Abstract
A pigment mutant strain of the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina BBS was isolated by plasposon mutagenesis. Nineteen open reading frame, most of which are thought to be genes involved in the biosynthesis of carotenoids, bacteriochlorophyll, and the photosynthetic reaction center, were identified surrounding the plasposon in a 22-kb-long chromosomal locus. The general arrangement of the photosynthetic genes was similar to that in other purple photosynthetic bacteria; however, the locations of a few genes occurring in this region were unusual. Most of the gene products showed the highest similarity to the corresponding proteins in Rubrivivax gelatinosus. The plasposon was inserted into the crtD gene, likely inactivating crtC as well, and the carotenoid composition of the mutant strain corresponded to the aborted spirilloxanthin pathway. Homologous and heterologous complementation experiments indicated a conserved function of CrtC and CrtD in the purple photosynthetic bacteria. The crtDC and crtE genes were shown to be regulated by oxygen, and a role of CrtJ in aerobic repression was suggested.
The photosynthetic apparatus in many purple bacteria consists of three kinds of pigment-protein complexes: two types of the light-harvesting antenna complex (LHI and LHII) absorb light and transfer the energy to the third complex, the reaction center (17). The pigment components of these complexes are essential for efficient photosynthetic performance.
In bacterial photosynthesis, carotenoids absorb light energy, participate in the assembly of the light-harvesting antenna complex (13), and protect the cells from photodamage (3).
Numerous pathways have been described for the biosynthesis of more than 100 known carotenoids in photosynthetic anoxygenic bacteria (30). The operons coding for the enzymes of specific carotenoid pathways in photosynthetic bacteria have been studied mainly in nonsulfur bacteria belonging to the α and β subclasses of proteobacteria (1, 10, 14, 16). Little is known about the genes of carotenoid biosynthesis in purple sulfur γ proteobacteria.
Oxygen and light are the main environmental factors affecting the transcription and assembly of the photosynthetic apparatus in Rhodobacter capsulatus and in Rhodobacter sphaeroides (19). In Rhodobacter sphaeroides the crt operons are repressed under aerobic conditions by PpsR (CrtJ is a counterpart in Rhodobacter capsulatus) (7, 20, 23). Thiocapsa roseopersicina BBS is a purple sulfur photosynthetic γ proteobacterium belonging to the family Chromatiaceae. It can be cultivated under photosynthetic anaerobic conditions and requires reduced sulfur compounds for growth. It also grows chemolithoautotrophically under dark, aerobic conditions. The members of the family Chromatiaceae have either spirilloxanthin (normal, unusual spirilloxanthin, and carotenal) or okenone carotenoid biosynthetic pathways (30). Spirilloxanthin was reported to be the major carotenoid in T. roseopersicina 1711 (DSM 217) (29). Here we describe the isolation and genetic analysis of a T. roseopersicina mutant strain with altered carotenoid content and the characterization of a 22-kb locus containing genes involved in pigment biosynthesis. The regulation of carotenoid biosynthesis genes in this purple sulfur bacterium is also discussed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used are listed in Table 1. T. roseopersicina strains were maintained in Pfennig's mineral medium (containing, per 1,000 ml, 20 g of NaCl, 1 g of KH2PO4, 1 g of MgCl2, 1 g of KCl, 1 g of NH4Cl, 2 g of Na2S2O3, 200 μl of vitamin B12 [100 μg/ml]), 1 ml of Fe-EDTA [3.3 g/liter], and 1 ml of microelement solution [2,975 mg of Na2-EDTA, 300 mg of H3BO4, 200 mg of CaCl2 · 6H2O, 100 mg of ZnSO4 · 7H2O, 30 mg of MnCl2 · 4H2O, 30 mg of Na2MoO4 · 2H2O, 20 mg of NiCl2 · 6H2O, and 10 mg of CuCl2 · 2H2O in 1,000 ml of H2O]); they were grown photoautotrophically and anaerobically in liquid cultures for 3 to 4 days (22). Plates were solidified with 7 g of Phytagel (Sigma) per liter, which was supplemented with acetate (2 g/liter) when selecting for transconjugants, and incubated for 2 weeks in anaerobic jars by using the GasPak (BBL) or AnaeroCult (Merck) systems. Cultures were illuminated with continuous light at 27 to 30°C (5). In the presence of oxygen the culture was supplemented with 5 g of d-glucose per liter and cultivated in dark under air. Escherichia coli strains were maintained on Luria-Bertani medium (27). Antibiotics were used at the following concentrations (micrograms per milliliter): for E. coli, streptomycin (50), ampicillin (100), kanamycin (50), and tetracycline (20); for T. roseopersicina, kanamycin (20) and streptomycin (5).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| T. roseopersicina strains | ||
| BBS | Wild type | 2 |
| RM26 | BBS crtD::Km | This work |
| E. coli strains | ||
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endAL supE44 thi-1 recA1 gyrA96, relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| S17-1(λpir) | 294 (recA pro res mod) Tpr Smr (pRP4-2-Tc::Mu-Km::Tn7) λpir | 8 |
| BL21(DE3) | E. coli B F−dcm ompT hsdS(rB− mB−) gal λ(DE3) | Novagen |
| Plasmids | ||
| pTnMod-OKm | Kmr; Tn5-based plasposon delivery plasmid with Kmr | 4 |
| pGEM T-Easy | Ampr; cloning vector | Promega |
| pBluescript SK(+) | Ampr; cloning vector | Stratagene |
| pBBRexSm2 | Smr; broad host range vector | B. Fodor unpublished |
| pRM261 | 3.5-kb BamHI fragment harboring the plasposon from RM26 | This work |
| pRM265 | 4.9-kb KpnI fragment harboring the plasposon from RM26 | This work |
| pRM268 | 18.8-kb XbaI fragment harboring the plasposon from RM26 | This work |
| pSOX | pBluescript KS(+); carries 1.2-kb SacI fragment of crtD from Rubrivivax gelatinosus | 18 |
| pSO24 | pBluescript KS(+); carries 1.8-kb SacI fragment of crtD-crtC from Rubrivivax gelatinosus | 18 |
| pRcrt3 | pBluescript SK(+); carries the ApaI-SacI fragment of promoterless crtD-crtC from Rubrivivax gelatinosus | This work |
| pRcrt4 | Derivative of pRcrt3; contains the promoter of crtD from T. roseopersicina | This work |
| pRcrt5 | pBBRexSm2 containing BamHI-KpnI fragment of pRcrt4 | This work |
| pTcrt3 | pBluescript SK(+); carries the wild-type BamHI-SacI fragment of the crtDC operon of T. roseopersicina | This work |
| pTcrt4 | pBBRexSm2 containing BamHI-SacI fragment of pTcrt4 | This work |
| pPHU235 | Broad-host-range lacZ fusion vector | 9 |
| pK18mobsacB | Kmr; sacB; RP4 oriT; ColE1 ori | 28 |
| pKlac2 | EcoRI-SalI fragment from pPHU235 in pK18mobsacB | This work |
| pCrtlac4 | pKlac2 with 1,067-bp PstI-XhoI fragment from pRM265 | This work |
| pCrtlac9 | pKlac2 with 1,067-bp PstI-XhoI fragment from pRM265 | This work |
| pET28::CrtJ | pET28 overexpression plasmid with crtJ gene | 23 |
| pCRTI | pGEM T-Easy; contains 444-bp fragment of crtI | This work |
| pPPSR | pGEM T-Easy; contains 929-bp fragment of ppsR | This work |
Plasposon mutagenesis.
Plasmid pTnMod-OKm (4) was introduced into a T. roseopersicina BBS recipient by conjugation (5), and mutants were selected for kanamycin resistance. From the kanamycin-resistant colonies, which were expected to contain the plasposon, a pigment mutant pale colony was chosen for further work.
Molecular biology techniques.
Standard recombinant DNA techniques were carried out as described previously (27) or according to the specifications of the manufacturers.
Isolation and analysis of the locus surrounding the plasposon.
DNA fragments were isolated from genomic DNA digested with BamHI, KpnI, and XbaI enzymes, self-ligated, and transformed into XL1-Blue MRF′ competent cells. A 21.7-kb region was subcloned and sequenced on both strands by primer walking with an automated Applied Biosystems 373 Stretch DNA sequencer.
Identification of the crtI and ppsR genes.
Multiple alignment of the known CrtI and PpsR proteins was performed, and conserved domains were chosen for designing PCR primers corresponding to the selected amino acid sequences as follows: MGLFVWY (amino acids [aa] 312 to 318) and AWFRPHN (aa 457 to 464) in the Rhodobacter capsulatus CrtI protein and ETRYRVL (aa 154 to 160) and LYVKLRR (aa 454 to 460) in the Rhodobacter capsulatus CrtJ (PpsR) enzyme. The presence of crtI and ppsR in the genome of T. roseopersicina was demonstrated by using PCR with the following primers: for crtI, crtio1 (5′ATGGGIYTITTYGTSTGGTA3′) and crtio2 (5′TTRTGSGGIGCRAACCASGC3′); for ppsR (crtJ), ppso1 (5′GAIACICGITAYCGNGTSCT3′) and ppso2 (5′CGICGIAGYTT SACRTASAG3′) (where S is C or G, R is A or G, and Y is C or T). The PCR products (444 bp for crtI and 929 bp for ppsR [crtJ]) were cloned into pGEM T-Easy vector and sequenced.
Bioinformatics tools.
Comparisons of DNA and protein sequences with the various databases were done with the FASTA and BLAST (N, P, and X) programs (www.ncbi.nih.nlm.gov). Multiple alignments were performed with the CLUSTALX program.
Constructions for complementation.
The plasmid for the homologous complementation of crtDC mutant strains was generated as follows. The 4.9-kbp BamHI-SacI fragment from the pRM261 clone (Table 1) containing the crtDC genes was cloned into the pBluescript SK(+) BamHI-SacI sites (pTcrt2). This region contained the plasposon inserted into the crtD gene (at nucleotide [nt] 16812 on the whole sequence). To restore the genomic sequence, a 526-bp region was amplified from the wild-type genome by using the following primers upstream and downstream from the plasposon insertion site: caro4 (5′GGACCGACGG TCTTCACGAT 3′; nt 17300 to 17325, reverse), and caro5 (5′GTCTGATGCA TGCCGCCTTC 3′; nt 16799 to 16818, forward). The PCR fragment was cloned and sequenced, and the 439-bp XhoI-SphI fragment of this clone replaced the corresponding region of the pTcrt2 construct, restoring the wild-type sequence (pTcrt3). The pBBRexSm2 vector was generated by cloning the polished 2,019-bp HindIII fragment of the pHP45Ω (24) vector harboring the streptomycin resistance cassette into the blunted SphI-EcoRV site of pBBR1ex vector. The pBBR1ex construct contained the EcoRV-SphI fragment of pET15b (Novagen) in pBBR1-MCS5 PvuI (polished)-SphI sites (12). The relevant features of pBBRexSm2, which will be a component of a vector set, are that it is a small, broad-host-range, mobilizable vector conferring streptomycin resistance to the host cells (B. Fodor et al., personal communication). The pTcrt4 construct was produced by cloning the 2.9-kbp BamHI-SacI fragment of pTcrt3 into BglII-SspI-digested pBBRexSm2. The plasmid for heterologous complementation of crtDC mutant strains, a 2,850-bp ApaI-SacI fragment carrying the promoterless crtDC genes of Rubrivivax gelatinosus, was assembled from the SacI fragment of the pSOX vector and the SacI-ApaI fragment of the pSO24 plasmid (18) in pBluescript SK(+) (pRcrt3) (Table 1). The 116-bp BamHI-HaeIII fragment of pRM261, containing the crtDC promoter from T. roseopersicina, was cloned into the BamHI-EcoRV sites of the pRcrt3 vector (pRcrt4). The whole operon was transferred into the pBBRexSm2 BglII-SspI sites (pRcrt5) as a BamHI-KpnI fragment after polishing of the noncompatible ends.
Construction of the crtD::lacZ and crtE::lacZ fusion strains.
The promoterless and slightly truncated lacZ gene coding for active enzyme was cloned from pPHU235 as an EcoRI-SalI fragment (9) into the EcoRI-SalI sites of the mobilizable suicide vector pK18mobsacB (pK18lac2). The blunted 1,071-bp PstI-XhoI fragment from pRM265 (containing a 247-bp region of the crtD gene, a 703-bp section of crtE gene, and the intergenic region of these genes) was inserted into the unique ScaI site of pK18lac2. Two plasmids containing the insert in different orientations were chosen: in one orientation (pCrtlac4), the crtD promoter drove the expression of the crtD::lacZ fusion gene, while in the other (pCrtlac9), the crtE promoter was active in producing the crtE::lacZ fused transcript. These plasmids were conjugated into T. roseopersicina BBS. The site of recombination was verified by PCR on genomic DNA with primers specific for the vector (reverse primer) and the crt genes (for the crtD fusion, caro5 [see above]; for crtE::lacZ, caro17 [5′TGCGAACCGACGCGACCTAA3′]). In both cases, fragments of the expected size were obtained, i.e., 1,282 bp for the crtD::lacZ fusion and 1,505 bp for the crtE::lacZ fusion.
Spectrophotometric analysis of the pigments.
Carotenoids were extracted from the cells (and from the dots in the thin-layer chromatography [TLC] plates) with acetone-methanol (7:2, vol/vol) as described previously (18). Spectral analysis was carried out with a UV2 Unicam spectrophotometer interfaced with a computer.
β-Galactosidase assay.
The β-galactosidase activities of the toluene-permeabilized cell extracts were assayed as described earlier (15). One Miller unit corresponded to 1 mmol of o-nitrophenyl-β-galactoside (Sigma-Aldrich) hydrolyzed per min, normalized to the optical density at 650 nm.
Overexpression and purification of CrtJ.
Plasmid pET28::CrtJ (23) harboring the Rhodobacter capsulatus crtJ gene was transformed into E. coli strain BL21(DE3) (Novagen), and CrtJ was expressed and purified as described previously (23).
Gel mobility retardation assay.
The 120-bp BamHI-HindIII fragment from pRcrt4 (see above), containing the putative CrtJ recognition sequence elements, was isolated and labeled with α-35S-dATP. The binding mixture contained 1 ng of radiolabeled DNA, 1 μg of poly(dI-dC), and various amounts of proteins in the binding buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 50 mM potassium acetate, 20% [vol/vol] glycerol). Each reaction mixture was then incubated for 30 min at 30°C and loaded onto a 6% nondenaturing polyacrylamide gel. The gel was electrophoresed at 70 V for 2 h, dried, and analyzed in a PhosphorImager (Molecular Dynamics).
Nucleotide sequence accession number.
The 21,710-bp sequence determined in this study has been deposited in GenBank under accession number AF528191.
RESULTS
Isolation of pigment mutant strains of T. roseopersicina.
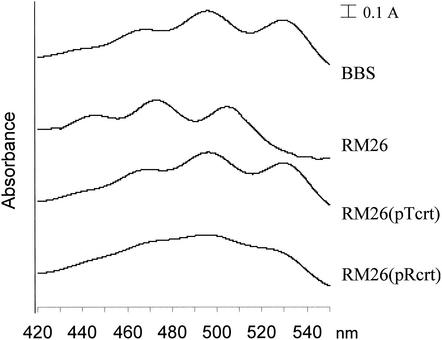
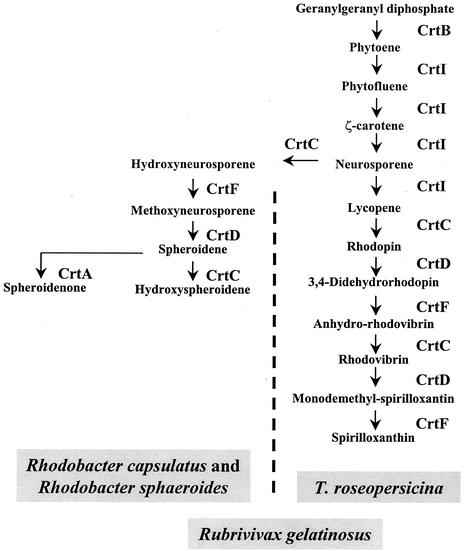
Plasposon-mediated mutagenesis was used to isolate colonies with altered pigmentation. A pale mutant colony (RM26) was chosen from a library of 4,000 kanamycin-resistant colonies. Spectral analysis of the extracted pigments showed that the absorbance peaks at 468, 496, and 528 nm (Fig. 1, BBS) disappeared and that new maxima appeared at 445, 472, and 504 nm (Fig. 1, RM26). These values coincided well with the published data for spirilloxanthin (465, 495, and 528 nm) and lycopene (442, 470, and 500 nm), respectively (18) (the actual values depend slightly on the solvent used). Separation of the extracted carotenoids by TLC on a silica gel revealed a single spot for the wild type and four to five spots with distinct mobilities for the mutant (data not shown). Each spot was cut out from the TLC plate, and their UV spectra were recorded after extraction. In each case, only peaks characteristic of lycopene could be observed (data not shown), indicating that these compounds were likely lycopene derivatives. Therefore, we concluded that wild-type T. roseopersicina synthesizes spirilloxanthin as the main carotenoid but that the carotenoid biosynthesis is aborted at lycopene in the RM26 mutant. The pathway previously described for spirilloxanthin biosynthesis is shown in Fig. 2 (11, 18, 30).
FIG. 1.
Absorption spectra of carotenoid extracts from wild-type T. roseopersicina, a crtD mutant, and complemented strains. BBS, wild type; RM26, crtD mutant; RM26(pTcrt) and RM26(pRcrt), RM26 complemented with crtDC of T. roseopersicina and Rubrivivax gelatinosus, respectively. The zero lines of the spectrums were shifted for better viewing. For details, see text.
FIG. 2.
Biosynthesis of speroidene and spirilloxanthin in photosynthetic bacteria. Rhodobacter sphaeroides and Rhodobacter capsulatus produce spheroidene (1, 14). In T. roseopersicina only spirilloxanthin can be detected, while in Rubrivivax gelatinosus both spheroidene and spirilloxanthin are synthesized (18).
Isolation and sequence analysis of genes involved in pigment biosynthesis.
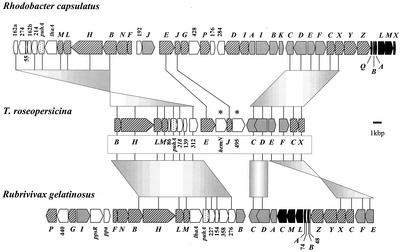
Overlapping restriction fragments of the genome of the RM26 containing the plasposon were isolated and cloned, and an almost-22-kb DNA region was sequenced. The in silico analysis of this contig resulted in the identification of genes coding for putative enzymes participating in bacteriochlorophyll and carotenoid biosynthesis (Fig. 3). The annotated open reading frames (ORFs) are listed in Table 2. Nineteen ORFs were identified; nine were involved in bacteriochlorophyll (two of them, bchB and bchX, were partial), four were involved in carotenoid biosynthesis, and the remaining six coded for putative proteins of the photosynthetic reaction center or heme biosynthesis or their function was not clear.
FIG. 3.
The structure of the photosynthetic gene cluster of T. roseopersicina compared to those of Rhodobacter capsulatus (1) and Rubrivivax gelatinosus (10). Genes and ORFs are represented as arrows pointing in the direction of their transcription. Solid arrows indicate the puf genes, coding for the reaction center and light-harvesting center; stippled arrow symbolize the gene of the reaction center H subunit (puhA). The hatched and gray arrows stand for ORFs assigned to bacteriochlorophyll and carotenoid biosynthesis, respectively. Genes showing unusual locations in T. roseopersicina are labeled with asterisks. Open arrows indicate ORFs with unknown functions.
TABLE 2.
Description of the ORFs identified
| Genea | Known or putative function of product | Length (aa) | nt (start-stop) | Top BLAST hits
|
|
|---|---|---|---|---|---|
| Gene product | % Identity/E value | ||||
| bchB | Light-independent prochlorophyllide reductase b subunit | 258 | 1-777 | Rubrivivax gelatinusus BchB | 72/e-107 |
| bchH | Mg-protoporphyrin IX chelatase H subunit | 1,245 | 752-4489b | Rubrivivax gelatinusus BchH | 66/0.0 |
| bchL | Light-independent prochlorophyllide reductase iron-sulfur ATP binding subunit | 294 | 4513-5397 | Rhodospirillum rubrum BchL | 66/e-105 |
| Rubrivivax gelatinusus BchL | 64/e-105 | ||||
| bchM | Mg-protoporphyrin methyltranserase | 233 | 5397-6098 | Rubrivivax gelatinusus BchM | 60/9e-76 |
| orf86 | Hypothetical protein | 86 | 6095-6355 | ||
| puhA | Photosynthetic reaction center H subunit | 255 | 6381-7148 | Thermochromatium tepidum PuhA | 72/e-108 |
| Rubrivivax gelatinusus PuhA | 49/e-63 | ||||
| orf218(G) | Hypothetical membrane protein | 218 | 7145-7801 | Rubrivivax gelatinusus ORF227 | 46/e-43 |
| orf139 | Hypothetical protein | 139 | 7853-8272 | Rubrivivax gelatinusus ORF154 | 39/9e-18 |
| orf312 | Hypothetical membrane protein | 312 | 8535-9473c | Rubrivivax gelatinusus ORF276 | 43/6e-58 |
| bchE | Mg-protoporphyrin IX monomethylester oxidative cyclase subunit | 551 | 9864-11519c | Heliobacillus mobilis BchE | 70/0.0 |
| hemN | O2-independent coproporphyrinogen III oxidase | 453 | 11533-12894 | Aquifex aeolicus HemN | 38/6e-77 |
| Rubrivavax gelatinusus HemN | 40/2e-76 | ||||
| bchJ (G) | 4-Vinyl reductase | 208 | 12870-13496b | Rhodobacter sphaeroides BchJ | 42/e-35 |
| orf543 | Long-chain fatty acid CoA ligase | 543 | 13330-14961b | Halobacterius sp. strain Lfl1 | 38/3e-52 |
| orf495 (G) | o-Succinyl-benzoic acid CoA ligase | 495 | 13474-14961b | MenE Listeria innocua | 34/4e-40 |
| crtC ⇐ | Hydroxyneurosporene dehydrogenase | 405 | 16294-15077b | Rubrivivax gelatinusus CrtC | 55/e-92 |
| crtD ⇐ | Methoxyneurosporene dehydrogenase | 498 | 17487-15991 | Rubrivivax gelatinusus CrtD | 54/e-150 |
| crtE | Geranylgeranyl pyrophosphate synthase | 288 | 17607-18473c | Rubrivivax gelatinusus CrtE | 55/6e-88 |
| crtF (G) | Hydroxyneurosporene methyltransferase | 371 | 18868-19982c | Rubrivivax gelatinusus CrtF | 49/e-89 |
| bchC (T) | 2-α-Hydroxyethyl bacteriochlorophyllide oxidase | 317 | 20092-21045 | Rubrivivax gelatinusus BchC | 61/e-106 |
| bchX | Bacteriochlorophyllide reductase subunit | 282 | 20871-21710 | Bradyrhizobium sp. BchX | 74/6e-79 |
| 225 | 21042-21710 | Rubrivivax gelatinusus BchX | 73/2e-76 | ||
G and T, GTG and TTG start codons, respectively. The arrows indicate the inverse orientation of the ORFs.
Overlaps with preceding ORF by more than 10 bp.
Gap longer than 50 bp.
The orientation of all ORFs was the same, with the exception of the crtC and crtD genes (Fig. 3). Several ORFs overlapped, and in few cases the genes were separated by gaps (Table 2.). Generally, the ORFs were preceded by more-or-less conserved ribosomal binding sites; four ORFs started with GTG, and one (bchC) probably started with TTG. Local arrangements of some photosynthetic genes, such as bchBHLM and puhA-orf218-orf138, were similar to that in Rhodobacter and Rubrivivax strains (Fig. 3). The arrangement of the crtCDEF-bchCX genes was the same in the case of Rhodobacter species but distinct from that in Rubrivivax gelatinosus, where the crtCD and crtEF genes are separated by many genes involved in the biosynthesis of bacteriochlorophyll and the photosynthetic center. Moreover, a few genes, like hemN and orf543/orf495 (“extra genes” in Fig. 3) inserted into this region; their locations in the other three photosynthetic bacteria were completely different. Most of the gene products showed the highest similarity to the corresponding proteins of Rubrivivax gelatinosus (10) (Table 2). In addition to CrtC, CrtD, CrtE, and CrtF, which could be identified in this locus, the spirilloxanthin pathway needed two additional enzymes, CrtB and CrtI (Fig. 2) (11, 30), but the corresponding genes were not found on this fragment. Hence degenerate primers were produced on the basis of the conserved regions of CrtB and CrtI of Rubrivivax and Rhodobacter species (see Materials and Methods). In the case of crtI the expected 444-bp fragment could be amplified and the deduced sequence showed the highest similarity (74%) to that of Rubrivivax gelatinosus CrtI (10). For CrtB, this approach did not succeed (data not shown).
The plasposon was inserted into the middle of the crtD gene (at nt 16812) (Table 2), coding for the methoxyneurosporene dehydrogenase. Downstream from the plasposon insertion site, the crtC gene was found in the same direction as crtD, and the two genes had a 304-bp overlap, so they are likely cotranscribed. Moreover, the orientation of the kanamycin resistance gene in the inserted plasposon is opposite to that in the crtDC genes, so the promoter of this gene cannot drive the expression of the crtC gene. Consequently, the mutation should have a polar effect as well, and we consider RM26 to be a crtDC mutant.
Complementation with crtDC genes.
Homologous complementation of the mutated crtDC genes restored the wild-type carotenoid composition [Fig. 1, RM26(pTcrt) spectrum]. CrtC was shown to be involved in the synthesis of hydroxyneurosporene from neurosporene in the spheroidene branch of carotenoid biosynthesis in Rhodobacter species (1, 14) and in the synthesis of both spheroidene and spirilloxanthin in Rubrivivax gelatinosus (18) (Fig. 2). CrtI has to catalyze three and four consecutive steps in the spheroidene and spirilloxanthin pathways, respectively (three-step and four-step phytoene desaturase) (Fig. 2). The spheroidene and spirilloxanthin pathways have a common origin, and they branch after the synthesis of ξ-carotene (Fig. 2). The next step is catalyzed by CrtC in the spheroidene pathway and by CrtI in the spirilloxanthin pathway. Downstream from this branching point the same enzyme set is used in both pathways, except for an additional step catalyzed by CrtA in the spheroidene pathway. Thus, the special properties of CrtC and or CrtI, which may be distinct in various species, determine the actual pathway taking place in the cells. In the crtIC mutant strain of Rhodobacter sphaeroides, the native four-step phytoene desaturase (CrtI) in trans was able to produce significant amount (13%) of lycopene in a crtC background (6). Lycopene is an intermediate of the spirilloxanthin route, which is normally not present in Rhodobacter sphaeroides. This suggested that CrtC might have a key role in determining the selection of the various carotenoid biosynthetic pathways. The isolated T. roseopersicina crtDC mutant also contained lycopene and its derivatives (Fig. 1, RM26). We addressed the question of whether the CrtC enzyme of Rubrivivax gelatinosus (in which both the spirilloxanthin and spheroidene pathways exist [Fig. 2]) can supplement the carotenoid pathway with the spheroidene branch in purple sulfur bacteria, since T. roseopersicina is able to synthesize spirilloxanthin only. The crtD gene from Rubrivivax gelatinosus S1 (18) was fused to the promoter of T. roseopersicina crtD and introduced into the RM26 mutant. In this construct the crtC gene was located downstream from the crtD gene, and they were thought to be cotranscribed (18), so in our construct the expression of both the crtD and crtC genes of Rubrivivax gelatinosus was driven by the T. roseopersicina crtD promoter. The spectral and TLC analyses of the pigments indicated the synthesis of spirilloxanthin (and the lycopene derivatives as in the case of the RM26 mutant), but intermediates of the spheroidene lineage could not be detected [Fig. 1, RM26(pRcrt) spectrum]. Moreover, the complementation was not as effective as with the homologous crtDC genes, and the spectrum is broadened, which might be caused by accumulated intermediates appearing in the spirilloxanthin biosynthesis as a consequence of reduced activity of the heterologous enzymes (S. Takaichi, personal communication).
Regulation of the crtD and crtE genes.
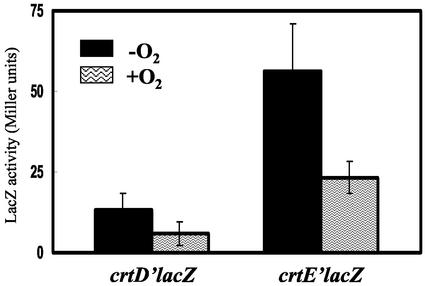
T. roseopersicina growing under oxygenic conditions has as pale a color as the RM26 mutant, suggesting that the carotenoid biosynthesis is repressed by molecular oxygen. To test this hypothesis, the regulation of the crtD and crtE genes was monitored with the aid of translational lacZ reporter gene fusions. The activity of LacZ produced from either the crtD or crtE promoter was measured in T. roseopersicina cells grown in the presence and absence of oxygen. The expression of both crt genes was repressed in the presence of oxygen (Fig. 4). The extent of the repression was the same in both cases (around 43%), but the promoter of the crtE gene seemed to be almost five times stronger. However, it could not be excluded that this effect derived from the fact that different sequences were fused to the lacZ gene (see Materials and Methods), resulting in dissimilar mRNA stabilities and consequently different LacZ activities (21). Since the crtC gene is believed to be cotranscribed with the crtD gene, this aerobic repression should regulate the expression of the crtC gene as well. The distance between the crtE and crtF genes is too large (395 bp) for such a conclusion to be made in this case.
FIG. 4.
Activity of LacZ expressed from the crtD and crtE promoters in T. roseopersicina BBS grown under aerobic and anaerobic conditions. For details, see Materials and Methods. Error bars indicate standard deviations.
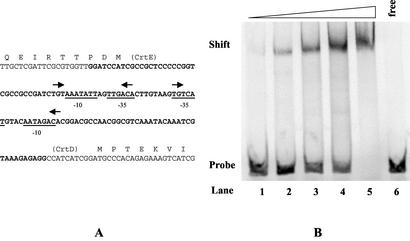
The regions upstream from the crtD and crtE genes have sequences similar to consensus σ70 promoters, which are typical of photosynthetic operons (Fig. 5A) (10). In Rhodobacter capsulatus and Rhodobacter sphaeroides, oxygen affected the expression of the crt and bch genes via a complex cascade to a repressor protein, named CrtJ in Rhodobacter capsulatus and PpsR in Rhodobacter sphaeroides (7, 20, 23). This factor (CrtJ) in Rhodobacter capsulatus recognized a palindrome TGT-N12-ACA sequence motif (23) which overlapped with the putative promoter. The consensus sequence could be found in two copies between the crtD and crtE genes of T. roseopersicina (Fig. 5A). In addition, a consensus sequence within the bchH gene was recognized, which might be irrelevant. To test whether these elements were really CrtJ (PpsR) binding motifs, CrtJ from Rhodobacter capsulatus (23) was overexpressed in E. coli and examined in a gel mobility retardation assay. The purified CrtJ protein bound strongly to the intergenic region of the crtD and crtE genes (Fig. 5B). The specificity of the interaction was confirmed by experiments in which specific and nonspecific cold competitors were added to the binding mixture. The disappearance of the band corresponding to the CrtJ-DNA complex required at least 1,000 times more nonspecific [poly(dI-dC)] molecules than specific competitor (data not shown), indicating the specific interaction of the labeled DNA probe and CrtJ. The presence of the repressor, PpsR, in T. roseopersicina was demonstrated by amplification and sequencing of an almost-1-kb region of the ppsR gene with degenerate primers, which were planned on the basis of the conserved regions of the known PpsR (CrtJ) proteins. The deduced amino acid sequence had 42% identity to the corresponding region of the PpsR protein in Bradyrhizobium sp. strain ORS278 (data not shown).
FIG. 5.
Binding of CrtJ to the promoter region of the crtD and crtE genes. (A) The intergenic region between the divergent crtE and crtD genes, where the relevant region of this fragment is displayed. The putative −10 and −35 promoter regions are underlined. The arrows indicate putative CrtJ palindrome recognition sites. (B) Gel retardation assay with the recombinant CrtJ. A 120-bp BamHI-HindIII fragment containing the crtE-crtD intergenic region (the region used is in boldface in panel A) was isolated and labeled with α-35S-dATP (see Materials and Methods). This labeled fragment was incubated with various amounts of Rhodobacter capsulatus CrtJ protein overexpressed and purified in E. coli and loaded onto a 6% native polyacrylamide gel (see Materials and Methods). Lane 6, control lane containing only the free DNA probe. Lanes 1 to 5, the DNA probe was incubated with increasing amounts (0.05, 0.11, 0.23, 0.45, and 0.9 μg, respectively) of pure CrtJ.
DISCUSSION
In accordance with the only literature source (29), spirilloxanthin was determined as the main carotenoid in the wild-type T. roseopersicina BBS. A mutant strain was produced, in which a plasposon was inserted into the crtD gene, disrupting the methoxyneurosporene dehydrogenase, and likely the expression of the CrtC was also affected. The carotenoid extract of this mutant revealed an absorption spectrum characteristic of lycopene (Fig. 1), but by TLC on a silica gel, four to five distinct bands could be separated (data not shown). It is possible that the functional CrtF in the RM26 mutant strain converted the lycopene to its nonnatural derivatives, since the expression of crtF was not influenced by the mutation. This is supported by the fact that the spectra of the isolated spots had absorption maxima characteristic of lycopene (data not shown). A similar situation has been described for Rhodospirillum rubrum, where a strain having a mutation in the rhodopin 3,4-desaturase gene was shown to contain not only rhodopin but its nonnatural derivatives produced by CrtF (11).
On a 22-kb locus surrounding the plasposon, 19 ORFs were identified (Fig. 3). Most of them code for putative proteins involved in bacteriochlorophyll and carotenoid biosynthesis. The majority of the putative gene products have higher identity to their counterparts in Rubrivivax gelatinosus than to those in Rhodobacter capsulatus or Rhodobacter sphaeroides (Table 2). This coincides with the relationship established from the 16S RNA analysis (16) and with the fact that Rubrivivax gelatinosus produces spirilloxanthin (18). However, the contig organization resembles that of the Rhodobacter species (10): the order of the crtCD-crtEF-bchCX genes is the same (Fig. 3). The arrangement of the pigment biosynthesis gene cluster has a few unusual features in T. roseopersicina. Most importantly, the crtB and crtI genes are missing from this region, although crtI is detected elsewhere in the genome. In T. roseopersicina, the scattering of functionally related genes appears to be a characteristic feature (26). Other differences between the species exist, such as the fact that hemN, involved in the heme biosynthesis, or the hypothetical coenzyme A (CoA) ligase gene (menE or lfl1) is located between bchE/bchJ and bchJ/crtC, respectively. These genes are localized outside the photosynthetic operon in Rubrivivax gelatinosus (10).
Introduction of the homologous crtDC genes into T. roseopersicina could restore the wild-type spirilloxanthin biosynthetic route in the mutant RM26. It has been reported that the CrtC and CrtD enzymes of Rubrivivax gelatinosus are involved in the biosynthesis of both spirilloxanthin and spheroidene (18). The two pathways branch after the synthesis of neurosporene: CrtC synthesizes hydroxyneurosporene, while CrtI produces lycopene (Fig. 2). Although these two genes are apparently present in T. roseopersicina, no carotenoid corresponding to the spheroidene pathway was detectable. The intriguing question that remained to be answered was what determines the branching selection of the carotenoid biosynthesis in bacteria having the enzymes for both pathways. In order to address this puzzle, heterologous complementation of the crtDC mutant T. roseopersicina strain with the crtDC genes of Rubrivivax gelatinosus was carried out. This apparently did not switch the carotenogenesis of T. roseopersicina toward the spheroidene pathway; absorption peaks corresponding to the spirilloxanthin pathway could be observed. TLC analysis of the pigment composition revealed only thee spots, which were attributed either to the wild type (spirilloxanthin) or to the RM26 mutant (lycopene and its derivatives) (data not shown). One possible explanation of the results is that in T. roseopersicina the CrtI protein, belonging to the four-step desaturases, may have very strong affinity to neurosporene and no free neurosporene remains in the cells for CrtC. Alternatively, it is also conceivable that in T. roseopersicina the spheroidene pathway is not functionally active.
The regulation of the crtD and crtE genes in T. roseopersicina was studied with promoter fusion constructs and gel mobility retardation assay. The expression of the operons is similarly affected by oxygen, which seems to be mediated by the repressor CrtJ (PpsR), which is known in purple nonsulfur bacteria (7, 20, 23). The consensus binding regions for CrtJ (PpsR) overlapping with the σ70 promoter-like sequences were identified upstream from the carotenoid biosynthesis genes crtD and crtE in both directions. Heterologously overexpressed CrtJ of Rhodobacter capsulatus binds to the crtD-crtE intergenic region of T. roseopersicina, suggesting an evolutionarily conserved mechanism for the anaerobic regulation. Remarkably, we could not detect any other consensus binding site of CrtJ in the 22-kb locus, although this was expected in the case of bchC, hemN, or bchE (19). Fnr is another redox regulator controlling the expression of the photosynthetic genes (19), but its consensus binding site was not found in this contig. The organization of the genes, gaps, overlapping regions, potential loops, and rare start codons might have a role in posttranscriptional events such as mRNA degradation (25) or translation, where the usage of rare start codons leads to reduced translational efficacy. These might result in altered expression levels of the various components, even with linked functions.
The similarity of the proposed genes and carotenogenesis and the regulation through PpsR (or CrtJ) of the crtDC and crtE genes suggest a strong relationship between the photosynthetic gene clusters of the purple sulfur bacteria from the γ subdivision and the purple nonsulfur bacteria from the β subdivision. The putative proteins of T. roseopersicina that are involved in pigment biosynthesis show higher identity to the corresponding enzymes of Rubrivivax gelatinosus than to those of the Rhodobacter species, although the arrangement of their genes suggests otherwise. One may thus speculate on the possibility of horizontal gene transfer from the Rhodobacter species to Rubrivivax gelatinosus through T. roseopersicina followed by genome rearrangements induced by environmental factors, such as oxidative stress (10, 17).
Acknowledgments
This work was funded by the EU 5th Framework Programme (QLK5-1999-01267 and ICA1-CT2000-70026).
We thank Chantal Astier (Centre de Génétique Moléculaire, Gif-sur-Yvette, France) for pSOX and pSO24, Paulette Vignais and Annette Colbeau (DBMS, CEA-CENG, Grenoble, France) for pPHU235, Carl E. Bauer (Indiana University, Bloomington) for pET28::CrtJ, Gerben J. Zylstra (Rutgers University, New Brunswick, N.J.) for pTnModOKm, and Andreas Schäfer (University of Bielefeld, Bielefeld, Germany) for pK18mobsacB.
REFERENCES
- 1.Armstrong, G. A., M. Alberti, F. Leach, and J. E. Hearst. 1989. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol. Gen. Genet. 216:254-268. [DOI] [PubMed] [Google Scholar]
- 2.Bogorov, L. V. 1974. About the properties of Thiocapsa roseopersicina BBS, isolated from estuaria of White Sea. Mikrobiologija 43:326-332. [PubMed] [Google Scholar]
- 3.Cogdell, R. J., and H. A. Frank. 1987. How carotenoids function in photosynthetic bacteria. Biochim. Biophys. Acta 895:63-79. [DOI] [PubMed] [Google Scholar]
- 4.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fodor, B., G. Rákhely, Á. T. Kovács, and K. L. Kovács. 2001. Transposon mutagenesis in purple sulfur photosynthetic bacteria: identification of hypF, encoding a protein capable of processing [NiFe] hydrogenases in α, β, and γ subdivisions of the proteobacteria. Appl. Environ. Microbiol. 67:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Asua, G., R. J. Cogdell, and C. N. Hunter. 2002. Functional assembly of the foreign carotenoid lycopene into the photosynthetic apparatus of Rhodobacter sphaeroides, achieved by replacement of the native 3-step phytoene desaturase with its 4-step counterpart from Erwinia herbicola. Mol. Microbiol. 44:233-244. [DOI] [PubMed] [Google Scholar]
- 7.Gomelsky, M., and S. Kaplan. 1995. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J. Bacteriol. 177:1634-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrero, M., V. Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hübner, P., J. C. Willison, P. M. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 173:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igarashi, N., J. Harada, S. Nagashima, K. Matsuura, K. Shimada, and K. V. P. Nagashima. 2001. Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J. Mol. Evol. 52:333-341. [DOI] [PubMed] [Google Scholar]
- 11.Komori, M., R. Ghosh, S. Takaichi, Y. Hu, T. Mizoguchi, Y. Koyama, and M. Kuki. 1998. A null lesion in the rhodopin 3,4-desaturase of Rhodospirillum rubrum unmasks a cryptic branch of the carotenoid biosynthetic pathway. Biochemistry 37:8987-8994. [DOI] [PubMed] [Google Scholar]
- 12.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, and R. M. Roop. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 13.Lang, H. P., and C. N. Hunter. 1994. The relationship between carotenoid biosynthesis and the assembly of the light-harvesting LH2 complex in Rhodobacter sphaeroides. Biochem. J. 298:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang, H. P., R. J. Cognell, S. Takaichi, and C. N. Hunter. 1995. Complete DNA sequence, specific Tn5 insertion map, and gene assignment of the carotenoid biosynthesis pathway of Rhodobacter sphaeroides. J. Bacteriol. 177:2064-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiment in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Nagashima, K. V. P., K. Shimada, and K. Matusuura. 1993. Phylogenetic analysis of photosynthetic genes of Rhodocyclus gelatinosus: possibility of horizontal gene transfer in purple bacteria. Photosynth. Res. 36:185-191. [DOI] [PubMed] [Google Scholar]
- 17.Ouchane, S., M. Picaud, C. Vernotte, and C. Astier. 1997. Photooxidative stress stimulates illegitimate recombination and mutability in carotenoid-less mutants of Rubrivivax gelatinosus. EMBO J. 16:4777-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouchane, S., M. Picaud, C. Vernotte, F. Reiss-Husson, and C. Astier. 1997. Pleiotropic effects of puf interposon mutagenesis on carotenoid biosynthesis in Rubrivivax gelatinosus. A new gene organization in purple bacteria. J. Biol. Chem. 272:1670-1676. [DOI] [PubMed] [Google Scholar]
- 19.Pemberton, J. M., I. M. Horne, and A. G. McEwan. 1998. Regulation of photosynthetic gene expression in purple bacteria. Microbiology 144:267-278. [DOI] [PubMed] [Google Scholar]
- 20.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which repress carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 176:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pessi, G., C. Blumer, and D. Haas. 2001. lacZ fusions report gene expression, don't they? Microbiology 147:1993-1995. [DOI] [PubMed] [Google Scholar]
- 22.Pfennig, N., and H. G. Trüper. 1991. The family Chromatiaceae, p. 3200-3221. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer, Berlin, Germany.
- 23.Ponnampalam, S. N., and C. E. Bauer. 1997. DNA binding characteristic of CrtJ. A redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 272:18391-18396. [DOI] [PubMed] [Google Scholar]
- 24.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 25.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353-370. [DOI] [PubMed] [Google Scholar]
- 26.Rákhely, G., A. Colbeau, J. Garin, P. M. Vignais, and K. L. Kovács. 1998. Unusual organization of the genes coding for HydSL, the stable (NiFe) hydrogenase in the photosynthetic bacterium Thiocapsa roseopersicina BBS. J. Bacteriol. 180:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, K. 1978. Biosynthesis of carotenoids, p. 729-750. In R. K. Clayton and W. R. Sistrom (ed.), The photosynthetic bacteria. Plenum Press, New York, N.Y.
- 30.Takaichi, S. 1999. Carotenoids and carotenogenesis in anoxygenic photosynthetic bacteria, p. 39-69. In H. A. Frank, A. J. Young, G. Britton, and R. J. Cognell (ed.), The photochemistry of carotenoids. Kluwer, Dordrecht, The Netherlands.