Abstract
Belonging to the subtilase family, the cell surface proteinase (CSP) PrtB of Lactobacillus delbrueckii subsp. bulgaricus differs from other CSPs synthesized by lactic acid bacteria. Expression of the prtB gene under its own promoter was shown to complement the proteinase-deficient strain MG1363 (PrtP− PrtM−) of Lactococcus lactis subsp. cremoris. Surprisingly, the maturation process of PrtB, unlike that of lactococcal CSP PrtPs, does not require a specific PrtM-like chaperone. The carboxy end of PrtB was previously shown to be different from the consensus anchoring region of other CSPs and exhibits an imperfect duplication of 59 amino acids with a high lysine content. By using a deletion strategy, the removal of the last 99 amino acids, including the degenerated anchoring signal (LPKKT), was found to be sufficient to release a part of the truncated PrtB into the culture medium and led to an increase in PrtB activity. This truncated PrtB is still active and enables L. lactis MG1363 to grow in milk supplemented with glucose. By contrast, deletion of the last 806 amino acids of PrtB led to the secretion of an inactive proteinase. Thus, the utmost carboxy end of PrtB is involved in attachment to the bacterial cell wall. Proteinase PrtB constitutes a powerful tool for cell surface display of heterologous proteins like antigens.
Lactobacillus delbrueckii subsp. bulgaricus (L. bulgaricus), a gram-positive, facultatively anaerobic, homofermentative lactic acid bacterium, plays an important role in the dairy industry because of its efficient and reliable utilization of milk constituents, mainly lactose and caseins, and its good resistance to low pH. Spontaneous mutants of L. bulgaricus, unable to grow on milk in the absence of peptide extract, were characterized by ISL3 transposition-induced deletions, including that of the prtB gene coding for the cell surface proteinase (CSP) (11). Actually, prtB is essential for L. bulgaricus growth in milk and is responsible for the first step of caseinolysis. Most L. bulgaricus strains are characterized by a high CSP activity resulting from the adaptation of this species to fast growth and rapid fermentation of milk (14).
So far, four different types of genes encoding CSPs of dairy lactic acid bacteria have been cloned and sequenced: prtB from L. bulgaricus, prtP from Lactococcus lactis and Lactobacillus casei, prtS from Streptococcus thermophilus, and prtH from Lactobacillus helveticus (8, 13, 18, 21, 28, 38). Comparative sequence analysis of CSPs revealed different domains associated with putative functions (35). CSPs are synthesized as long and inactive preproproteins (∼2,000 residues). For the N terminus of CSP, eight domains have been predicted (Fig. 1A). (i) The predomain (∼30 residues) corresponds to a signal sequence required for secretion and is removed by a specific signal peptidase during translocation through the cytoplasmic membrane. (ii) The prodomain (∼150 residues) is essential for enzyme maturation and is removed by autoproteolytic cleavage. (iii) The catalytic domain (∼500 residues, including a variable insert of about 150 residues) shows the highest similarity between CSPs and belongs to the superfamily of subtilisin-like serine proteinases, often referred to in abbreviated form as subtilases. (iv) The A domain (∼400 residues) is specific to CSP and characterized by a beta-sheet structure, but its function is not yet known. The last four domains vary among the different known CSPs and have been well characterized in PrtPs. (v) The B domain (∼500 residues) should have stabilizing functions but seems not to be essential for proteolysis, as it is absent in some CSPs. (vi) The H domain (up to 200 residues) constitutes a long helix forming a stalk-like structure able to position the A and B domains outside the bacterial cell. (vii) The W domain (∼100 residues) is predicted to be a hydrophilic domain spanning the cell wall. (viii) The AN domain (∼40 residues) is characterized by a sorting and anchoring motif (LPXTG) followed by a hydrophobic putative membrane-spanning alpha-helix and a short charged tail. This last domain is involved in anchoring many cell surface proteins from gram-positive bacteria to the cell wall via a covalent link with peptidoglycan (26).
FIG. 1.
Schematic representations of preproproteinase PrtP from L. lactis (A) and PrtB from L. bulgaricus and four truncated forms (PrtBδ99, PrtBδ168, PrtBδ247, and PrtBδ806) (B). The different domains of the proteinases and the amino acids (D, H, N, and S for PrtB) involved in the active site are indicated. Small bent arrows correspond to primers designed for gene amplification. The thick horizontal bar in the catalytic domain indicates the peptide (amino acids 280 to 467) used for preparation of the anti-PrtB serum. The dashed thick horizontal bars within the C-terminal region correspond to the two imperfect repeats of 59 residues surrounding the degenerated sorting signal LPKKT. The motifs of the B domain are indicated (B2 to B5) to position the truncated PrtBδ806. Amino acids are numbered starting from the amino end of the mature proteinase.
The PrtB of L. bulgaricus strongly differs from other CSPs in its specificity of cleavage and the structure of the long C-extension domains (∼1,100 residues) (13, 23). The cleavage specificity of CSP is mainly dependent on two regions: a substrate binding site located in the catalytic domain and a short region of the A domain (35, 36). Therefore, differences in sequences between these regions of PrtPs and PrtB could explain the differences in cleavage of beta-casein (14). In addition, the different C terminus of PrtB raises the possibility of a mechanism of attachment to the cell envelope that is different from the covalent anchoring of lactococcal PrtPs to the peptidoglycan via the LPXTG motif.
The present paper describes expression of the prtB gene of L. bulgaricus in the plasmid-free strain MG1363 (PrtP− PrtM−) of L. lactis subsp. cremoris (9), because no transformation procedure with L. bulgaricus led to the recovery of transformants with entire plasmid. Expression of the prtB gene complements the proteinase-negative phenotype of strain MG1363. Deletions were performed in the 3′-end region of the prtB gene, and L. lactis MG1363 cells synthesizing entire or truncated PrtBs were analyzed to determine maturation, cellular localization, stability, and cleavage specificity towards beta-casein of parental and mutant PrtB proteins.
MATERIALS AND METHODS
Bacteria and plasmid.
L. bulgaricus NCDO 1489 was maintained at 40°C in MRS broth (Difco Laboratories, Sparks, Md.). L. lactis subsp. cremoris (NCDO 712) and its plasmid-free derivative strain MG1363 (9) were maintained at 30°C in M17 broth (Difco Laboratories) supplemented with 1% glucose (GM17). Plasmid pNZ124 (29) was used to clone prtB and truncated prtB genes. The version used, pNZ124.1, lacked 390 nucleotides from 61 nucleotides downstream of the repA gene to 12 nucleotides upstream of the cm gene. Chloramphenicol was added to growth media at a concentration of 5 μg/ml to maintain plasmids (pNZ124.1 and derivatives) in L. lactis MG1363.
Molecular techniques and cell transformation.
L. bulgaricus chromosomal DNA was purified by the spooling method (5). The 6-kb-long prtB gene (GenBank accession no. L48487) and its truncated versions were amplified by using the forward primer 2162, containing a BamHI site (located 230 bp upstream of the start codon), and the reverse primer 2164, exhibiting an XbaI site (located 160 bp downstream of the stop codon). The truncated versions of the prtB gene were amplified by using different reverse primers located within the prtB gene (Table 1). Primers were obtained from Microsynth (Balgach, Switzerland). DNA amplification was performed in a total volume of 50 μl containing 25 nmol of each deoxynucleoside triphosphate, 15 pmol of each primer, 0.5 U of Taq polymerases (Expand Template PCR system; Boehringer, Mannheim, Germany), 0.05 U of Pfu polymerase (Stratagene, La Jolla, Calif.), and the appropriate buffer for 30 cycles. A DNA thermocycler (Perkin-Elmer, Foster City, Calif.) was used, and each cycle consisted of a denaturing step at 95°C for 10 s, a primer-annealing step at 55°C for 30 s, and an extension step at 68°C for 1 min per kilobase to be amplified. The amplification products were digested with BamHI and XbaI enzymes and ligated in BamHI/XbaI-cut pNZ124.1. Following ligation, plasmids were introduced by electroporation into L. lactis MG1363 (19).
TABLE 1.
Primers used to generate full-length or truncated prtB genes cloned in plasmid pNZ124.1
| Primer | Sequence | pNZ124.1 derivative with full-length or truncated prtB genes | Proteinase expressed |
|---|---|---|---|
| 2162a | 5′-TTTTGTGGATCCTTAACTTCATAGCACG | ||
| 2164 | 5′-ATATTATCTAGAATTGAATAGATTGCC | pMD112 | PrtB |
| 9049 | 5′-CTAGTCTAGACCAACTACCTTGCC | pMD116 | PrtBδ99b |
| G059 | 5′-CATGTCTAGATTACGTTCGCCATTGCC | pMD142 | PrtBδ168 |
| G060 | 5′-CATGTCTAGAAGTCAGCTTAACTTCGC | pMD143 | PrtBδ247 |
| 9048 | 5′-CTAGTCTAGACCATCCTTGACTGGC | pMD115 | PrtBδ806 |
Forward primer; all others are reverse primers.
The numbers in the designations indicate the number of deleted amino acids at the carboxy end of the proteinase.
Bacterial growth in milk.
Lactococci were precultured in sterilized 10% reconstituted skim milk (120°C, 30 min) containing 0.1% yeast extract and 1% glucose. Bacterial growth in pasteurized 10% skim milk (80°C, 30 min) containing 1% glucose was followed by conductivity measurement (rapid automated bacterial impedance technique [RABIT]; Don Witley Scientific Ltd., Shipley, West Yorkshire, England).
Proteinase activity and beta-casein hydrolysis.
Bacteria were grown to an optical density at 600 nm of 1.0 (mid-logarithmic phase) in MRS or GM17 with chloramphenicol (5 μg/ml). The procedure of Hill and Gasson (17) was modified as follows. Bacterial cultures (7 × 108 cells) were collected, washed, and concentrated by centrifugation (7,000 × g 10 min, 6°C) in 3 ml of the incubation buffer (100 mM HEPES [pH 6.5] containing 10 mM CaCl2). Cellular lysis of Lactobacillus cells was significantly increased by the presence of 10 mM CaCl2, which was omitted for treatments of Lactobacillus cells. Culture supernatants corresponding to the first centrifugation of bacterial cultures were concentrated by ultrafiltration on an XM50 membrane (Amicon; Millipore) at 6°C and under a pressure of 2.0 kg/cm2. These concentrated preparations of excreted proteinases were washed in two successive dilutions and ultrafiltrations with the incubation buffer under the same conditions as described above.
Dephosphorylated beta-casein (13 μg) was incubated with 3 ml of a bacterial suspension (7 × 108 cells or the equivalent culture supernatant) as described previously (14). Aliquots (150 μl) were taken at different times (0, 5, 15, and 60 min) and immediately mixed with 32 μl of the solubilization buffer (125 mM Tris-HCl [pH 6.8] containing 2% sodium dodecyl sulfate [SDS] and 100 mM 2-mercaptoethanol). These mixtures were incubated at 80°C for 10 min and centrifuged (7,000 × g rpm, 10 min, 6°C). An aliquot (50 μl) of each supernatant was mixed with 10 μl of 30% glycerol containing 0.04% bromophenol blue. These preparations of peptides resulting from the beta-casein hydrolysis were heated at 100°C for 2 min before SDS-polyacrylamide gel electrophoresis (PAGE) (22) on 15% polyacrylamide gels. The remaining beta-casein and peptides were stained with Coomassie brilliant blue R250 as described previously (14). A BenchMark prestained protein ladder (Invitrogen/Life Technologies, Cergy Pontoise, France) was used for the determination of molecular mass.
Immunoblot analysis of bacteria-associated and excreted PrtB.
Bacterial cells that were developed in GM17 containing 5 μg of chloramphenicol/ml until an optical density at 600 nm of 1.0 was reached were collected by centrifugation (7,000 × g 10 min, 6°C) and immediately frozen at −80°C. Culture supernatants were mixed (vol/vol) with acetone and incubated for 15 min at −20°C. Precipitated proteins were harvested by centrifugation (15 min, 15,000 × g, 4°C), washed with cold 70% ethanol, and suspended in 1/15 of the original volume of the loading buffer (125 mM Tris, 100 mM β-mercaptoethanol, 2% SDS, 0.1% bromophenol blue, 20% glycerol [pH 6.8]). Bacterial pellets were resuspended in the same volume of the loading buffer. Samples (5 μl) from both preparations (bacterial cells and culture supernatants) were not treated at 100°C before loading on SDS-8% polyacrylamide gels, and electrophoresis was carried out with the electrophoresis buffer (50 mM Tris, 384 mM glycin, 0.1% SDS). BenchMark prestained protein ladder was used as the molecular mass standard. Proteins were then electrotransferred overnight (0.2 mA/cm2) onto reinforced cellulose nitrate membranes (Schleicher & Schuell, Dassel, Germany). Immunoblotting was carried out by the method of Harlow and Lane (16) using polyclonal rabbit anti-PrtB antiserum (1:2,000). Anti-rabbit IgG-peroxidase conjugate (1:4,000; Sigma-Aldrich, L'isle-d'Abeau Chesnes, France) was used as a secondary antibody. Revelation was performed with a chemiluminescent substrate (BM chemiluminescence Western blotting substrate; Roche Molecular Biochemicals, Meylan, France), with visualization after a 3-s exposure time.
Preparation of polyclonal rabbit anti-PrtB antiserum.
A fragment comprising amino acids 280 to 467 of L. bulgaricus mature cell wall proteinase was overexpressed in Escherichia coli and used for production of antiserum in rabbits (1).
Sequence analysis.
The search of specific patterns was done through the SWISS-PROT and TrEMBL databases by using the ScanProsite program.
RESULTS
Heterologous expression of the prtB gene in L. lactis.
The prtB gene encoding the CSP of L. bulgaricus and its promoter region were isolated by DNA amplification using the primers 2162, which introduces a BamHI restriction site at the 5′ end, and 2164, which introduces an XbaI site at the 3′ end (Fig. 1B). This amplification product was cloned into the lactococcal plasmid pNZ124.1, and the resulting plasmid, pMD112, was transferred into L. lactis MG1363. This PrtP− PrtM− Lac− strain is a plasmid-free derivative of the strain NCDO 712 that harbors the prtP prtM genes and the lac operon on the pLP712 plasmid (10), allowing growth in milk by fermentation of caseins and lactose.
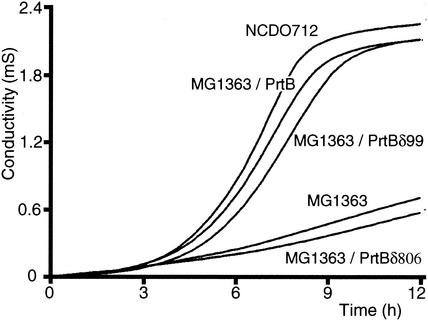
The growth characteristics of the different lactococcal strains in milk complemented with glucose were monitored by direct measurement of impedance (Fig. 2). Compared with L. lactis NCDO 712, L. lactis MG1363 developed much more slowly. The presence of the pMD112 plasmid harboring prtB in L. lactis MG1363 cells restores growth similar to that of the parental L. lactis NCDO 712. These results indicate that the prtB gene can be heterologously expressed under its own promoter in L. lactis and that the proteinase PrtB is exported and biologically active enough to generate peptides from casein, which can be transported by the L. lactis uptake systems and processed in the following steps of peptide metabolism.
FIG. 2.
Growth kinetics in milk of L. lactis NCDO 712, L. lactis MG1363, and L. lactis MG1363 expressing the full-length proteinase of L. bulgaricus, PrtB, and the truncated forms PrtBδ99 and PrtBδ806.
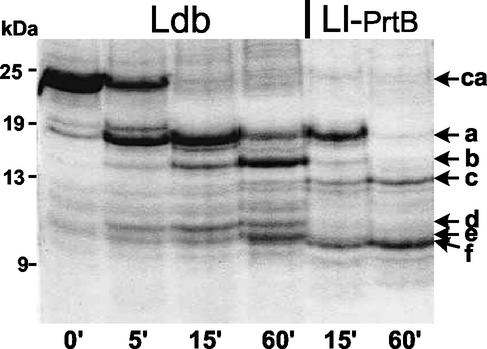
Peptides resulting from hydrolysis of beta-casein by PrtB associated with L. bulgaricus or L. lactis MG1363 expressing PrtB were analyzed by SDS-PAGE (Fig. 3). The former PrtB generated two major products of hydrolysis (a and b) and three minor ones (c, d, and e). Under the same conditions of hydrolysis, the cleavage specificity of PrtB associated with lactococcal cells appeared to be different. Peptide b was only a minor product, and peptide c was the intermediate in the hydrolysis process. Peptides d and e were not intermediates, but peptide f was generated and accumulated. The kinetics of hydrolysis were also different, as the major hydrolysis product (a) disappeared much more rapidly. These results show that PrtB associated with L. lactis MG1363 cells seems to have acquired different characteristics of beta-casein cleavage.
FIG. 3.
SDS-PAGE analysis of beta-casein (ca) hydrolyzed for the times indicated (in minutes) by L. bulgaricus (Ldb) and L. lactis MG1363 expressing PrtB (Ll-PrtB). Hydrolysis products (a to f) and molecular mass standards in kilodaltons are indicated.
Analysis of the C terminus of the CSP PrtB.
The domains of the PrtB carboxy end were shown to have an organization different from that of other CSPs. The region downstream of the B domain (from residue 1370) does not correspond to a classical H domain and is characterized by only two regions of about 10 amino acids with a predicted alpha-helix structure (Fig. 4). The transwall domain (W domain), which is also hydrophilic but very rich in lysine residues (32%) compared with the equivalent domain of PrtPs (8%), starts at around residue 1530. It is characterized by several copies of a 4-amino-acid repeat, KSDX (with X indicating any amino acid). In the two proteinase-deficient strains of L. delbrueckii subsp. delbrueckii (ATCC 9649 and NCFB701744), several copies of this repeat have been shown to be duplicated or deleted (12). This suggests that since this repeated structure spans the cell wall, it is susceptible to length variation by strand slippage during replication, which is a good way to account for differences in the thickness of cell walls during horizontal transfer of this proteinase.
FIG. 4.
Amino acid sequence of the H and W domains located at the carboxy end of L. bulgaricus PrtB. The last amino acids of truncated proteinases (PrtBδ247, PrtBδ168, and PrtBδ99) are boxed. Dashed bars underline both alpha-helix structures of the W domain. The 4-amino-acid repeats (KSDX) are underlined (thin lines), and lysine residues are shown in boldface type. Brackets surround the 59-amino-acid imperfect repeats, and identical amino acids are underlined (thick lines). The LPKKT degenerated sorting signal is indicated by a horizontal bracket above the sequence.
The utmost carboxy end of PrtB displays an imperfect duplication of 59 amino acids, beginning with KKT (residues 1630 to 1688 and 1693 to 1751) (Fig. 4). This duplication led to a series of modifications of the consensus sequence found in several cell envelope-associated proteins of gram-positive bacteria. A degenerated sorting signal (LPKKT) is found between these lysine-rich duplicated regions (Fig. 4). Downstream of the LPKKT motif, there is no hydrophobic alpha-helix of 18 to 20 amino acids and no charged tail (the last five to seven amino acids). These structures were shown to be necessary for interactions with the cytoplasmic membrane: they are involved in stopping the translocation of the polypeptide and allow the proteolytic cleavage and covalent binding of the proteinase to the cell wall peptidoglycan by the sortase (26).
C terminus-truncated PrtBs and effect on bacterial growth in milk.
To determine how the carboxy end of PrtB is involved in binding of the proteinase to the cell wall, three deletions of this region were generated (Fig. 1B). For the shortest deletion (PrtBδ99), 99 C-terminal amino acids were deleted, including one repeat of 59 residues, the LPKKT motif, and 35 amino acids upstream. Two other deletions of 168 and 247 amino acids eliminated both imperfect repeats of 59 residues (PrtBδ168) and the part of the W domain corresponding to the KSDX repeats (PrtBδ247), respectively. These deletions were generated by DNA amplification using forward primer 2162, located upstream of the prtB promoter, and three reverse primers, 9049, G059, and G060 (Table 1). The different amplified fragments were cloned, leading to plasmids pMD116, pMD142, and pMD143 (Fig. 1B and 4). A longer deletion of 806 amino acids, removing almost all of the B domain and following domains (PrtBδ806), was generated by using reverse primer 9048 (pMD115) (Table 1) (Fig. 1B). The different plasmids were transferred into L. lactis MG1363, and the recombinant strains were assayed for their growth in milk supplemented with glucose (Fig. 2). The growth kinetics of L. lactis MG1363 synthesizing truncated PrtBδ99, PrtBδ168, and PrtBδ247 were similar to that of L. lactis MG1363 producing the full-length PrtB, and similar bacterial biomasses were reached in all cases. Therefore, the removal of the degenerated anchoring LPKKT motif and most of the lysine-rich C-terminal domain does not significantly affect the maturation and export processes of PrtB or proteinase activity. In contrast, L. lactis MG1363 expressing PrtBδ806 did not grow in milk, suggesting an absence of the CSP activity (Fig. 2).
Biosynthesis and cellular location of parental and truncated PrtBs.
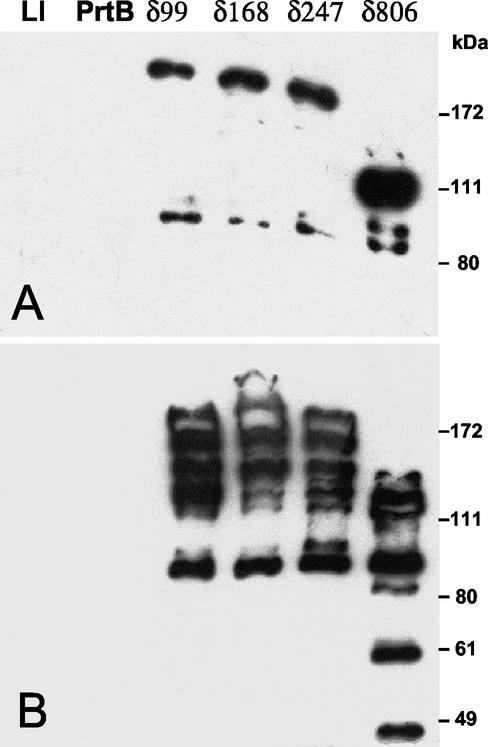
The different truncated PrtBs associated with the bacterial cell were analyzed by Western blot analysis of whole bacterial cells (Fig. 5A) by using antibodies raised against a region of the catalytic domain of PrtB (residues 280 to 467) (Fig. 1B). The results show that L. lactis MG1363 harboring the cloning vector pNZ124.1 did not reveal any signal with anti-PrtB serum, thus confirming the specificity of the serum for PrtB from L. bulgaricus. The full-length PrtB is efficiently bound to the lactococcal cell wall and cannot be loosened under the electric conditions used in SDS-PAGE, as no signal could be detected with cells expressing the full-length PrtB (Fig. 5A), whereas the four truncated and mature PrtBs (PrtBδ99, PrtBδ168, PrtBδ247, and PrtBδ806) can be loosened from bacterial cells upon electrophoresis with the expected sizes (179, 172, 163, and 101 kDa, respectively). It is interesting that PrtBδ806 is found at a much higher level than the other truncated proteinases (Fig. 5A).
FIG. 5.
Immunodetection of cells (A) or culture supernatants (B) of L. lactis MG1363 (Ll) synthesizing the full-length PrtB or its truncated forms (PrtBδ99, PrtBδ168, PrtBδ247, and PrtBδ806). PrtB and truncated derivatives were revealed by using anti-PrtB serum. Molecular mass markers in kilodaltons are indicated.
Analysis of the culture supernatants under the same conditions revealed that part of the truncated PrtBs produced was excreted and submitted to strong proteolysis (Fig. 5B). With the largest truncated PrtB, PrtBδ806, smaller degradation products (<49 kDa) were detected, which probably reflects a higher level of instability associated with a different folding of this proteinase, revealing new cleavage sites. It must be kept in mind that only products carrying the short epitope of 188 residues (located in the catalytic domain) (Fig. 1B) could be detected; consequently, proteolysis is probably undervalued. All these results prove that the C-terminal repeats of 59 residues are involved in attachment to the cell wall and that the capacity of attachment is drastically reduced by the absence of these duplicated sequences.
Caseinolytic activity and substrate specificity of truncated PrtBs.
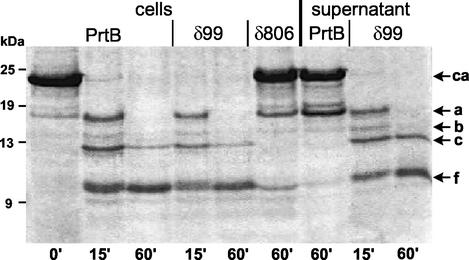
The proteinase activity of L. lactis MG1363 cells producing the different forms of PrtB was assessed by monitoring the kinetics of beta-casein hydrolysis by whole cells (Fig. 6). Cells synthesizing full-length PrtB or PrtBδ99 degraded the beta-casein with identical cleavage specificity and at a similar rate. This result is in agreement with the rapid growth in milk of the respective L. lactis MG1363 recombinants. Cells synthesizing PrtBδ168 and PrtBδ247 hydrolyzed beta-casein similarly to PrtBδ99 (data not shown). By contrast, cells producing large amounts of PrtBδ806 (Fig. 5) converted, even after 60 min of incubation, only a very small proportion of beta-casein to peptides a and f (Fig. 6); these results are in agreement with the absence of bacterial growth in milk. Thus, the proteolytic activity was conserved for all truncated forms, where the N-terminal W domain was sequentially deleted, whereas removal of the H domain and a part of the B domain inactivated proteolytic activity.
FIG. 6.
SDS-PAGE analysis of beta-casein (ca) hydrolyzed for the times indicated (in minutes) by whole cells of L. lactis MG1363 producing full-length PrtB, PrtBδ99, or PrtBδ806. Beta-casein was also hydrolyzed by culture supernatants of L. lactis MG1363 synthesizing full-length PrtB or PrtBδ99.
The different culture supernatants were also tested for casein proteolysis. Almost no activity was obtained from the supernatant of cells synthesizing the full-length PrtB: only a small proportion of beta-casein was converted to fragment a (Fig. 6), which is in agreement with the lack of immunodetection of proteinase in PrtB's supernatant (Fig. 5B). On the contrary, the PrtBδ99 supernatant, shown to contain large amounts of partially degraded forms of PrtBδ99 (Fig. 5B), was as active as the corresponding recombinant cells (Fig. 6). Therefore, at least two times more proteolytic activity was produced by cells expressing truncated PrtB than by cells expressing full-length PrtB. The same results were obtained with PrtBδ168 and PrtBδ247 supernatants, but PrtBδ806 supernatant showed no proteolytic activity (data not shown). All these results taken together suggest that excretion of truncated PrtBs is associated with an increase of the proteinase activity, which could result from a different folding of truncated PrtBs or an increase of their biosynthesis.
DISCUSSION
The prtB gene encoding the cell wall-associated proteinase PrtB of L. bulgaricus was successfully expressed in L. lactis MG1363 (PrtP− PrtM−) and fully complements the proteinase-negative phenotype of the recipient cells. Thus, PrtB is matured and associated to the lactococcal cell surface in spite of a C extension (domains A, B, H, and W) very different from that of lactococcal PrtPs and many other cell surface proteins of gram-positive bacteria (27). These latter proteins are covalently anchored to the cell wall via a sorting and anchoring motif (LPXTG), which is followed by two characteristic regions, a hydrophobic transmembrane domain and a short tail of five to seven charged residues. In PrtB, a degenerated LPKKT motif is surrounded by two imperfect repeats of 59 residues. The high content of positive charges in the C terminus suggests interactions with the negatively charged teichoic acids of the cell wall. Taken together, our results suggest that the mechanism of PrtB attachment to the cell wall probably implicates electrostatic forces, as was proposed for WprA, a cell wall-associated protease of Bacillus subtilis (24). The success of the heterologous expression of prtB in L. lactis could have been favored by a composition of the cell wall peptidoglycan that is similar in both species (Lys-D-Arg [31]).
Deletion of the major part of the duplicated region at the utmost C terminus led to the partial release of the deleted PrtB into the culture medium. Furthermore, this release is correlated with a high proteolysis of this proteinase. Removing up to 250 residues did not modify the rate of casein digestion or the stability of the truncated PrtBs. The entire W domain is then essential for stable binding to the cell wall, but not for PrtB activity. In addition, the biosynthesis of proteinase PrtB was increased by loosening its binding to the cell wall. Therefore, this overproduction suggests that PrtB biosynthesis could be regulated by a limited number of anchoring sites in the cell wall. Thus far, only a limited increase in the cell wall proteinase activity has been obtained with the prtB gene cloned in a high-copy-number plasmid. This raises the hypothesis that the accumulation of numerous cell wall-associated PrtB molecules could decrease cell viability. However, increase in PrtB activity does not confer a faster growth rate of bacterial cells in milk. Thus, the first step of caseinolysis performed by the CSP PrtB is probably not a factor limiting the development of lactic acid bacteria in milk.
Removal of the last 800 residues of PrtB led to a 100-kDa inactive proteinase, which can still be found associated with the cell wall. This suggests that at least a part of the B domain is involved in stabilization of the catalytic domain of PrtB, similar to the situation described with lactococcal PrtP (3). The function of the A and B domains has not yet been unambiguously defined. These domains display several patterns of amino acids found in all sequenced CSPs (35). In a search of the SWISS-PROT and TrEMBL databases, these patterns grouped by two or three were not found in any proteins other than CSPs and the C5a peptidase precursor of Streptococcus agalactiae.
In contrast to lactococcal PrtPs, PrtB is processed and matured to an active form without the help of an extracytoplasmic PrtM-like chaperone. Only a very short pseudo-prtM gene coding for a few amino acids has been identified upstream of the prtB gene (13), and no prtM-like gene has been identified downstream of prtB (unpublished data). A similar situation is found with the cell wall-associated protease PrtA, which contributes to the virulence of Streptococcus pneumoniae (2). In L. lactis, it cannot be ruled out that another general extracytoplasmic chaperone could be implicated in PrtB folding. Actually, L. lactis synthesizes at least two extracytoplasmic chaperones, PrtM and PmpA, homologous to the PrsA chaperone of B. subtilis that is essential for the correct folding and secretion of several exoenzymes (6, 20). Nevertheless, another hypothesis is favored. The prodomains of several proteinases of prokaryotes and eukaryotes, and more particularly of B. subtilis subtilisin, have been shown to function as intramolecular chaperones (IMCs) (33). IMCs facilitate the correct folding of their associated proteins but are not required for the stability of folded proteinases. After completion of folding, the IMCs are then removed by the active folded proteinases. If the overall sequence identity of IMCs is low, the prodomains of proteinases belonging to the subtilisin family are characterized by two well-conserved motifs, N1 and N2, containing the hydrophobic core residues. A box of the motif N2 (positions 119 to 146) is quite well conserved in the proregion of PrtB, and 30% identity was observed with subtilisin E motif 2 (Table 2). In PrtP, motifs 1 and 2 are severely degenerated. This prompted us to propose the prodomain of PrtB as a class I propeptide that confers a foldase activity sufficient for proteinase folding (34). On the other hand, the single prodomain of the lactococcal PrtP is unable to directly catalyze folding without the help of the PrtM chaperone (15) and may be classified as a class II propeptide.
TABLE 2.
Comparison of sequences surrounding the conserved motifs N1 and N2 of the propeptides from proteinases belonging to the subtilisin family
| Proteinase (species) | Motif N1a | Motif N2 |
|---|---|---|
| Subtilisin E (B. subtilis) | KK YIVGF KQT | AAAATL DEKAVKELKKDPS- VAVY EED |
| Elastase (B. subtilis) | EK YLIGF KEQ | VLSVEL DPEDVDALELDPA- IAYI EED |
| Proteinase B (S. cerevisiae) | NR YIIVF KRG | GYIGYF TQEIVDLIRQNPL- VDFV ERD |
| PrtB (L. bulgaricus) | KK SYSKF QEA | AASVKK IEQASDQVKDGQEK VIKQ VEEI |
| PrtP (L. lactis) | NQ AIATQ LAA | YSSTAE IQQETNKVIAAQAS VKAA VEQV |
Bold letters indicate residues similar to those of subtilisin E in the different motifs.
L. lactis, widely used in food fermentation, is considered a food-grade microorganism (“GRAS,” generally recognized as safe). As a noncolonizing bacterium that is nevertheless able to survive in the gastrointestinal tract, it constitutes an appropriate vehicle for the delivery of antigens in the gastrointestinal tract for mucosal immunization (4, 7, 25, 30, 37). The different mucosa (intestinal, nasal, and vaginal) are the major sites of aggressions by pathogens and food allergens. Thus, live bacterial vaccines could constitute an easy-to-use, cheap, and powerful response to common illnesses. PrtB is an ideal tool for the display of various antigens at the bacterial cell surface. The combination of efficient secretion and a cell wall anchoring system, which can be modulated, provides the needed variability of antigen delivery (cell surface display or secretion) in various lactic acid bacteria. Proteins of interest were already genetically inserted in the catalytic domain of the proteinase PrtB. This system was successfully used for the expression of different antigens, such as beta-lactoglobulin and immunoglobulin E mimotopes, as tools for the modulation of tolerance and allergy (1, 32).
Acknowledgments
We thank Veronique Delaire for excellent technical assistance, David Vilanova for help with bioinformatics, Harald Bruessow for critical reading of the manuscript, and Elizabeth Prior for reviewing the manuscript.
REFERENCES
- 1.Bernasconi, E., J. E. Germond, M. Delley, R. Fritsche, and B. Corthesy. 2002. Lactobacillus bulgaricus proteinase expressed in Lactococcus lactis is a powerful carrier for cell wall-associated and secreted bovine beta-lactoglobulin fusion proteins. Appl. Environ. Microbiol. 68:2917-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bethe, G., R. Nau, A. Wellmer, R. Hakenbeck, R. R. Reinert, H. P. Heinz, and G. Zysk. 2001. The cell wall-associated serine protease PrtA: a highly conserved virulence factor of Streptococcus pneumoniae. FEMS Microbiol. Lett. 205:99-104. [DOI] [PubMed] [Google Scholar]
- 3.Bruinenberg, P. G., W. M. de Vos, and R. J. Siezen. 2000. Deletion of various carboxy-terminal domains of Lactococcus lactis SK11 proteinase: effects on activity, specificity, and stability of the truncated enzyme. Appl. Environ. Microbiol. 66:2859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatel, J. M., P. Langella, K. Adel-Patient, J. Commissaire, J. M. Wal, and G. Corthier. 2001. Induction of mucosal immune response after intranasal or oral inoculation of mice with Lactococcus lactis producing bovine beta-lactoglobulin. Clin. Diagn. Lab. Immunol. 8:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delley, M., B. Mollet, and H. Hottinger. 1990. DNA probe for Lactobacillus delbrueckii. Appl. Environ. Microbiol. 56:1967-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drouault, S., J. Anba, S. Bonneau, A. Bolotin, S. D. Ehrlich, and P. Renault. 2002. The peptidyl-prolyl isomerase motif is lacking in PmpA, the PrsA-like protein involved in the secretion machinery of Lactococcus lactis. Appl. Environ. Microbiol. 68:3932-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enouf, V., P. Langella, J. Commissaire, J. Cohen, and G. Corthier. 2001. Bovine rotavirus nonstructural protein 4 produced by Lactococcus lactis is antigenic and immunogenic. Appl. Environ. Microbiol. 67:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Espla, M. D., P. Garault, V. Monnet, and F. Rul. 2000. Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 66:4772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasson, M. J. 1983. Genetic transfer systems in lactic acid bacteria. Antonie Leeuwenhoek 49:275-282. [DOI] [PubMed] [Google Scholar]
- 10.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Germond, J. E., L. Lapierre, M. Delley, and B. Mollet. 1995. A new mobile genetic element in Lactobacillus delbrueckii subsp. bulgaricus. Mol. Gen. Genet. 248:407-416. [DOI] [PubMed] [Google Scholar]
- 12.Germond, J. E., L. Lapierre, M. Delley, B. Mollet, G. E. Felis, and F. Dellaglio. 2003. Evolution of the bacterial species Lactobacillus delbrueckii: a partial genomic study with reflections on prokaryotic species concept. Mol. Biol. Evol. 20:93-104. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, C., D. Atlan, B. Blanc, R. Portalier, J. E. Germond, L. Lapierre, and B. Mollet. 1996. A new cell surface proteinase: sequencing and analysis of the prtB gene from Lactobacillus delbruekii subsp. bulgaricus. J. Bacteriol. 178:3059-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert, C., B. Blanc, J. Frot-Coutaz, R. Portalier, and D. Atlan. 1997. Comparison of cell surface proteinase activities within the Lactobacillus genus. J. Dairy Res. 64:561-571. [Google Scholar]
- 15.Haandrikman, A. J., J. Kok, H. Laan, S. Soemitro, A. M. Ledeboer, W. N. Konings, and G. Venema. 1989. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J. Bacteriol. 171:2789-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Hill, S., and M. Gasson. 1986. A quantitative screening procedure for the detection of casein hydrolysis by bacteria, using sodium dodecyl sulphate polyacrylamide gel electrophoresis. J. Dairy Res. 53:625-629. [Google Scholar]
- 18.Holck, A., and H. Naes. 1992. Cloning, sequencing and expression of the gene encoding the cell-envelope-associated proteinase from Lactobacillus paracasei subsp. paracasei NCDO 151. J. Gen. Microbiol. 138:1353-1364. [DOI] [PubMed] [Google Scholar]
- 19.Holo, H. Y., and I. F. Nes. 1989. High frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs, M., J. B. Andersen, V. Kontinen, and M. Sarvas. 1993. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol. Microbiol. 8:957-966. [DOI] [PubMed] [Google Scholar]
- 21.Kok, J., K. J. Leenhouts, A. J. Haandrikman, A. M. Ledeboer, and G. Venema. 1988. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl. Environ. Microbiol. 54:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Laloi, P., D. Atlan, B. Blanc, C. Gilbert, and R. Portalier. 1991. Cell-wall-associated proteinase of Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397: differential extraction, purification and properties of the enzyme. Appl. Microbiol. Biotechnol. 36:196-204. [DOI] [PubMed] [Google Scholar]
- 24.Margot, P., and D. Karamata. 1996. The wprA gene of Bacillus subtilis 168, expressed during exponential growth, encodes a cell-wall-associated protease. Microbiology 142:3437-3444. [DOI] [PubMed] [Google Scholar]
- 25.Mercenier, A., H. Muller-Alouf, and C. Grangette. 2000. Lactic acid bacteria as live vaccines. Curr. Issues Mol. Biol. 2:17-25. [PubMed] [Google Scholar]
- 26.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 27.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pederson, J. A., G. J. Mileski, B. C. Weimer, and J. L. Steele. 1999. Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J. Bacteriol. 181:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson, K., L. M. Chamberlain, K. M. Schofield, J. M. Wells, and R. W. Le Page. 1997. Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat. Biotechnol. 15:653-657. [DOI] [PubMed] [Google Scholar]
- 31.Roissart, H., and F. M. Luquet (ed.). 1994. Bactéries lactiques, vol. I. Lorica, Uriage, France.
- 32.Scheppler, L., M. Vogel, A. W. Zuercher, M. Zuercher, J. E. Germond, S. M. Miescher, and B. M. Stadler. 2002. Recombinant Lactobacillus johnsonii as a mucosal vaccine delivery vehicle. Vaccine 20:2913-2920. [DOI] [PubMed] [Google Scholar]
- 33.Shinde, U., and M. Inouye. 1993. Intramolecular chaperones and protein folding. Trends Biochem. Sci. 18:442-446. [DOI] [PubMed] [Google Scholar]
- 34.Shinde, U., and M. Inouye. 2000. Intramolecular chaperones: polypeptide extensions that modulate protein folding. Semin. Cell Dev. Biol. 11:35-44. [DOI] [PubMed] [Google Scholar]
- 35.Siezen, R. J. 1999. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Leeuwenhoek 76:139-155. [PubMed] [Google Scholar]
- 36.Siezen, R. J., P. G. Bruinenberg, P. Vos, I. Alen-Boerrigter, M. Nijhuis, A. C. Alting, F. A. Exterkate, and W. M. de Vos. 1993. Engineering of the substrate-binding region of the subtilisin-like, cell-envelope proteinase of Lactococcus lactis. Protein Eng. 6:927-937. [DOI] [PubMed] [Google Scholar]
- 37.Steidler, L., K. Robinson, L. Chamberlain, K. M. Schofield, E. Remaut, R. W. Le Page, and J. M. Wells. 1998. Mucosal delivery of murine interleukin-2 (IL-2) and IL-6 by recombinant strains of Lactococcus lactis coexpressing antigen and cytokine. Infect. Immun. 66:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vos, P., G. Simons, R. J. Siezen, and W. M. de Vos. 1989. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J. Biol. Chem. 264:13579-13585. [PubMed] [Google Scholar]