Abstract
In recent years, culture-independent methods have been used in preference to traditional isolation techniques for microbial community analysis. However, it is questionable whether uncultured organisms from a given sample are important for determining the impact of anthropogenic stress on indigenous communities. To investigate this, soil samples were taken from a site with patchy metal contamination, and the bacterial community structure was assessed with a variety of approaches. There were small differences in microscopic epifluorescence bacterial counts. Denaturing gradient gel electrophoresis (DGGE) profiles of 16S rRNA gene fragments (16S-DGGE) amplified directly from soil samples were highly similar. A clone library generated from the most contaminated sample revealed a diverse bacterial community, which showed similarities to pristine soil communities from other studies. However, the proportion of bacteria from the soil samples that were culturable on standard plate-counting media varied between 0.08 and 2.2%, and these values correlated negatively with metal concentrations. The culturable communities from each sample were compared by 16S-DGGE of plate washes and by fatty acid profiling of individual isolates. Each approach indicated that there were considerable differences between the compositions of the culturable communities from each sample. DGGE bands from both culture-based and culture-independent approaches were sequenced and compared. These data indicated that metal contamination did not have a significant effect on the total genetic diversity present but affected physiological status, so that the number of bacteria capable of responding to laboratory culture and their taxonomic distribution were altered. Thus, it appears that plate counts may be a more appropriate method for determining the effect of heavy metals on soil bacteria than culture-independent approaches.
The use of microbial communities to ascertain the impact caused by anthropogenic stress in natural habitats is increasing. However, there is considerable discussion as to the most valid approach (5). While it has been stated that the assessment of community diversity with nucleic acid-based methods is more appropriate than with those based on culturing organisms (27), there has been little work to qualify this supposition. It is often shown that culture-independent methods realize greater complexity than traditionally found (38), but the relevance of this, in terms of ecosystem function, is rarely discussed (24). Obviously, the use of culture-independent approaches will remove the bias imposed by isolation of bacteria on laboratory media but, conversely, will fail to take into account differences in cellular activity (21). As many bacteria can exist in dormant forms (17), it is important to measure activity so that ecologically relevant bacteria are assessed and not inactive cells that do not contribute to ecosystem function.
Culture-independent methods are receiving particular attention because it is commonly held that only a small proportion of bacteria present in any environment will form colonies on general laboratory media. Although methods and media that improve on the proportion of bacteria that can be cultured from environmental samples are now coming to light (15, 29, 39), this study was based on bacteria isolated by traditional plate culture methods and, as such, focuses on what are termed readily culturable bacteria. The reason for the discrepancy between culturable and direct microscopic counts of bacteria is not fully understood (14). However, there are several pieces of evidence which, when combined, suggest that the readily culturable component of soil microbial communities may be the most important in terms of both biomass and activity.
It has been shown that there are positive correlations between activity and cell size (3), cell size and culturability (1, 2), and activity and culturability (33). Thus, while they may carry a large proportion of the genetic material, the numerically dominant component of soil bacterial communities (consisting of very small cells [15]) does not contribute greatly to biomass or metabolic activity (1). In addition, differences in the proportion of bacteria that are isolated from soil samples from increasing depths have been attributed to differences in bacterial activity (29). Given these pieces of evidence, it could be hypothesized that the readily culturable component of soil bacteria may be those that are the greatest contributors to their ecosystem.
We recently identified a relationship between the number of colonies that developed on generalized media and heavy metal contamination in soils from an industrial site and found that these plate counts were a useful indicator of the impact associated with the contamination (8). Further work determined that the number of bacteria counted by epifluorescence microscopy was not affected by the contamination (7). As the culturable portion of the bacterial community appeared to be affected to a greater extent by the metal contamination than the community as a whole, it was decided to investigate the basis for this differential response.
The diversity of the cultured and total communities was not examined in the previous work (7, 8), and therefore no comment could be made on the selectivity of each of the approaches. Therefore, the aim of this work was to determine the link between the diversity of the culturable portion of the communities and that obtained by direct amplification of rRNA genes from soil samples. In this study, denaturing gradient gel electrophoresis (DGGE) (26) was used to compare the diversity of 16S rRNA gene amplicons obtained directly from soil samples (total community) with those of the bacterial isolates from the same samples (plate wash). In this way, it was hoped that the importance of culturability for determining the impact of heavy metal contamination could be elucidated.
MATERIALS AND METHODS
Soil samples.
Soil cores (20-cm depth by 6-cm diameter) were taken in October 1999 from a sandy site in Scotland (United Kingdom) that had a history of patchy contamination with heavy metals because waste from an explosives factory was burnt there for over 20 years until the late 1950s (7, 8). The samples used in this study were 25 m apart from a northeast to northwest transect across the site, and samples were numbered 11 to 15, where sample 11 was from the most northeasterly site (7). Cores were mixed in plastic bags, and subsamples (10 g) were stored (where necessary) at 4°C in sterile universal containers. All subsequent samples for microbiological and molecular analyses were taken from these. The metal contents of the samples were determined by microwave digestion followed by inductively coupled plasma analysis (IRIS; Thermo Electron Corp., Waltham, Mass.) (36). Approximately 3 to 4 g of soil sample was placed in foil packets and weighed, and the packets were then kept at 80°C and reweighed daily until no further weight loss was detected. The proportion of water in each soil sample was then calculated. Quantitative data obtained were corrected for these dry-weight estimations.
Isolation and identification of culturable organisms.
Triplicate samples (1 g) were taken from each core subsample, homogenized in 10 ml of 0.85% (wt/vol) saline, and serially diluted (10-fold) in the same. Aliquots (100 μl) were spread on high-nutrient-concentration tryptic soy broth agar (TSBA; Difco) and lower-nutrient R2A medium for heterotrophs (Difco) (28). The number of colonies forming on each medium was counted 2 days and 5 days after inoculation after incubation at 20°C. Dilution plates that carried between 50 and 500 colonies were counted on day 2. The same plates were also counted on day 5, and these plates were used for all further work.
From each sample (5 cores times 3 replicates = 15), 45 colonies were picked at random from the R2A plates onto fresh TSBA plates. Once the purity of each of these cultures had been verified, they were stored as glycerol stocks at −80°C as described previously (9). Each isolate that survived the subculture step was identified by fatty acid methyl ester (FAME) analysis with the Microbial Identification System (MIS; Microbial Identification Inc., Delaware) as described previously (35). After picking colonies, the remaining bacterial biomass was removed from the counted plates (R2A) and transferred to microcentrifuge tubes for subsequent DNA extraction.
Total bacterial counts.
Subsamples (1 g) were taken from each core for assessing the total number of bacteria. To this was added 9 ml of filter-sterilized saline (0.85% NaCl) and 1 ml of filter-sterilized formalin (37% formaldehyde). The fixed samples were then stored at 4°C for up to 1 week. Essentially, soil suspensions were stained with 5-(4,6-dichlorotriazine-2-yl)aminofluorescein (DTAF) as described by Bloem (4). The slides were examined by epifluorescence microscopy with a Zeiss microscope. Objects shaped like bacteria were counted under 800× magnification. The number of fields of view required to count 300 bacterium-shaped objects was recorded, and the number of bacteria per gram of soil was then calculated.
DNA extraction methods.
DNA was extracted from the bacterial biomass from plates by the cetyltrimethylammonium bromide (CTAB) method (9). This method was adapted to extract DNA from soil. Soil (250 mg) was added to 250 mg of acid-washed glass beads (0.4 to 0.52 mm in diameter; Stratagene, Texas) in a 2-ml microcentrifuge tube. Buffer A (1 ml of 100 mM Tris-HCl-100 mM EDTA [pH 8.0]-100 mM phosphate buffer [pH 8.0]-1.5 M NaCl-1% CTAB) (40) was added with 175 μg of proteinase K and 1 mg of lysozyme. This was then agitated on a platform shaker at maximum speed for 20 min. Subsequently, 120 μl of 20% sodium dodecyl sulfate was added, and the samples were incubated at 65°C for 30 min. The samples were then centrifuged at 2,800 × g for 2 min. The supernatant was transferred to a fresh tube, and the pellet was reextracted with 300 μl of buffer A and centrifuged again. The combined supernatants were extracted with an equal volume of chloroform-isoamyl alcohol (24:1, vol/vol). The aqueous phase was precipitated with 0.6 volume of isopropanol at −20°C for 30 min. The pellet was washed in 300 μl of 70% (vol/vol) ethanol and air dried before resuspending in 100 μl of 10 mM Tris (pH 8.5).
Due to the high metal content of the samples, the DNA preparations were purified by gel electrophoresis, which also served to separate humic acids from the DNA. Aliquots (50 μl) were run on a 0.7% agarose gel at 60 V for 2 h. DNA bands of approximately 10 to 30 kb were excised from the gel, and the DNA was extracted with the Qiaquick gel extraction kit (Qiagen Ltd., United Kingdom). Essentially, the gel slice was solubilized and bound to glass particles in solution at <pH 7.5. It was then washed and eluted in 10 mM Tris (pH 8.5). This was then used directly for subsequent PCR amplification.
PCR amplification of extracted DNA.
Two approaches were used for the amplification of 16S rRNA gene fragments suitable for analysis by denaturing gradient gel electrophoresis. Either the DGGE primers GC-341F (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG) and 534R (5′-ATTACCGCGGCTGCTGG) (26) were used to directly amplify from the DNA samples, or almost full-length 16S rRNA gene sequences were amplified with the primers 63F (CAG GCC TAA CAC ATG CAA GTC) and 1387R (GGG CGG WGT GTA CAA GGC) (22). In each case, the PCR mix consisted of deoxynucleoside triphosphates at 200 μM each, 0.25 μM each primer, 3 mM MgCl2, 1× PCR buffer, and 1 U of HotStarTaq DNA polymerase (Qiagen Ltd., United Kingdom) in a total volume of 50 μl. Approximately 200 ng of target DNA was added to each reaction. The following cycle conditions were used for the DGGE primer pair: 95°C for 15 min (for enzyme activation and target denaturation), followed by 20 cycles of 95°C for 1 min, 65°C (reduced by 0.5°C each cycle) for 45 s, and 72°C for 1 min; 10 cycles of 95°C for 1 min, 55°C for 45 s, and 72°C for 1 min; and a final extension at 72°C for 5 min. The conditions for the amplification of full-length 16S rRNA genes were: 95°C for 15 min (for enzyme activation and target denaturation), followed by 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min 30 s, and a final extension at 72°C for 5 min.
DGGE analysis.
The DCode System for DGGE (Bio-Rad Laboratories Ltd., Hertfordshire, United Kingdom) was used. Samples were loaded onto 10% polyacrylamide-bisacrylamide (37.5:1) gels with denaturing gradients from 35 to 60% (where 100% is 7 M urea and 40% [vol/vol] deionized formamide) in 1× TAE electrophoresis buffer. Each gel also included at least two marker lanes consisting of fragments amplified from isolated bacteria or cloned directly from amplified 16S rRNA gene fragments, both from the site being studied. The phylogenetic affiliations were determined by FAME for isolates and comparative sequence analysis for cloned fragments as Arthrobacter sp., Bacillus sp., Cytophaga sp., Nevskia sp., Nocardia sp., Pseudomonas sp., Ralstonia sp., and Rhodoplanes sp. Electrophoresis was performed at 180 V at a temperature of 58°C for 4.5 h. Gels were then stained with SBYR Gold (Cambridge BioScience, United Kingdom) in 1× TAE for 30 min at room temperature and visualized under UV illumination. Bands of interest were excised, and DNA was eluted with an equal volume of diffusion buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA [pH 8.0], and 0.1% [wt/vol] sodium dodecyl sulfate) at 50°C for 30 min.
The resulting solution (2 μl) was used as target DNA for a subsequent PCR amplification with primers 341F (CCTACGGGAGGCAGCAG) and 534R (26). The purity and correct running position of each fragment was confirmed by further DGGE. When the confirmation was ambiguous, reamplified PCR products were cloned into the pGEM-T Easy vector (Promega) as described by the manufacturer. Clones with the correct migration on DGGE gels were sequenced. These short fragments were compared to sequences in the Ribosomal Database Project (RDP) database (22), and the closest match of known phylogenetic affiliation was used to assign the bands to taxonomic groups.
Clone library and sequence analysis.
Almost full-length16S rRNA genes were amplified from sample 13 with the 63F and 1387R primers under the conditions described above. The mixed PCR products from three replicate PCRs were cloned into the pGEM-T Easy vector (Promega) in duplicate as described by the manufacturer. Single colonies containing inserts were selected at random, and the inserts were sequenced in the forward and reverse directions. These sequences were aligned with the closest matches found in the Ribosomal Database Project (22) with the ClustalW function of the BioEdit package (13). Neighbor-joining phylogenetic trees were constructed with the Molecular Evolutionary Genetics Analysis package (MEGA version 2.1) (19) with the Jukes-Cantor algorithm, and the robustness of the phylogeny was tested by bootstrap analysis with 500 iterations.
Data analysis.
All bacterial counts were log10 (x + 1) transformed to normalize before analysis of variance. Significant differences between means were determined by calculation of minimum significant differences (10). Dendrograms for comparison of DGGE banding patterns were constructed with the unweighted pair group method with arithmetic averages following the pairwise calculation of Dice coefficients according to the presence and absence of matching bands (6). Coverage was calculated as described previously (23).
Accession numbers and nomenclature.
The 39 rRNA gene clones sequenced in this study were submitted to GenBank with accession numbers AY102308 to AY102345 and AF531538. The sequences of the bands excised from the DGGE gels were given accession numbers AF531506 to AF531537. The accession numbers of database entries used to construct trees and identify sequences are noted in tables and figures where appropriate. The taxonomic names used here are those given in the 2002 Taxonomic Outline for the Procaryotes (12).
RESULTS
Physicochemical analysis of soil samples.
The physicochemical characteristics of each of the soil samples are shown in Table 1. The metal concentrations divided the samples into two groups, with 11, 12, and 13 having particularly high concentrations of copper, lead, and zinc. The only metal for which samples 14 and 15 were higher than the other three was arsenic. Despite the high metal concentrations in samples 12 and 13, the pH was higher than in the other samples.
TABLE 1.
Physicochemical analysisa of soil samples
| Physiochemical variable | pH or concn (mg/kg) in soil sample no:
|
||||
|---|---|---|---|---|---|
| 11 | 12 | 13 | 14 | 15 | |
| pH | 6.8 | 7.4 | 7.4 | 6.1 | 6.6 |
| TOC | 8,100 | 10,100 | 5,700 | 6,400 | 12,900 |
| As | 41 | 43 | 38 | 82 | 83 |
| Cd | 26 | 23 | 17 | 4 | 6 |
| Cr | 212 | 221 | 269 | 89 | 64 |
| Cu | 10,300 | 13,100 | 14,400 | 628 | 516 |
| Hg | 18 | 43 | 23 | 3 | <1 |
| Ni | 142 | 148 | 140 | 65 | 97 |
| Pb | 18,800 | 19,300 | 12,800 | 1,210 | 925 |
| Zn | 16,300 | 14,000 | 9,640 | 671 | 906 |
Total organic carbon (TOC) and metal concentrations are given in milligrams per kilogram.
Size of bacterial communities.
Plate count analysis revealed a large discrepancy in the number of bacteria that could be cultured from the five soil samples. Numbers in samples 12 and 13 were significantly lower than in the other samples for both TSBA and R2A (Table 2). Although there were differences between the microscopic direct counts, the relative difference was very small (fourfold) compared with the difference between the plate counts (100-fold). Thus, significant differences in the percentage of culturable bacteria in each sample were observed (Table 2). These values corresponded to the extent of the heavy metal contamination in the samples. That is, culturability was significantly reduced in the samples with the greatest metal content.
TABLE 2.
Mean numbers of bacteria in soil samples (per gram dry weight) determined by plate counts and epifluorescence microscopya
| Variable (medium/method) | Value in soil sample no:
|
||||
|---|---|---|---|---|---|
| 11 | 12 | 13 | 14 | 15 | |
| Log10 CFU (TSBA) | 6.10 c | 5.31 d | 5.07 d | 6.47 b | 7.26 a |
| Log10 CFU (R2A) | 6.17 b | 5.62 c | 5.45 c | 6.60 b | 7.45 a |
| Log10 bacteria (DTAF) | 8.57 ab | 8.51 b | 8.52 b | 8.90 a | 9.09 a |
| % culturable (R2A/ DTAF × 100) | 0.40 b | 0.13 c | 0.08 c | 0.51 b | 2.27 a |
Values within rows which have a common letter suffix did not differ at a probability of 5%. These were determined from calculations of minimum significant differences following analysis of variance.
Diversity of cultured bacteria.
Many isolates (n = 501) from the five soil samples survived subculturing and were identified by FAME analysis. Due to the low similarity indices obtained for some isolates (less that 0.2 in some cases) and the inherent limitations of fatty acid profiling, these data were used to divide the isolates into broad taxonomic groupings. The relative abundance of the taxonomic groups is shown in Table 3. A total of 20 genera were tentatively identified from the five samples and used to assign each of the isolates to broad taxonomic groups: these were Arthrobacter, Bacillus, Brevibacterium, Brochothrix, Comamonas, Cytophaga, Deinococcus, Enterobacter, Hafnia, Micrococcus, Mycobacterium, Nocardia, Pseudomonas, Rathayibacter, Rhodococcus, Salmonella, Serratia, Staphylococcus, Variovorax, and Xanthomonas. It can be seen that there were large differences between samples in the relative abundance of the different taxonomic groups (Table 3). These differences appeared to correlate with the amount of metal in the samples. In particular, Firmicutes (mainly Bacillus spp.) had a higher relative abundance in the most contaminated samples (12 and 13), whereas the Gammaproteobacteria (mainly Pseudomonas spp. and a Xanthomonas sp.) increased in relative abundance in the least-contaminated samples (14 and 15). With species identity provided by the MIS, the coverage of these isolate collections was calculated to be 82.8%.
TABLE 3.
Mean relative abundance distribution of taxonomic groups (identified by FAME profiling) within the collection of isolates (n = 501) from soil samplesa
| Group | Mean abundance, % (SE) in sample no:
|
||||
|---|---|---|---|---|---|
| 11 | 12 | 13 | 14 | 15 | |
| Bacteroidetes | 25.9 (4.5) | 0 | 1.9 (0.9) | 1.0 (1.0) | 0 |
| Actinobacteria | 15.7 (5.9) | 15.7 (0.8) | 11.1 (3.9) | 3.1 (1.3) | 3.4 (2.2) |
| Firmicutes | 45.4 (6.5) | 73.2 (6.2) | 69.4 (2.8) | 51.5 (5.3) | 44.5 (6.9) |
| Proteobacteria | |||||
| Alphaproteobacteria | 0 | 0 | 0 | 0 | 0 |
| Betaproteobacteria | 4.6 (1.0) | 0 | 6.5 (3.4) | 0 | 1.7 (0.8) |
| Gammaproteobacteria | 5.6 (3.0) | 6.3 (4.8) | 10.2 (4.6) | 40.2 (1.7) | 50.4 (4.9) |
| Deltaproteobacteria | 0 | 0 | 0 | 0 | 0 |
| Deinococcus-Thermus | 0.9 (0.5) | 0 | 0 | 0 | 0 |
| Unknown | 1.9 (2.4) | 4.9 (2.1) | 0.9 (0.5) | 4.1 (1.1) | 0 |
Numbers in parentheses indicate the standard error (n = 3). Due to the loss of some isolates on subculturing, there were different numbers of isolates for each replicate.
Diversity of short 16S rRNA gene amplicons from DGGE gels.
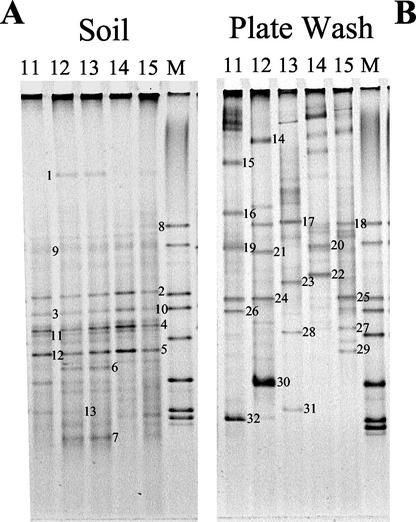
DGGE indicated that when with direct amplification of rRNA genes from soil, the bacterial community structure appeared to be similar (but not identical) in all samples (Fig. 1A). However, it should be noted that the apparent diversity was relatively low in all samples. In contrast, the profiles from the cultured bacteria (plate washes) were very different from one another and from their corresponding total community profile (Fig. 1B).
FIG. 1.
DGGE gels showing diversity of 16S rRNA gene fragments amplified from (A) DNA extracted directly from soil (total community) and (B) DNA extracted from biomass accumulated on agar plate (plate wash). M, marker lane consisting of 16S rRNA gene fragments from (listed top to bottom) Pseudomonas sp., Ralstonia sp., Bacillus sp., Cytophaga sp., Rhodoplanes sp., Arthrobacter sp., Nevskia sp., and Nocardia sp. Lanes are labeled with sample numbers. Numbers on gels are to the right of bands that were excised and sequenced and correspond to the list in Table 4.
The 32 bands labeled in Fig. 1 were excised from the gels and subsequently sequenced. These gave short sequences, but they were processed to give their approximate phylogenetic affiliation based on around 180 bases of the V3 region of the 16S rRNA gene. This allowed the structures of the amplified and cultured communities to be compared. Sequencing of these bands confirmed that the major dense bands from soil DNA were identical in all lanes (e.g., bands 2, 3, 4, 5, 9, 10, 11, and 12 were common to all lanes) and that several fainter bands were also common to all five lanes, indicating that the communities were dominated by the same phylotypes in all samples. Those bands that migrated similar distances were very similar to one another (e.g., bands 3 and 10 and bands 5 and 12, Table 4). However, additional bands were observed in the most contaminated samples, 12 and 13 (bands 1, 6, 7, and 13, Deltaproteobacteria and Actinobacteria). Band 8 (Gammaproteobacteria) was only observed in the least-contaminated sample (sample 15).
TABLE 4.
Assignment of taxonomic groups to band sequences extracted from a DGGE gel based on ∼180 bp and the closest sequence match of known phylogenetic affiliation
| Banda | Laneb | Identityc | Taxonomic group | Similarityd |
|---|---|---|---|---|
| 1 | S12,13 | Nitrospina environmental clone (AF173822) | Deltaproteobacteria | 0.959 (287) |
| 2 | S11-15 | Comamonas sp. (AF149849) | Betaproteobacteria | 1.000 (253) |
| 3 | S11-15 | Pseudomonas coronafaciens (Z76660) | Gammaproteobacteria | 0.924 (279) |
| 4 | S11-15 | Denitrobacter permanens (Y12639) | Betaproteobacteria | 0.947 (230) |
| 5 | S11-15 | Serratia marcescens (AF124035) | Gammaproteobacteria | 0.975 (307) |
| 6 | S12-13 | Streptomycoides glaucoflavus (AB006155) | Actinobacteria | 0.894 (160) |
| 7 | S12-13 | Thermobispora bispora (U83911) | Actinobacteria | 0.925 (135) |
| 8 | S15 | Pseudomonas putida (AF095892) | Gammaproteobacteria | 0.994 (303) |
| 9 | S11-15 | Pseudomonas fluorescens (D86003) | Gammaproteobacteria | 0.993 (302) |
| 10 | S11-15 | Pseudomonas coronafaciens (Z76660) | Gammaproteobacteria | 0.925 (284) |
| 11 | S11-15 | Mesorhizobium loti (X67230) | Alphaproteobacteria | 0.985 (301) |
| 12 | S11-15 | Rahnella sp. (U90758) | Gammaproteobacteria | 0.991 (223) |
| 13 | S11-12 | Propionibacterium acnes (M61903) | Actinobacteria | 0.993 (307) |
| 14 | P12 | Pseudomonas putida (D85995) | Gammaproteobacteria | 0.994 (303) |
| 15 | P11 | Flavobacterium xylanivorum (AF162266) | Bacteroidetes | 0.974 (312) |
| 16 | P11 | Flavobacterium columnare (M58781) | Bacteroidetes | 0.968 (309) |
| 17 | P13 | Bacillus fusiformis (L14013) | Firmicutes | 0.988 (315) |
| 18 | P15 | Pseudomonas putida (D86000) | Gammaproteobacteria | 0.953 (289) |
| 19 | P11 | Flavobacterium columnare (M58781) | Bacteroidetes | 0.974 (300) |
| 20 | P14 | Flavobacterium columnare (M58781) | Bacteroidetes | 0.951 (294) |
| 21 | P12 | Pseudomonas sp. (AJ002801) | Gammaproteobacteria | 0.961 (301) |
| 22 | P14 | Flavobacterium johnsoniae (M59053) | Bacteroidetes | 0.922 (304) |
| 23 | P13 | Pseudomonas sp. (AB030085) | Gammaproteobacteria | 0.913 (260) |
| 24 | P11-12 | Bacillus cereus (Z84577) | Firmicutes | 0.987 (300) |
| 25 | P15 | Bacillus thuringiensis (AF157112) | Firmicutes | 0.959 (280) |
| 26 | P11 | Sporocytophaga cauliformis (M93151) | Bacteroidetes | 0.992 (270) |
| 27 | P15 | Mesorhizobium loti (X67230) | Alphaproteobacteria | 0.961 (300) |
| 28 | P13 | Stentrophomonas maltophilia (X95924) | Gammaproteobacteria | 0.961 (296) |
| 29 | P15 | Rahnella sp. (U90758) | Gammaproteobacteria | 0.924 (320) |
| 30 | P12 | Arthrobacter globiformis (X80736) | Actinobacteria | 1.000 (303) |
| 31 | P13 | Magnetospirillum sp. (AF050531) | Alphaproteobacteria | 0.985 (304) |
| 32 | P11 | Propionibacterium acnes (M61903) | Actinobacteria | 0.978 (298) |
Band, number as indicated on Fig. 1.
Prefix, P indicates plate wash, and prefix S indicates soil extraction. Numbers indicate sample numbers.
Closest match to band sequence obtained by comparison with RDP release 8.1 (22). Numbers in parentheses indicate the GenBank accession number.
Similarity calculated with the RDP similarity matrix facility (22) between the database entry and band sequence. Numbers in parentheses indicate the number of alignment positions used.
Bands 14 to 32 were from the plate washes and were dominated by bands corresponding to Firmicutes and the Bacteroidetes. However, it appears that some of the dominant bands from the soil DNA were also found in the plate washes. These were Mesorhizobium loti (bands 11 and 27), Propionibacterium acnes (bands 13 and 32), Pseudomonas putida (bands 8 and 18), and a Rahnella sp. (bands 12 and 29). The majority of these matches came from plate wash sample 15, the culturability of which was significantly higher than that of other samples. Of these, only those types similar to Mesorhizobium loti (bands 11 and 27) were also seen in the clone library (a13113).
With the information above, it was possible to determine the similarity of the profiles based on the presence or absence of bands with the Dice coefficient. A dendrogram was constructed (data not shown) which illustrated a high degree of similarity between the total soil DNA banding profiles (Fig. 1A), which supported the above information, showing that plate wash sample 15 was most similar to the total soil profiles.
Phylogenetic analysis of 16S rRNA gene clone library.
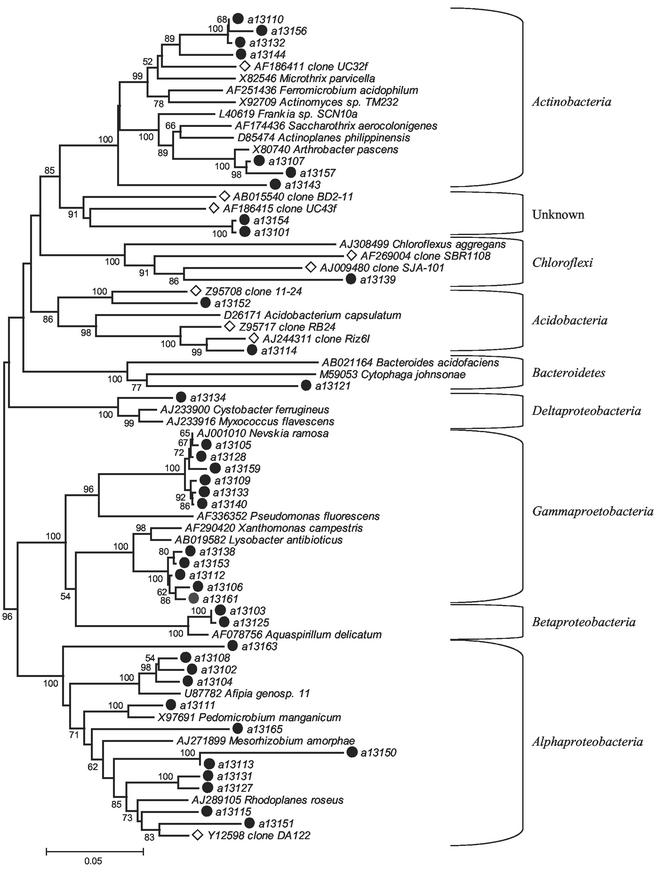
Given that DGGE revealed that the diversity of 16S rRNA gene amplicons from directly extracted DNA was similar for all five samples, a clone library was constructed from sample 13, which had the greatest heavy metal content. Thirty-nine 16S rRNA gene clones of over 1,300 bp amplified from DNA extracted directly from this soil were isolated and sequenced. Clones with similarity to the Actinobacteria, Bacteroidetes, Acidobacteria, and Chloroflexi phyla and to four classes within the Proteobacteria phylum were identified. Based on a similarity of >97%, indicating identical clones, coverage was 30.7%. A phylogenetic reconstruction of these clone sequences, together with sequences from their closest relatives from the RDP database is shown in Fig. 2. The distribution of phyla was very different from that of the collection of isolates from sample 13 identified by FAME profiling. In particular, no Bacillus species were identified in the clone library. Instead, the clone library was dominated by Alphaproteobacteria, which were not evident in the collection of isolates.
FIG. 2.
Neighbor-joining tree showing phylogeny of 16S rRNA gene sequences cloned directly from total soil DNA from sample 13 and selected at random. Sequences were aligned with ClustalW, and distances were calculated with the Jukes and Cantor algorithm. Sequences marked • are those from this study, and those marked ◊ are from cloned fragments from other studies. Bar indicates 5% sequence divergence. Aligned sequences were 1,547 bp in length. Bootstrap values above 50% are shown.
DISCUSSION
The relative change in epifluorescence counts of bacteria between the samples was small compared to the change in the proportion of those bacteria that formed colonies on laboratory media (Table 2). This proportion was negatively correlated with the amount of metal contamination in the sample (Table 1), and such an effect has been observed before (28). The observation that the total bacterial population was largely unaffected by the metal inputs was corroborated because DGGE profiles from directly extracted DNA showed the same dominant bands (Fig. 1A). Although differences in DGGE profiles from metal-contaminated soils have been seen previously (18), the differences are often minor.
The 16S rRNA gene clone library generated from sample 13 contained diverse sequences from a variety of phylogenetic groups (Fig. 2). However, a large proportion of the clones (30%) were identified as Alphaproteobacteria, and although related to the genera Afipia, Bradyrhizobium, Hyphomicrobium, Mesorhizobium, and Rhodoplanes, no close relatives were found in the sequence databases, so they were probably from uncultured groups. The dominance of Alphaproteobacteria in clone libraries from relatively pristine soils has been observed previously (24), implying that the metals in this study did not have a great effect on diversity. Of particular note was the striking similarity between the clone sequences from this study and those amplified from the oilseed rape rhizoplane (16). As in our study, the rhizoplane clone library was dominated by Alphaproteobacteria, with smaller proportions of clones from the Bacteroidetes, Betaproteobacteria, Gammaproteobacteria, and Actinobacteria.
Sample 13 came from an area of soil that was heavily contaminated with metals and had been devoid of vegetation for many years (7, 8). The similarity of the oil seed rape rhizoplane diversity and the diversity that we found was remarkable, as the soils, DNA extraction methods, and PCR primers were all different. This apparently contradicts the theory that different primer combinations will affect the diversity found (31), but it does not rule out the possibility that the diversity observed in these samples could have been increased by the use of additional PCR primers.
The Chloroflexi and Acidobacteria clusters (Fig. 2) also contained sequences that were distinct from those of any cultured organisms in the database, and these groups are common in other soil clone libraries (11, 24, 32). In addition, four sequences were found (a13101, a13143, a13154, and a13163) that were deep branching and did not appear to fall within any of the defined divisions or subdivisions and so may represent novel lineages. However, given that the theoretical coverage of the clone library was only 30%, it is probable that the community is more diverse than indicated here. This suggests that bacterial communities in a variety of soils may be very similar when assessed by molecular methods.
In contrast to the clone library, the collection of isolates from sample 13 was dominated by Bacillus spp. from the Firmicutes (Tables 3 and 5) but did not contain Alphaproteobacteria or Deltaproteobacteria, probably because members of these groups are generally slow-growing (25), or they may have specific physiological requirements and therefore may have been lost during subculture. This contrast between plated and total communities is a common phenomenon. The inability to detect Firmicutes in clone libraries despite their presence on plates has been observed previously (16). When Firmicutes were found in clone libraries, they were at a much lower proportion than in plated communities (30). Several reasons have been put forward to explain this, such as PCR bias, the difficulty of lysing spores, and the ease with which these bacteria grow on plates (34).
TABLE 5.
Comparison of distribution of phylogenetic groups of Bacteria detected in soil samples by different methodologies
| Phylogenetic group | Distribution (%)
|
|||
|---|---|---|---|---|
| Culture-based approach
|
Culture-independent approach
|
|||
| FAME | DGGE | Clone librarya | DGGE | |
| Bacteroidetes | 6 | 30 | 3 | 0 |
| Actinobacteria | 10 | 10 | 15 | 14 |
| Firmicutes | 55 | 20 | 0 | 0 |
| Proteobacteria | ||||
| Alphaproteobacteria | 0 | 10 | 31 | 11 |
| Betaproteobacteria | 3 | 0 | 5 | 23 |
| Gammaproteobacteria | 23 | 30 | 28 | 48 |
| Deltaproteobacteria | 0 | 0 | 3 | 4 |
| Deinococcus-Thermus | <1 | 0 | 0 | 0 |
| Chloroflexi | 0 | 0 | 3 | 0 |
| Acidobacteria | 0 | 0 | 5 | 0 |
| Unclassified | 2 | 0 | 7 | 0 |
| Sample size (n) | 501 | 20 | 39 | 44 |
Data for sample 13 only.
A number of differences were seen between the different approaches used (Table 5) that should ideally have yielded the same results. First, few common sequences appeared to be shared between DGGE bands and the clone library. This was surprising but perhaps indicates the difficulty of identifying clones based on <200 bases or the use of different primer sets, which is thought to affect the perceived diversity (31). Alignment of short segments of the cloned fragments with the DGGE products indicated that there might be greater similarity than indicated based purely on database matches. For instance, band 11 shows similarity to clone a13115 (92.3%), band 31 to a13111 (93.0%), and band 30 to clone a13107 (93.4%). Second, the identities of isolates determined by FAME analysis did not accurately correspond to the identities of the bands from the plate wash DGGE. Again, this may be due to the short length of sequence data available or the limitations of the MIS database and may have been overcome by sequencing 16S rRNA genes from the isolates.
Apart from the single Deinococcus isolate (Deinococcus-Thermus phylum), there were no groups that were identified by culture-based approaches but not by culture-independent means. However, the collated information in Table 5 indicates that the overall trends were similar. One major discrepancy was identified, the absence of Alphaproteobacteria in the collection of isolates identified by FAME analysis. This may be explained by the high proportion (∼25%) of isolates that were lost during subculture. Therefore, the DGGE analysis of biomass from plate washes may be beneficial for the detection of organisms that cannot be readily identified by other culture methods.
The subpopulation of bacteria isolated on laboratory media was significantly different from the community identified by analysis of 16S rRNA genes. Although the dominant bands of the total community assessed by DGGE were similar for all samples (Fig. 1A), different subsets from each sample formed colonies on plates (Fig. 1B, Table 3). This could reflect the variation in metal contamination between the soil samples (Table 1). The implication is that the metals had a detrimental effect on certain Bacteria by affecting their ability to replicate on laboratory media. This hypothesis is tenable because culturability is related to physiological status (17), and catabolic diversity may be reduced by metal stress (37). Such data point to the alteration in cellular physiology that results in the loss of viability seen at the most contaminated sites.
We suggest that the proportion of microscopically counted bacteria that form colonies on laboratory media is an ecologically relevant parameter. Essentially, this study indicates that the effect of the different heavy metal concentrations on the dominant bands observed in DGGE analysis of DNA extracted directly from soil was minimal compared to the effects on the culturable portion of the community. Therefore, the most appropriate measures to determine the effect of the contamination on “soil health” will be to assess activity rather than the presence or absence of different cell types (20). Although culture-dependent approaches to microbial community analysis are often criticized for their selectivity, it is perhaps this discrimination that makes them useful for determining the impact of anthropogenic activity.
Readily culturable bacteria are probably the largest, most active prokaryotes in a given sample (1, 2) and so provide a useful, rapid assessment of biological responses to heavy metal pollution. While this study adds weight to the argument that culturable bacteria represent the ecologically relevant portion of the soil bacterial community (14), it is apparent that further studies are required to clarify this. It is also possible that recent advances in the design of solid media and culture methods for soil bacteria (15, 29) will make plate counting even more useful for assessing the impact of pollution.
Acknowledgments
This work was funded by ICI Technology Research Programme SRF 9703 and United Kingdom Natural Environment Research Council CONNECT grant GR3/C0026.
We thank the following people for technical assistance: Barry Neish for FAME analysis and epifluorescence microscopy, George Best and Marcus Trett for sampling, and Robert Crawford and Janet Harris for physicochemical analysis of soil samples. The support of the DNA sequencing facilities at Cardiff University and Imperial College at Silwood Park is greatly appreciated.
REFERENCES
- 1.Bakken, L. R. 1997. Culturable and nonculturable bacteria in soil, p. 47-61. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker, New York, N.Y.
- 2.Bakken, L. R., and R. A. Olsen. 1987. The relationship between cell-size and viability of soil bacteria. Microb. Ecol. 13:103-114. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, L., C. Courties, P. Servais, M. Troussellier, M. Petit, and P. LeBaron. 2000. Relationships among bacterial cell size, productivity, and genetic diversity in aquatic environments with cell sorting and flow cytometry. Microb. Ecol. 40:148-158. [DOI] [PubMed] [Google Scholar]
- 4.Bloem, J. 1995. Fluorescent staining of microbes for total direct counts, p. 4.1.8:1-4.1.8:12. In A. D. L. Akkermans (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Chapman, P. M. 1999. The role of soil microbial tests in ecological risk assessment. Hum. Ecol. Risk Assess. 5:657-660. [Google Scholar]
- 6.Eichner, C. A., R. W. Erb, K. N. Timmis, and I. Wagner-Döbler. 1999. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl. Environ. Microbiol. 65:102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis, R. J., J. G. Best, J. C. Fry, P. Morgan, B. Neish, M. W. Trett, and A. J. Weightman. 2002. Similarity of microbial and meiofaunal community analyses for mapping ecological effects of heavy metal contamination in soil. FEMS Microb. Ecol. 40:113-122. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, R. J., B. Neish, M. W. Trett, J. G. Best, A. J. Weightman, P. Morgan, and J. C. Fry. 2001. Comparison of microbial and meiofaunal community analyses for determining impact of heavy metal contamination. J. Microbiol. Methods 45:171-185. [DOI] [PubMed] [Google Scholar]
- 9.Ellis, R. J., I. P. Thompson, and M. J. Bailey. 1999. Temporal fluctuations in the pseudomonad population associated with sugar beet leaves. FEMS Microb. Ecol. 28:345-356. [Google Scholar]
- 10.Fry, J. C. 1993. One-way analysis of variance, p. 1-40. In J. C. Fry (ed.), Biological data analysis. Oxford University Press, Oxford, United Kingdom.
- 11.Furlong, M. A., D. R. Singleton, D. C. Coleman, and W. B. Whitman. 2002. Molecular and culture-based analyses of prokaryote communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 68:1265-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrity, G. M., M. Winters, A. W. Kuo, and D. B. Searles (ed). 2002. Taxonomic outline of the procaryotes. In Bergey's manual of systematic bacteriology, 2nd ed., release 2. Springer-Verlag, New York, N.Y.
- 13.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Hattori, T., H. Mitsui, H. Haga, N. Wakao, S. Shikano, K. Gorlach, Y. Kasahara, A. El-Beltagy, and R. Hattori. 1997. Advances in soil microbial ecology and the biodiversity. Antonie Leeuwenhoek 72:21-28. [DOI] [PubMed] [Google Scholar]
- 15.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser, O., A. Pühler, and W. Selbitschka. 2001. Phylogenetic analysis of microbial diversity in the rhizosphere of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb. Ecol. 42:136-149. [DOI] [PubMed] [Google Scholar]
- 17.Kell, D. B., A. S. Kaprelyants, D. H. Weichart, C. R. Harwood, and M. R. Barer. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Leeuwenhoek 73:169-187. [DOI] [PubMed] [Google Scholar]
- 18.Kozdrój, J., and J. D. van Elsas. 2000. Response of the bacterial community to root exudates in soil polluted with heavy metals assessed by molecular and cultural approaches. Soil Biol. Biochem. 32:1405-1417. [Google Scholar]
- 19.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 20.Lawlor, K., B. P. Knight, V. L. Barbosa-Jefferson, P. W. Lane, A. K. Lilley, G. I. Paton, S. P. McGrath, S. M. O'Flaherty, and P. R. Hirsch. 2000. Comparison of methods to investigate microbial populations in soils under different agricultural management. FEMS Microb. Ecol. 33:129-137. [DOI] [PubMed] [Google Scholar]
- 21.LeBaron, P., P. Servais, H. Agogué, C. Courties, and F. Joux. 2001. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active and inactive cells in aquatic systems? Appl. Environ. Microbiol. 67:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (ribosome database project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microb. Ecol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 24.McCaig, A. E., S. J. Grayston, J. I. Prosser, and L. A. Glover. 2001. Impact of cultivation on characterisation of species composition of soil bacterial communities. FEMS Microb. Ecol. 35:37-48. [DOI] [PubMed] [Google Scholar]
- 25.Mitsui, H., K. Gorlach, H.-j. Lee, R. Hattori, and T. Hattori. 1997. Incubation time and media requirements of culturable bacteria from different phylogenetic groups. J. Microbiol. Methods 30:103-110. [Google Scholar]
- 26.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovreås, L., and V. Torsvik. 1998. Microbial diversity and community structure in two different agricultural soil communities. Microb. Ecol. 36:303-315. [DOI] [PubMed] [Google Scholar]
- 28.Roane, T. M., and I. L. Pepper. 2000. Microbial responses to environmentally toxic cadmium. Microb. Ecol. 38:358-364. [DOI] [PubMed] [Google Scholar]
- 29.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent studies. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 30.Sandaa, R. A., V. Torsvik, and O. Enger. 2001. Influence of long-term heavy-metal contamination on microbial communities in soil. Soil Biol. Biochem. 33:287-295. [Google Scholar]
- 31.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söderberg, K. H., and E. Bååth. 1998. Bacterial activity along a young Barley root measured by the thymidine and leucine incorporation techniques. Soil Biol. Biochem. 30:1259-1268. [Google Scholar]
- 34.Stackebrandt, E., W. Liesack, and B. M. Goebel. 1993. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 7:232-236. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, I. P., M. J. Bailey, R. J. Ellis, and K. J. Purdy. 1993. Subgrouping of bacterial populations by cellular fatty acid composition. FEMS Microb. Ecol. 102:75-84. [Google Scholar]
- 36.Trett, M. W., U. Calvo, S. J. Forster, J. D. Hutchinson, R. L. Feil, S. P. Trett, and G. Best. 2000. Terrestrial meiofauna and contaminated land assessment. Environ. Sci. Technol. 34:1594-1602. [Google Scholar]
- 37.Wenderoth, D. F., E. Stackebrandt, and H. H. Reber. 2001. Metal stress selects for bacterial ARDRA-types with a reduced catabolic diversity. Soil Biol. Biochem. 33:667-670. [Google Scholar]
- 38.Yang, C.-H., D. E. Crowley, J. Borneman, and N. T. Keen. 2001. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 98:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zengler, K., G. Toledo, M. Rappé, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]