenke and Alice Y. Chang for excellent technical assistance, Samuel Gandy for antibody Ab14, and Richard Roth for antibody 9B12.
Footnotes
Address reprint requests to Dennis J. Selkoe, Center for Neurologic Diseases, HIM 730, 77 Ave Louis Pasteur, Boston, MA 02115. E-mail: dselkoe@rics.bwh.harvard.edu.
Supported by the National Institutes of Health (grant AG12479 to W.F., S.M., M.A.L., D.J.S.), the Mayo Alzheimer’s Disease Research Center (to C.E. and E.E.), the Alzheimer’s Association (to C.E. and E.E.), and the Smith Fellowship (to E.E.).
References
- St. George-Hyslop PH. Genetic factors in the genesis of Alzheimer’s disease. Ann NY Acad Sci. 2000;924:1–7. doi: 10.1111/j.1749-6632.2000.tb05552.x. [DOI] [PubMed] [Google Scholar]
- Savage MJ, Trusko SP, Howland DS, Pinsker LR, Mistretta S, Reaume AG, Greenberg BD, Siman R, Scott RW. Turnover of amyloid beta-protein in mouse brain and acute reduction of its level by phorbol ester. J Neurosci. 1998;18:1743–1752. doi: 10.1523/JNEUROSCI.18-05-01743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, Mutter LC, Nietz J, Quinn KP, Sacchi KL, Seubert PA, Shopp GM, Thorsett ED, Tung JS, Wu J, Yang S, Yin CT, Schenk DB, May PC, Altstiel LD, Bender MH, Boggs LN, Britton TC, Clemens JC, Czilli DL, Dieckman-McGinty DK, Droste JJ, Fuson KS, Gitter BD, Hyslop PA, Johnstone EM, Li WY, Little SP, Mabry TE, Miller FD, Audia JE. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Kurochkin IV, Goto S. Alzheimer’s β-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett. 1994;345:33–37. doi: 10.1016/0014-5793(94)00387-4. [DOI] [PubMed] [Google Scholar]
- McDermott JR, Gibson AM. Degradation of Alzheimer’s β-amyloid protein by human and rat brain peptidases: involvement of insulin-degrading enzyme. Neurochemical Res. 1997;22:49–56. doi: 10.1023/a:1027325304203. [DOI] [PubMed] [Google Scholar]
- Qiu WQ, Ye Z, Kholodenko D, Seubert P, Selkoe DJ. Degradation of amyloid β-protein by a metalloprotease secreted by microglia and other neural and non-neural cells. J Biol Chem. 1997;272:6641–6646. doi: 10.1074/jbc.272.10.6641. [DOI] [PubMed] [Google Scholar]
- Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang J, Podlisny M, Rosner MR, Safavi A, Hersh LB, Selkoe DJ. Insulin degrading enzyme regulates the level of monomeric amyloid β-protein extracellularly via degradation and oligomerization. J Biol Chem. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- Vekrellis K, Ye Z, Qiu WQ, Walsh D, Hartley D, Chesneau V, Rosner MR, Selkoe DJ. Neurons regulate extracellular levels of amyloid beta-protein via proteolysis by insulin-degrading enzyme. J Neurosci. 2000;20:1657–1665. doi: 10.1523/JNEUROSCI.20-05-01657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell S, Nalbantoglu J, Crine P. Neutral endopeptidase can hydrolyze β-amyloid (1-40) but shows no effect on β-amyloid precursor protein metabolism. Peptides. 1995;16:647–652. doi: 10.1016/0196-9781(95)00021-b. [DOI] [PubMed] [Google Scholar]
- Iwata N, Tsubuki S, Takaki Y, Watanabe K, Sekiguchi M, Hosoki E, Kawashima-Morishima M, Lee HJ, Hama E, Sekine-Aizawa Y, Saido TC. Identification of the major Abeta1-42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat Med. 2000;6:143–150. doi: 10.1038/72237. [see comments] [DOI] [PubMed] [Google Scholar]
- Eckman EA, Reed DK, Eckman CB. Degradation of the Alzheimer’s amyloid beta peptide by endothelin-converting enzyme. J Biol Chem. 2001;276:24540–24548. doi: 10.1074/jbc.M007579200. [DOI] [PubMed] [Google Scholar]
- Tucker HM, Kihiko M, Caldwell JN, Wright S, Kawarabayashi T, Price D, Walker D, Scheff S, McGillis JP, Rydel RE, Estus S. The plasmin system is induced by and degrades amyloid-beta aggregates. J Neurosci. 2000;20:3937–3946. doi: 10.1523/JNEUROSCI.20-11-03937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma MD, Da Silva JS, Crassaerts K, Delacourte A, De Strooper B, Dotti CG. Brain plasmin enhances APP alpha-cleavage and Abeta degradation and is reduced in Alzheimer’s disease brains. EMBO Rep. 2000;1:530–535. doi: 10.1093/embo-reports/kvd107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Igarashi A, Kamata M, Nakagawa H. Angiotensin-converting enzyme degrades Alzheimer amyloid beta-peptide (A beta); retards A beta aggregation, deposition, fibril formation; and inhibits cytotoxicity. J Biol Chem. 2001;276:47863–47868. doi: 10.1074/jbc.M104068200. [DOI] [PubMed] [Google Scholar]
- Iwata N, Tsubuki S, Takaki Y, Shirotani K, Lu B, Gerard NP, Gerard C, Hama E, Lee HJ, Saido TC. Metabolic regulation of brain Abeta by neprilysin. Science. 2001;292:1550–1552. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- Eckman EA, Watson M, Marlow L, Sambamurti K, Eckman CB. Alzheimer’s disease beta-amyloid peptide is increased in mice deficient in endothelin-converting enzyme. J Biol Chem. 2003;278:2081–2084. doi: 10.1074/jbc.C200642200. [DOI] [PubMed] [Google Scholar]
- Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci USA. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19:608–624. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- Goldfine ID, Williams JA, Bailey AC, Wong KY, Iwamoto Y, Yokono K, Baba S, Roth RA. Degradation of insulin by isolated mouse pancreatic acini. Evidence for cell surface protease activity. Diabetes. 1984;33:64–72. doi: 10.2337/diab.33.1.64. [DOI] [PubMed] [Google Scholar]
- Seta KA, Roth RA. Overexpression of insulin degrading enzyme: cellular localization and effects on insulin signaling. Biochem Biophys Res Commun. 1997;231:167–171. doi: 10.1006/bbrc.1997.6066. [DOI] [PubMed] [Google Scholar]
- Bennett RG, Duckworth WC, Hamel FG. Degradation of amylin by insulin-degrading enzyme. J Biol Chem. 2000;275:36621–36625. doi: 10.1074/jbc.M006170200. [DOI] [PubMed] [Google Scholar]
- Kurochkin IV. Insulin-degrading enzyme: embarking on amyloid destruction. Trends Biochem Sci. 2001;26:421–425. doi: 10.1016/s0968-0004(01)01876-x. [DOI] [PubMed] [Google Scholar]
- Sudoh S, Frosch MP, Wolf BA. Differential effects of proteases involved in intracellular degradation of amyloid beta-protein between detergent-soluble and -insoluble pools in CHO-695 cells. Biochemistry. 2002;41:1091–1099. doi: 10.1021/bi011193l. [DOI] [PubMed] [Google Scholar]
- Lue LF, Kuo YM, Roher AE, Brachova L, Shen Y, Sue L, Beach T, Kurth JH, Rydel RE, Rogers J. Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer’s disease. Am J Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Beyreuther K, Bush AI, Masters CL. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann Neurol. 1999;46:860–866. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Song E, Kihiko-Ehmann M, Goodman JP, Jr, Pyrek JS, Estus S, Hersh LB. Insulysin hydrolyzes amyloid beta peptides to products that are neither neurotoxic nor deposit on amyloid plaques. J Neurosci. 2000;20:8745–8749. doi: 10.1523/JNEUROSCI.20-23-08745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DG, Leverenz JB, McMillan PJ, Kulstad JJ, Ericksen S, Roth RA, Schellenberg GD, Jin LW, Kovacina KS, Craft S. Reduced hippocampal insulin-degrading enzyme in late-onset Alzheimer’s disease is associated with the apolipoprotein E-epsilon4 allele. Am J Pathol. 2003;162:313–319. doi: 10.1016/s0002-9440(10)63822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis MG, Go RC, Vekrellis K, Selkoe DJ, Saunders AJ, Tanzi RE. Evidence for genetic linkage of Alzheimer’s disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Allen FA, Jr, Goetz CG, Mastaglia F, Stajich JM, Gibson RA, Middleton LT, Saunders AM, Scott BL, Small GW, Nicodemus KK, Reed AD, Schmechel DE, Welsh-Bohmer KA, Conneally PM, Roses AD, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet. 2002;70:985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Saunders A, Mullin K, Sampson A, Moscarillo T, Becker D, Velicelebi G, Wanger S, Blacker D, Tanzi R. Further assessment of novel Alzheimer’s disease loci on chromosomes 10 and 9. Neurobiol Aging. 2002;23:S324. (Abstract) [Google Scholar]
- Ait-Ghezala G, Abdullah L, Crescentini R, Crawford F, Town T, Singh S, Richards D, Duara R, Mullan M. Confirmation of association between D10S583 and Alzheimer’s disease in a case-control sample. Neurosci Lett. 2002;325:87–90. doi: 10.1016/s0304-3940(02)00243-4. [DOI] [PubMed] [Google Scholar]
- Abraham R, Myers A, Wavrant-DeVrieze F, Hamshere ML, Thomas HV, Marshall H, Compton D, Spurlock G, Turic D, Hoogendoorn B, Kwon JM, Petersen RC, Tangalos E, Norton J, Morris JC, Bullock R, Liolitsa D, Lovestone S, Hardy J, Goate A, O’Donovan M, Williams J, Owen MJ, Jones L. Substantial linkage disequilibrium across the insulin-degrading enzyme locus but no association with late-onset Alzheimer’s disease. Hum Genet. 2001;109:646–652. doi: 10.1007/s00439-001-0614-1. [DOI] [PubMed] [Google Scholar]
- Boussaha M, Hannequin D, Verpillat P, Brice A, Frebourg T, Campion D. Polymorphisms of insulin degrading enzyme gene are not associated with Alzheimer’s disease. Neurosci Lett. 2002;329:121–123. doi: 10.1016/s0304-3940(02)00586-4. [DOI] [PubMed] [Google Scholar]
- Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze FW, Crook R, Hamshere M, Abraham R, Tunstall N, Rice F, Carty S, Lillystone S, Kehoe P, Rudrasingham V, Jones L, Lovestone S, Perez-Tur J, Williams J, Owen MJ, Hardy J, Goate AM. Susceptibility locus for Alzheimer’s disease on chromosome 10. Science. 2000;290:2304–2305. doi: 10.1126/science.290.5500.2304. [DOI] [PubMed] [Google Scholar]
- Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Science. 2000;290:2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, Mohlke KL, Silander K, Kohtamaki K, Chines P, Balow J, Jr, Birznieks G, Chang J, Eldridge W, Erdos MR, Karanjawala ZE, Knapp JI, Kudelko K, Martin C, Morales-Mena A, Musick A, Musick T, Pfahl C, Porter R, Rayman JB. The Finland-United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet. 2000;67:1174–1185. [PMC free article] [PubMed] [Google Scholar]
- Wiltshire S, Hattersley AT, Hitman GA, Walker M, Levy JC, Sampson M, O’Rahilly S, Frayling TM, Bell JI, Lathrop GM, Bennett A, Dhillon R, Fletcher C, Groves CJ, Jones E, Prestwich P, Simecek N, Rao PV, Wishart M, Bottazzo GF, Foxon R, Howell S, Smedley D, Cardon LR, Menzel S, McCarthy MI. A genomewide scan for loci predisposing to type 2 diabetes in a U. K. population (the Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet. 2001;69:553–569. doi: 10.1086/323249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamohamed S, Demissie S, Volcjak J, Liu C, Heard-Costa N, Liu J, Shoemaker CM, Panhuysen CI, Meigs JB, Wilson P, Atwood LD, Cupples LA, Herbert A. Polymorphisms in the insulin-degrading enzyme gene are associated with type 2 diabetes in men from the NHLBI Framingham Heart Study. Diabetes. 2003;52:1562–1567. doi: 10.2337/diabetes.52.6.1562. [DOI] [PubMed] [Google Scholar]
- Meigs JB, Panhuysen CI, Myers RH, Wilson PW, Cupples LA. A genome-wide scan for loci linked to plasma levels of glucose and HbA(1c) in a community-based sample of Caucasian pedigrees: the Framingham Offspring Study. Diabetes. 2002;51:833–840. doi: 10.2337/diabetes.51.3.833. [DOI] [PubMed] [Google Scholar]
- Fakhrai-Rad H, Nikoshkov A, Kamel A, Fernstrom M, Zierath JR, Norgren S, Luthman H, Galli J. Insulin-degrading enzyme identified as a candidate diabetes susceptibility gene in GK rats. Hum Mol Genet. 2000;9:2149–2158. doi: 10.1093/hmg/9.14.2149. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L. New frontiers in Alzheimer’s disease genetics. Neuron. 2001;32:181–184. doi: 10.1016/s0896-6273(01)00476-7. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, Podlisny MB, Joachim CL, Vickers EA, Lee G, Fritz LC, Oltersdorf T. β-amyloid precursor protein of Alzheimer disease occurs as 110–135 kilodalton membrane-associated proteins in neural and nonneural tissues. Proc Natl Acad Sci USA. 1988;85:7341–7345. doi: 10.1073/pnas.85.19.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlisny MB, Citron M, Amarante P, Sherrington R, Xia W, Zhang J, Diehl T, Levesque G, Fraser P, Haass C, Koo EHM, Seubert P, St. George-Hyslop P, Teplow DB, Selkoe DJ. Presenilin proteins undergo heterogeneous endoproteolysis between Thr291 and Ala299 and occur as stable N- and C-terminal fragments in normal and Alzheimer brain tissue. Neurobiol Dis. 1997;3:325–337. doi: 10.1006/nbdi.1997.0129. [DOI] [PubMed] [Google Scholar]
- Petanceska SS, Nagy V, Frail D, Gandy S. Ovariectomy and 17beta-estradiol modulate the levels of Alzheimer’s amyloid beta peptides in brain. Exp Gerontol. 2000;35:1317–1325. doi: 10.1016/s0531-5565(00)00157-1. [DOI] [PubMed] [Google Scholar]
- Duff K, Eckman C, Zehr C, Yu X, Prada C-M, Perez-Tur J, Hutton M, Buee L, Harigaya Y, Yager D, Morgan D, Gordon MN, Holcomb L, Refolo L, Zenk B, Hardy J, Younkin S. Increased amyloid-β42(43) in brains of mice expressing mutant presenilin 1. Nature. 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- Sun X, Sato S, Murayama O, Murayama M, Park JM, Yamaguchi H, Takashima A. Lithium inhibits amyloid secretion in COS7 cells transfected with amyloid precursor protein C100. Neurosci Lett. 2002;321:61–64. doi: 10.1016/s0304-3940(01)02583-6. [DOI] [PubMed] [Google Scholar]
- Johnson-Wood K, Lee M, Motter R, Hu K, Gordon G, Barbour R, Khan K, Gordon M, Tan H, Games D, Lieberburg I, Schenk D, Seubert P, Mconlogue L. Amyloid precursor protein processing and Aβ42 deposition in a transgenic mouse model of Alzheimer disease. Proc Natl Acad Sci USA. 1997;94:1550–1555. doi: 10.1073/pnas.94.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Ray WJ, Ostaszewski BL, Rahmati T, Kimberly WT, Wolfe MS, Zhange J, Goate AM, Selkoe DJ. Presenilin complexes with the C-terminal fragments of amyloid precursor protein at the sites of amyloid β-protein generation. Proc Natl Acad Sci USA. 2000;97:9299–9304. doi: 10.1073/pnas.97.16.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Kakizaki M, Masaki N. Spontaneous diabetes produced by selective breeding of normal Wistar rats. Proc Jpn Acad. 1975;51:80–85. [Google Scholar]
- Koo EH, Squazzo S. Evidence that production and release of amyloid β-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- Leissring MA, Lu A, Condron MM, Teplow DB, Stein RL, Farris W, Selkoe DJ. Kinetics of amyloid {beta}-protein degradation determined by novel fluorescence- and fluorescence polarization-based assays. J Biol Chem. 2003;278:37314–37320. doi: 10.1074/jbc.M305627200. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T, Loike JD, Brionne TC, Lu E, Anankov R, Yan F, Silverstein SC, Husemann J. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat Med. 2003;9:453–457. doi: 10.1038/nm838. [DOI] [PubMed] [Google Scholar]
- Rogers J, Strohmeyer R, Kovelowski CJ, Li R. Microglia and inflammatory mechanisms in the clearance of amyloid beta peptide. Glia. 2002;40:260–269. doi: 10.1002/glia.10153. [DOI] [PubMed] [Google Scholar]
- Schulingkamp RJ, Pagano TC, Hung D, Raffa RB. Insulin receptors and insulin action in the brain: review and clinical implications. Neurosci Biobehav Rev. 2000;24:855–872. doi: 10.1016/s0149-7634(00)00040-3. [DOI] [PubMed] [Google Scholar]
- Ott A, Stolk RP, van Harskamp F, Pols HA, Hofman A, Breteler MM. Diabetes mellitus and the risk of dementia: the Rotterdam Study. Neurology. 1999;53:1937–1942. doi: 10.1212/wnl.53.9.1937. [DOI] [PubMed] [Google Scholar]
- Leibson CL, Rocca WA, Hanson VA, Cha R, Kokmen E, O’Brien PC, Palumbo PJ. Risk of dementia among persons with diabetes mellitus: a population-based cohort study. Am J Epidemiol. 1997;145:301–308. doi: 10.1093/oxfordjournals.aje.a009106. [DOI] [PubMed] [Google Scholar]
- Luchsinger JA, Tang MX, Stern Y, Shea S, Mayeux R. Diabetes mellitus and risk of Alzheimer’s disease and dementia with stroke in a multiethnic cohort. Am J Epidemiol. 2001;154:635–641. doi: 10.1093/aje/154.7.635. [DOI] [PubMed] [Google Scholar]
- Curb JD, Rodriguez BL, Abbott RD, Petrovitch H, Ross GW, Masaki KH, Foley D, Blanchette PL, Harris T, Chen R, White LR. Longitudinal association of vascular and Alzheimer’s dementias, diabetes, and glucose tolerance. Neurology. 1999;52:971–975. doi: 10.1212/wnl.52.5.971. [DOI] [PubMed] [Google Scholar]
- Peila R, Rodriguez BL, Launer LJ. Type 2 diabetes, APOE gene, and the risk for dementia and related pathologies: the Honolulu-Asia Aging Study. Diabetes. 2002;51:1256–1262. doi: 10.2337/diabetes.51.4.1256. [DOI] [PubMed] [Google Scholar]
- Stolk RP, Breteler MM, Ott A, Pols HA, Lamberts SW, Grobbee DE, Hofman A. Insulin and cognitive function in an elderly population. The Rotterdam Study. Diabetes Care. 1997;20:792–795. doi: 10.2337/diacare.20.5.792. [DOI] [PubMed] [Google Scholar]
- Razay G, Wilcock GK. Hyperinsulinaemia and Alzheimer’s disease. Age Ageing. 1994;23:396–399. doi: 10.1093/ageing/23.5.396. [DOI] [PubMed] [Google Scholar]
- Kuusisto J, Koivisto K, Mykkanen L, Helkala EL, Vanhanen M, Hanninen T, Kervinen K, Kesaniemi YA, Riekkinen PJ, Laakso M. Association between features of the insulin resistance syndrome and Alzheimer’s disease independently of apolipoprotein E4 phenotype: cross sectional population based study. BMJ. 1997;315:1045–1049. doi: 10.1136/bmj.315.7115.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft S, Newcomer J, Kanne S, Dagogo-Jack S, Cryer P, Sheline Y, Luby J, Dagogo-Jack A, Alderson A. Memory improvement following induced hyperinsulinemia in Alzheimer’s disease. Neurobiol Aging. 1996;17:123–130. doi: 10.1016/0197-4580(95)02002-0. [DOI] [PubMed] [Google Scholar]