Abstract
The metalloregulatory protein MerR, which exhibits high affinity and selectivity toward mercury, was exploited for the construction of microbial biosorbents specific for mercury removal. Whole-cell sorbents were constructed with MerR genetically engineered onto the surface of Escherichia coli cells by using an ice nucleation protein anchor. The presence of surface-exposed MerR on the engineered strains enabled sixfold-higher Hg2+ biosorption than that found in the wild-type JM109 cells. Hg2+ binding via MerR was very specific, with no observable decline even in the presence of 100-fold excess Cd2+ and Zn2+. The Hg2+ binding property of the whole-cell sorbents was also insensitive to different ionic strengths, pHs, and the presence of metal chelators. Since metalloregulatory proteins are currently available for a wide variety of toxic heavy metals, our results suggest that microbial biosorbents overexpressing metalloregulatory proteins may be used similarly for the cleanup of other important heavy metals.
Mercury is highly toxic to the nervous system, particularly the developing nervous system of a fetus or young child (11). Because of the lack of knowledge about mercury's toxicity and potential environmental impact, in the past, factory effluents typically associated with chlor-alkali plants, battery facilities, and military and medical wastes were commonly released into the surrounding areas (17). The best-documented cases of severe mercury poisoning were from Minamata Bay, Japan, in 1956 and Iraq in 1971.
Conventional treatments to remove Hg2+ from contaminated sources are often inadequate to reduce Hg2+ concentrations to acceptable regulatory standards. Current interest is focused on developing microbial-based biosorbents for the efficient removal of mercury (6). Both naturally occurring metal-binding peptides such as metallothioneins (MTs) and synthetic peptides such as synthetic phytochelatins (ECs [e.g., EC20]) (1-3, 23) have been expressed onto the surface of bacterial cells for improved uptake and biosorption of mercury. However, one major problem associated with these cysteine-rich peptides is their lack of specificity, which may cause difficulty in the specific recovery and recycling of mercury (4).
Many bacteria develop resistance to heavy metals by inducing the expression of an array of resistance proteins (16). Besides the high affinity of these metalloregulatory proteins, a clear advantage is their specificity. One example is the 15.8-kDa regulatory protein MerR, used for controlling the expression of enzymes responsible for mercury detoxification (18). The binding affinity of MerR is several orders of magnitude higher for mercury than for other heavy metals (4, 19).
In this work, we present a new method for selective removal of mercury by generating microbial biosorbents with surface-exposed MerR. The resulting Escherichia coli strain is endowed with the ability to bind mercury with high affinity and selectivity similar to that exhibited by MerR.
MATERIALS AND METHODS
Strains, plasmids, media, and general procedures.
E. coli strain JM109 {Δ(lac-proAB) glnV44 e14− gyrA96 recA1 relA1 endA1 thi hsdR17 [F′ traD36 proA+B+ lacIq Δ(lacZ)M15]} was used in this study. Plasmid pUNIM, a pUC18Not (9) derivative, was used to express an ice nucleation protein (INP)-MerR fusion on the cell surface. Cultures were grown in Luria-Bertani medium supplemented with 100 μg of ampicillin per ml at 37°C to an optical density at 600 nm (OD600) of 0.6 when 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce the expression of the fusion proteins.
Construction of INP-MerR fusion.
The INP-MerR fusion was constructed as follows. The merR fragment was PCR amplified from plasmid pT7KB (5) with the primers merR1 (5′ CCGGGATCCTATGGAAAACAATTTGGAGA 3′) and merR2 (5′ CAGCTGCAGCCCTAAGGCATAGCCGAACC 3′). The amplified fragment was digested with BamHI and PstI, gel purified, and subcloned into a similarly digested pUNI, which contains an EcoRI-BamHI INP fragment inserted into pUC18Not, to generate pUNIM. The resulting construct allows expression of MerR on the surface of E. coli.
To probe the surface localization of MerR, a hexahistidine tag was added to the C-terminal part of the INP-MerR fusion. The merR fragment was reamplified with a new reverse primer, merR3 (5′ ATTCTGCAGCTAATGATGATGGTGGTGGTGATAAGGCATAGCCGAACCTGCCAAGCTT 3′), coding for six histidines at the C terminus. The resulting plasmid, pUNIMH, coding for the INP-MerR-H6 fusion, was prepared as described above.
Cell fractionation.
After overnight induction, cells were harvested and resuspended in 50 mM Tris-Cl buffer (pH 7.4). Cells were disrupted by a French pressure cell at 16,000 lb/in2 (SLM Instruments, Inc.). After two passes through a French press, cell extracts were centrifuged for 10 min at 10,000 rpm to remove the cell debris. The cell extract was then ultracentrifuged at 115,000 × g (Beckman) for 1 h to separate the membrane and soluble fractions. The pellet obtained (membrane fraction) was resuspended in 50 mM Tris buffer (pH 7.4).
Western blot analysis.
Samples (10 μl) of concentrated cells (OD600 = 10) were mixed with loading buffer (15) and boiled for 10 min. Samples were run on a 12.5% (wt/vol) acrylamide sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Proteins were then transferred to a nitrocellulose support before incubation with INP-specific antibodies (14). Western blot analysis was performed with a Bio-Rad Immun-Blot GAR-AP kit (Bio-Rad, Hercules, Calif.). Prestained broad-range molecular weight markers were used to estimate protein molecular weights.
Immunofluorescence microscopy.
Following overnight incubation, cells were harvested and resuspended (OD600 = 0.5) in phosphate-buffered saline (PBS) buffer with 3% bovine serum albumin. Intact cells were then incubated with mouse anti-His6 antisera (1:5,000) for 8 h at 4°C. Cells were washed extensively, resuspended in PBS containing a secondary goat anti-mouse immunoglobulin G (IgG) antibody conjugated with Alexa Fluor 488 (Molecular Probes, Eugene, Oreg.) at a 1:500 dilution, and incubated overnight at 4°C. Prior to microscopy, cells were washed five times with PBS. Photographs were taken with a fluorescence microscope (Olympus).
Mercury binding experiments.
Overnight cultures were harvested, washed twice with 50 mM Tris-Cl buffer (pH 7.4), and resuspended to a final OD600 of 1.0 in the same buffer containing 5 μM HgCl2. Samples were removed after various incubation times, and the amount of bound Hg2+ was determined. For mercury analysis, cells were washed twice with saline, dried, and digested with concentrated nitric acid. Total mercury was analyzed by cold vapor atomic absorption spectrophotometry by using a mercury analyzer (Coleman model 50B). The binding capacity was determined by incubating cells with different concentrations of Hg2+. The selectivity of the engineered cells for Hg2+ was investigated by performing the Hg2+ binding experiments in the presence of various amounts of cadmium and zinc ions. The effect of ionic strength was determined by measuring mercury binding in Tris-Cl buffer (pH 7.4) with sodium chloride concentrations of between 0 and 400 mM. The effect of pH was determined by performing the Hg2+ binding experiments in citric-phosphate-borate buffers at pHs between 3 and 11.
RESULTS
Surface expression of MerR using an INP anchor.
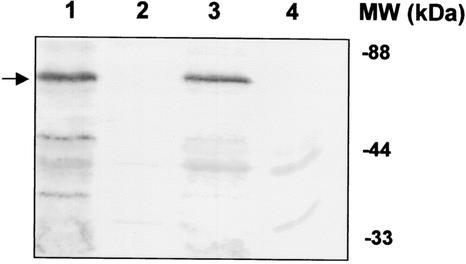
INP is an outer membrane protein that acts as a template for ice nucleation (3, 21, 22). Since INP and the truncated version of INP containing only the N- and C-terminal portions have been used to target a wide range of monomeric and dimeric proteins onto the surface of E. coli (21) and Moraxella sp. (22) without any adverse effects on cell growth and integrity, this strategy was adopted for targeting the dimeric MerR to the surface of E. coli. Plasmid pUNIM, carrying the INP-MerR fusion, was used for genetic immobilization of MerR. Expression of full-length INP-MerR fusion proteins was confirmed by blotting with INP antisera. A protein band of ∼65 kDa corresponding to the correct size of INP-MerR was detected from cells carrying pUNIM (Fig. 1, lane 1). No such fusion protein was detected in cells carrying pUC18Not (Fig. 1, lane 4). The localization of INP-MerR in the membrane fraction was also demonstrated by immunoblotting (Fig. 1, lane 3).
FIG. 1.
Expression of INP-MerR in different cellular fractions. Total cell proteins were separated by SDS-PAGE (12.5% [wt/vol] polyacrylamide) and transferred to a nitrocellulose membrane. Western blot analysis with anti-INP sera was performed at a 1:3,000 dilution. Lanes 1, 2, and 3 represent the total protein, soluble fraction, and membrane fraction of JM109/pUNIM. Lane 4 represents the total protein of JM109/pUC18Not. The desired fusion proteins are marked with an arrow. MW, molecular mass.
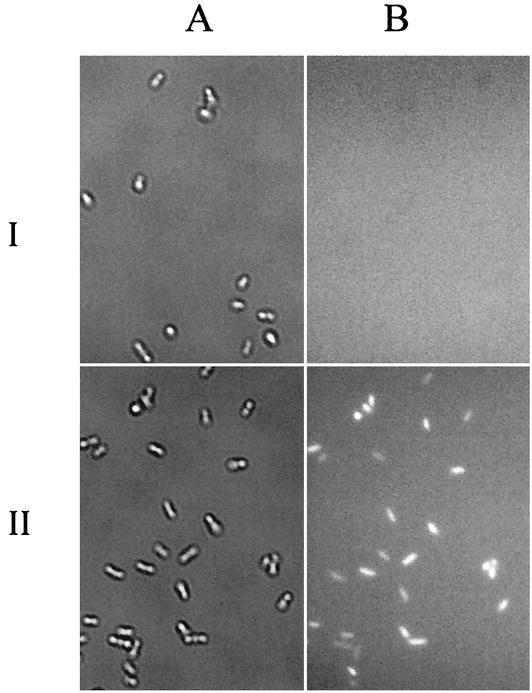
The surface localization of the MerR domain was verified by immunofluorescence labeling. For ease of detection, a hexahistidine tag was added to the C terminus of the INP-MerR fusion (pUNIMH), which can be easily detected with an anti-His antibody. Immunofluorescence labeling of cells expressing the INP-MerR-H6 fusion was performed by first probing with the hexahistidine antisera, followed by incubation with an Alexa Fluor 488-conjugated goat anti-mouse IgG as the second antibody. The results are shown in Fig. 2. While cells carrying pUC18Not were not labeled, cells carrying pUNIMH were brightly labeled fluorescent, confirming the presence of INP-MerR-H6 fusion on the cell surface.
FIG. 2.
Phase-contrast micrographs (A) and immunofluorescence micrographs (B) of E. coli JM109 cells harboring pUC18Not (I) or pUNIMH (II). Cells were probed with anti-(His)6 sera and fluorescently stained with a goat anti-mouse IgG conjugated with Alexa Fluor 488.
Whole-cell binding of mercury.
To investigate whether the surface-displayed MerR protein retains the ability to bind mercury, overnight cultures of JM109/pUNIM and JM109/pUC18Not were resuspended to a final OD600 of 1.0 in Tris buffer (pH 7.4) containing 5 μM Hg2+, and the amount of bound Hg2+ was determined after 1 h. Cells displaying MerR on the surface accumulated a significant level of Hg2+ (17.3 nmol/mg [dry weight]); all added Hg2+ was removed within 1 h (data not shown). For comparison, JM109 cells transformed with pUC18Not yielded a sixfold-lower level of accumulation (3.1 nmol/mg [dry weight]). These results demonstrate that MerR retains its mercury binding characteristics even when displayed on the cell surface.
To confirm the binding of Hg2+ to the surface-exposed MerR, the amount of bound Hg2+ was also determined for the cytoplasmic and membrane cell fractions. Consistent with the expected localization of MerR, over 75% of the accumulated Hg2+ was associated with the membrane fraction of JM109/pUNIM cells. In contrast, less than 20% of the bound Hg2+ was associated with the membrane fraction for JM109/pUC18Not cells. These results again confirm that surface-displayed MerR is mainly responsible for whole-cell binding of mercury.
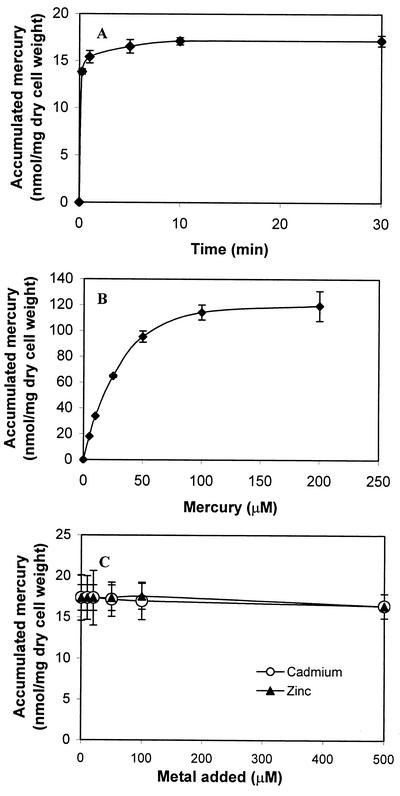
To determine the mercury-binding rate for cells harboring pUNIM, a time course study was conducted. As shown in Fig. 3A, 90% of the total Hg2+ was removed within the first 2 min. This rapid initial binding rate suggests an instantaneous binding of Hg2+ by the surface-exposed MerR, followed by slower nonspecific binding to other cell surface components (2).
FIG. 3.
Hg2+ binding by E. coli strain JM109 carrying pUNIM. (A) Time profile of mercury uptake by resting cells harboring pUNIM. Resting cells (0.265 mg [dry weight]) were resuspended in Tris buffer (pH 7.4) containing 5 μM Hg2+ and incubated for the indicated lengths of time. (B) Hg2+ binding isotherm for JM109/pUNIM cells. Hg2+ binding was determined at various concentrations after 1 h of incubation. (C) Selectivity of Hg2+ binding in the presence of competing cadmium and zinc ions. JM109/pUNIM cells were incubated with 5 μM Hg2+ and various concentrations of competing heavy metals. Hg2+ binding was determined after 1 h of incubation. The data shown are the mean values (± standard deviation) obtained from three independent experiments.
To determine the maximum binding capacity of cells with surface-exposed MerR, whole-cell binding was determined over a range of Hg2+ concentrations (Fig. 3B). At lower concentrations (<20 μM), 100% of the added Hg2+ was removed within 1 h. The binding level quickly saturated at higher concentrations and reached a plateau at around 120 nmol/mg (dry weight). The maximum binding capacity is approximately two times lower than the value reported for cells with surface-exposed EC20 (2). It has been reported that the mercury binding stoichiometry of one Hg2+ per MerR dimer (20) is significantly lower than the value of 20 for EC20 (2), which could explain the differences in the Hg2+ binding capacity.
MerR is typically a dimer, and its high-affinity mercury binding domain consists of three conserved cysteine residues at positions C82, C117, and C126, which form a highly specific, trigonal Hg2+ coordination site involving C117 and C126 from one subunit and C82 from the other (12). Although cells displaying MerR bind mercury with high affinity, it is unclear whether the surface-displayed MerR retains its dimeric conformation and selectivity toward mercury. To demonstrate the selectivity of surface-exposed MerR toward mercury, whole-cell binding of Hg2+ was performed in the presence of up to 100-fold molar excess of competing heavy metals (Fig. 3C) such as Cd2+ and Zn2+. In both cases, only mercury was removed in notable amounts, and the extent of mercury binding was minimally affected by the competing heavy metals. These results are consistent with those obtained with purified MerR, which binds Hg2+ preferentially even in the presence of a 1,000-fold excess of Zn2+ and Cd2+ (20), suggesting that the striking specificity of MerR appears to be preserved even when displayed on the cell surface.
Evaluation of mercury biosorption.
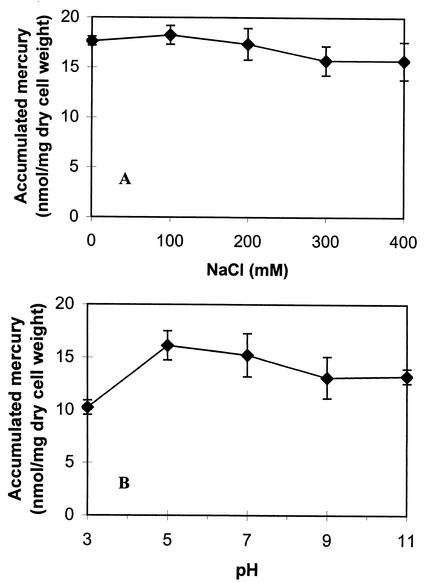
To demonstrate the utility of the whole-cell sorbents in contaminated waters, the effects of different environmental factors such as ionic strength, pH, and chelators on mercury binding were evaluated. Na+ is an ion commonly found in contaminated waters, and levels of 40 mM have been reported in coal mine tailings and acid mine waters (10). To investigate whether the presence of Na+ has any effect on whole-cell binding of mercury, Hg2+ binding experiments were performed at increasing concentrations of sodium chloride. As shown in Fig. 4A, no significant decrease in mercury binding was observed with Na+ concentrations up to 400 mM. This observation contrasts with the biosorption behavior reported for an Hg2+-resistant strain of Pseudomonas aeruginosa, PU21(Rip64) (7), in which a concentration of 150 mM sodium chloride can reduce Hg2+ binding by over 90%. The insensitivity of our whole-cell sorbents to the presence of NaCl is again consistent with the specific nature of the MerR moiety toward Hg2+.
FIG. 4.
Evaluation of mercury binding by JM109 cells harboring pUNIM. (A) Effect of sodium ion on mercury binding. Resting cells (0.265 mg [dry weight]) were resuspended in Tris buffer (pH 7.4) containing 5 μM Hg2+, and the indicated concentrations of NaCl were added. Hg2+ binding was determined after 1 h of incubation. (B) pH profile of mercury binding. Resting cells (0.265 mg [dry weight]) were resuspended in citric-phosphate-borate buffer containing 5 μM Hg2+ with pHs ranging from 3 to 11. Hg2+ binding was determined after 1 h of incubation. The data shown are the mean values (± standard deviation) obtained from three independent experiments.
In addition to earth metals, complexing agents and chelators are frequently found in contaminated waters (8). These agents are able to form tight complexes with metals and decrease their bioavailability. The effect of metal chelators on Hg2+ binding was investigated. The addition of up to 10 mM EDTA did not affect the accumulated Hg2+ levels (data not shown). The fact that biosorbents with surface-displayed MerR bind Hg2+ significantly stronger than EDTA (Kd = 10−25 M) (19) suggests that whole-cell sorbents can be used for specific removal of Hg2+ from soil or particulates, which normally adsorb Hg2+ strongly.
The effect of pH is another major factor that greatly influences metal binding by affecting bioavailability through metal speciation, sequestration, and/or mobility (13). Most biosorbents reported to date are highly pH sensitive; in some cases, even a change of pH by 1 unit results in a 50% reduction in mercury binding (7). Figure 4B depicts the pH profile of mercury binding by cells with surface-displayed MerR. The level of mercury binding remained virtually the same from pHs 5 to 11 and decreased by 30% only at pH 3. The resistance of the whole-cell sorbent to pH variation and to the presence of other competing heavy metals, earth metals, and EDTA suggests that this whole-cell sorbent may be a useful tool for the removal of Hg2+ in contaminated wastewaters.
DISCUSSION
Eukaryotes limit the concentrations of reactive free metal ions by intracellular sequestration. Cysteine-rich MTs and PCs are the main metal-sequestering molecules used for cellular immobilization of metal ions. Expression of MTs and protein analogs of phytochelatins (ECs) have been employed as biological chelators to increase the immobilization of heavy metals by bacterial cells. Although these cysteine-rich peptides have high affinity for a wide range of heavy metals, they lack the required selectivity to enable the specific removal and recycling of the desired metal (4).
To provide high specificity and affinity, one could exploit what nature can offer. Many bacteria acquire resistance to heavy metals by triggering the production of transport proteins and enzymes that can actively metabolize and inactivate the toxic effects of these metals (16). These active mechanisms of resistance are highly specific and are only triggered in the presence of the metal of interest. The highly specific nature of these resistance mechanisms is the result of a cleverly designed genetic circuit that is tightly controlled by a specific metalloregulatory protein. To provide sensitive resistance, these metalloregulatory proteins also possess high affinity in the submicromolar range.
The high affinity and selectivity of a mercury-specific metalloregulatory protein, MerR, were exploited for the selective removal and recycling of Hg2+. The dimeric MerR was successfully targeted onto the cell surface and provided increased Hg2+ binding in the virtual absence of uptake. The selectivity of MerR is significantly better than that of other cysteine-rich peptides due to the tricoordinate arrangement of cysteines in the binding pocket of MerR, which allows very specific binding of Hg2+. While no decline in Hg2+ binding was observed for cells with surface-expressed MerR in the presence of 100-fold excess Cd2+, a more than 20% decline in Hg2+ binding was observed with whole cells expressing EC20 on the surface even at 20-fold excess (2). The use of MerR in place of other MTs or ECs also offers improved affinity, because no effect on Hg2+ binding was observed even in the presence of 10 mM EDTA, a concentration 10 times higher than the value reported for the extraction of metal ions from MTs (23). The resistance of the whole-cell sorbents to various environmental conditions such as ionic strength and pH suggests the potential of this strategy for the removal and recovery of Hg2+ from contaminated water, soil, or sediment.
Acknowledgments
This work was supported by grants from the University of California Toxic Substances Research and Teaching Program and the Environmental Protection Agency (R827227).
REFERENCES
- 1.Bae, W., W. Chen, A. Mulchandani, and R. Mehra. 2000. Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol. Bioeng. 70:518-523. [DOI] [PubMed] [Google Scholar]
- 2.Bae, W., R. K. Mehra, A. Mulchandani, and W. Chen. 2001. Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Appl. Environ. Microbiol. 67:5335-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, W., A. Mulchandani, and W. Chen. 2002. Cell surface display of synthetic phytochelatins using ice nucleation protein for enhanced heavy metal bioaccumulation. J. Inorg. Biochem. 88:223-227. [DOI] [PubMed] [Google Scholar]
- 4.Bontidean, I., C. Berggren, G. Johansson, E. Csorgi, B. Mattiasson, J. R. Lloyd, K. J. Jakeman, and N. L. Brown. 1998. Detection of heavy metal ions at femtomolar levels using protein-based biosensors. Anal. Chem. 70:4162-4169. [DOI] [PubMed] [Google Scholar]
- 5.Brown, W. C., and J. L. Campbell. 1993. A new cloning vector and expression strategy for genes encoding proteins toxic to Escherichia coli. Gene 127:99-103. [DOI] [PubMed] [Google Scholar]
- 6.Byrnes-Brower, J., R. L. Ryan, and M. Pazirandeh. 1997. Comparison of ion-exchange resins and biosorbents for the removal of heavy metals from plating factory wastewater. Environ. Sci. Technol. 31:2910-2914. [Google Scholar]
- 7.Chang, J., J. Hongf, O. A. Ogunseitan, and B. H. Olsen. 1994. Biosorption of mercury by the inactivated cells of Pseudomonas aeruginosa PU21 (Rip64). Biotechnol. Bioeng. 44:999-1006. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S., and D. B. Wilson. 1997. Genetically engineered bacteria and their potential for Hg2+ bioremediation. Biodegradation 8:97-103. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gang, M. A., and D. Langmuir. 1974. Controls on heavy metals in surface and ground waters affected by coal mine drainage, p. 39-69. In Proceedings of the 5th Symposium on Coal Mine Drainage Research. National Coal Mine Association, Washington, D.C.
- 11.Harada, M. 1995. Minamata disease: methylmercury poisoning in Japan caused by environmental pollution. Crit. Rev. Toxicol. 25:1-24. [DOI] [PubMed] [Google Scholar]
- 12.Helmann, J. D., B. T. Vallard, and C. T. Walsh. 1990. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science 247:946-948. [DOI] [PubMed] [Google Scholar]
- 13.Krishnaswamy, R., and D. B. Wilson. 2000. Construction and characterization of an Escherichia coli strain genetically engineered for Ni(II) bioaccumulation. Appl. Environ. Microbiol. 66:5383-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwak, Y.-D., S.-K. Yoo, and E.-J. Kim. 1999. Cell surface display of human immunodeficiency virus type 1 gp120 on Escherichia coli by using ice nucleation protein. Clin. Diagn. Lab. Immunol. 6:499-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 17.Nriagu, J. O., and J. M. Pacyna. 1989. Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature 333:34-139. [DOI] [PubMed] [Google Scholar]
- 18.O'Halloran, T. V., B. Frantz, M. K. Shin, D. M. Ralston, and J. G. Wright. 1989. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell 56:119-129. [DOI] [PubMed] [Google Scholar]
- 19.Ralston, D. M., and T. V. O'Halloran. 1990. Ultrasensitivity and heavy-metal selectivity of the allosterically modulated MerR transcription complex. Proc. Natl. Acad. Sci. USA 87:3846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shewchuk, L. M., G. L. Verdine, H. Nash, and C. T. Walsh. 1989. Mutagenesis of the cysteines in the metalloregulatory protein MerR indicates that a metal-bridged dimer activates transcription. Biochemistry 28:6140-6145. [DOI] [PubMed] [Google Scholar]
- 21.Shimazu, M., A. Mulchandani, and W. Chen. 2001. Cell surface display of organophosphorus hydrolase using ice nucleation protein. Biotechnol. Prog. 17:76-80. [DOI] [PubMed] [Google Scholar]
- 22.Shimazu, M., A. Mulchandani, and W. Chen. 2001. Simultaneous degradation of organophosphorus pesticides and p-nitrophenol by a genetically engineered Moraxella sp. with surface-expressed organophosphorus hydrolase. Biotechnol. Bioeng. 76:318-324. [DOI] [PubMed] [Google Scholar]
- 23.Sousa, C., P. Kotrba, T. Ruml, A. Cebolla, and V. de Lorenzo. 1998. Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. J. Bacteriol. 180:2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]