Abstract
We isolated a methanogen from deep in the sediments of the Nankai Trough off the eastern coast of Japan. At the sampling site, the water was 950 m deep and the sediment core was collected at 247 m below the sediment surface. The isolated methanogen was named Nankai-1. Cells of Nankai-1 were nonmotile and highly irregular coccoids (average diameter, 0.8 to 2 μm) and grew with hydrogen or formate as a catabolic substrate. Cells required acetate as a carbon source. Yeast extract and peptones were not required but increased the growth rate. The cells were mesophilic, growing most rapidly at 45°C (no growth at ≤10°C or ≥55°C). Cells grew with a maximum specific growth rate of 2.43 day−1 at 45°C. Cells grew at pH values between 5.0 and 8.7 but did not grow at pH 4.7 or 9.0. Strain Nankai-1 grew in a wide range of salinities, from 0.1 to 1.5 M Na+. The described phenotypic characteristics of this novel isolate were consistent with the in situ environment of the Nankai Trough. This is the first report of a methanogenic isolate from methane hydrate-bearing sediments. Phylogenetic analysis of its 16S rRNA gene sequence indicated that it is most closely related to Methanoculleus marisnigri (99.1% sequence similarity), but DNA hybridization experiments indicated a DNA sequence similarity of only 49%. Strain Nankai-1 was also found to be phenotypically similar to M. marisnigri, but two major phenotypic differences were found: strain Nankai-1 does not require peptones, and it grows fastest at a much higher temperature. We propose a new species, Methanoculleus submarinus, with strain Nankai-1 as the type strain.
Marine methane hydrates are an ice-like material in which methane molecules are trapped within cages of the crystalline lattice of water molecules. Methane hydrates form at temperatures of up to about 15 or 16°C when the partial pressure of methane is very high. This occurs in many deep marine sediments, where temperatures are low and the hydrostatic pressure of the water keeps methane in solution at high partial pressures (29). Gas hydrates are globally distributed along coastal margins, trapping enormous volumes of methane, estimated at about twice the amount of all other known fossil fuel reserves (1). Methane hydrates have been estimated to contain roughly 4,000 times today's atmospheric content of methane (4). With most of the methane trapped in known hydrate formations being of biogenic origin, this represents a significant new source of natural gas from biological methanogenesis.
Natural gas hydrates typically occur along coastal margins where organic matter accumulates. Microbial numbers are high in these zones: 1.5 × 109 cells g−1 of hydrate-bearing sediments and 1.0 × 106 cells ml−1 within the hydrates themselves (17). Metabolic studies of these sediments, based on incubations with stable isotopes of substrate and subsequent measurement of the isotope-containing product, have demonstrated methanogenesis, sulfate reduction, and methane oxidation (8). Phylogenetic analysis of DNA extracted from marine sediments in a methane hydrate zone has revealed the presence of a diversity of prokaryotic species, including members of the orders Methanobacteriales and Methanosarcinales (21). Bidle et al. (3) targeted the α subunit of the gene for methyl coenzyme M methylreductase, an enzyme that is unique to and ubiquitous among methanogens (32), and found three unique methylreductase sequences. Lanoil et al. (17) established the physical association of Bacteria and Archaea with intact hydrates by direct microscopic cell counts with DNA staining and subsequent phylogenetic analysis. Methanogens specifically associated with the intact hydrate in this study were related to the genus Methanosaeta.
Methanogens are distinguished from all other known living organisms in that their major catabolic product is methane (34). The fact that most of the methane bound within marine hydrates is of biogenic origin has targeted methanogens as keystone members of the methane hydrate community (1).
We report here the first methanogen isolated from deep marine sediments that contain methane hydrates. We propose the new species name Methanoculleus submarinus for this novel organism.
MATERIALS AND METHODS
Source of inoculum.
The Japanese island arc system is surrounded by deep trenches resulting from the subduction boundaries of the Eurasia and Philippine Sea plates (S. Kuramoto, J. Hiramura, M. Joshima, and Y. Okuda, Int. Symp. Methane Hydrates Resour. Future, abstr. SII-1, 1998). The forearc basin of the Nankai Trough is landward of one of the deep trenches of this arc system. In November 1997, cores were collected by Japan Petroleum Exploration Company Limited personnel to a depth of 250 m below the seafloor. The water depth at this site was 950 m. A seismic study of the sediments indicated a bottom-simulating reflector at 290 ± 10 m below the seafloor (Y. Tsuju, A. Gurutani, S. Matsuura, and K. Kanamori, Int. Symp. Methane Hydrates Resour. Future, abstr. SI-1, 1998); bottom-simulating reflectors often occur at the phase boundary between methane hydrates and the free methane plus water that occurs below them. We selected a depth within the methane hydrate zone (just above this bottom-simulating reflector). Our sample was from a section of a core (BH-1) from a depth of 247 m below the sediment surface. The methane gas within the sediment samples from this drilling is depleted of 13C (−95.5 to −63.9‰), indicating that this methane is biogenic (A. K. B. Waseda, M. Yagi, R. Matsumoto, H. Lu, Y. Hiroki, and T. Fujii, Int. Symp. Methane Hydrates Resour. Future, abstr. SII-3, 1998).
Culture techniques and culture media.
The anaerobic culture techniques of Hungate (12), as modified by Sowers and Noll (31), were used in this study. All of the culture media used in this study were based on MSH medium (22), which is a medium containing minerals including a total salinity near that of seawater, a bicarbonate-CO2 buffer system, and 2 g each of yeast extract and Trypticase peptones per liter. Unless otherwise indicated, the gas phase was 100% CO2, the pH was adjusted to a final pH of 6.5, and the incubation temperature was 37°C. MSH mineral medium is the same as MSH medium but with organic constituents (yeast extract, Trypticase peptones, and mercaptoethane sulfonate, a reducing agent) omitted. Generally, 80 mM sodium formate was added to the media prior to autoclaving as a catabolic substrate. When H2 was used as a catabolic substrate, formate was omitted and H2 was added as an overpressure (2-atm gauge) after cultures were inoculated.
Measurement of growth.
Growth was estimated from the accumulation of methane in the headspace gas, as measured by gas chromatography (18) and taking into account the methane produced during the growth of the inoculum (25). The specific growth rate was calculated by fitting the Gompertz equation (37) to these data. The optimal temperature for growth was determined from the specific growth rates of cultures incubated at various temperatures. We fitted the square-root equation (26, 27) to these data.
Electron microscopy.
Exponential-phase cultures were prepared for electron microscopy by centrifuging and resuspending cells in 2.5% glutaraldehyde in 0.2 M sodium cacodylate buffer (pH 7.2) at room temperature for 30 min. After two buffer washes, cells were fixed in 1% osmium tetroxide in the same buffer for 1 h at 4°C, washed in distilled water, dehydrated in an ethanol series, and embedded in low-viscosity epoxy resin for sectioning. Sections on Formvar-coated grids were poststained with uranyl acetate and lead citrate and viewed and photographed with a Zeiss EM-10CA transmission electron microscope. To prepare the osmotically fragile cells for observation of flagella, cells were placed on a grid that was floated on glutaraldehyde for fixation. The cells were then desalted on water before negative staining.
Phylogenetic analysis.
The 16S rRNA gene (rDNA) sequence was determined after extraction of DNA from a cell pellet of strain Nankai-1 with a DNeasy Tissue Kit (Qiagen, Inc., Valencia, Calif.). Approximately 1,500 nucleotides of the 16S rDNA were amplified from the genomic DNA by PCR. PCR mixtures (50 μl) contained 50 mM KCl, 10 mM Tris-HCl, 0.1% Triton X-100, 1.5 mM MgCl2, deoxynucleoside triphosphates (200 μM each dATP, dCTP, dGTP, and dTTP), 0.05% Igepal CA-630 (Sigma, St. Louis, Mo.), 0.4 μM forward primer 4F (Table 1), 0.4 μM reverse primer 1492R (Table 1), 5 U of Taq DNA polymerase, and approximately 100 ng of Nankai-1 DNA. Negative controls (DNA template replaced with sterile water) were always used. PCR products were purified with an UltraClean PCR Clean-up DNA purification kit (MoBio Laboratories, Inc., Solana Beach, Calif.). The purified PCR product was sequenced directly with the ABI Big Dye Terminator protocol and an ABI Prism 310 genetic analyzer (Applied Biosystems Inc., Foster City, Calif.). Approximately 20 ng of template DNA was used in a 10-μl reaction mixture with 1.6 pmol of the sequencing primers per reaction mixture (Table 1). The final sequence was assembled with the ABI Prism AutoAssembler software package. The Basic Local Alignment Search Tool (BLAST version 2.0) was used to determine sequence similarity between Nankai-1 (1,395 bases) and sequences in the National Center for Biotechnology Information database (available online [http://www.ncbi.nlm.nih.gov]). Because the portion of the sequences corresponding to the 4F and 1492R primer sites was originally generated by PCR amplification, this portion of the sequence was not included in phylogenetic comparisons.
TABLE 1.
Oligonucleotide primers used for PCR amplification and sequencing reactions of the Nankai-1 16S rDNAa
| Primer use(s) and name | Direction | 5′ → 3′ Sequence |
|---|---|---|
| PCR and sequencing | ||
| 4F | Forward | TCC GGT TGA TCC TGC CRG |
| 1492R | Reverse | GGT TAC CTT GTT ACG ACT |
| Sequencing | ||
| 515F | Forward | GTG CCA GCM GCC GCG GTA |
| 765F | Forward | TAG ATA CCC SSG TAG TCC |
| 906F | Forward | GAA ACT TAA AKG AAT TG |
| 1100F | Forward | GGC AAC GAG CGM GAC CC |
| 690Ra | Reverse | TCT ACG CAT TTC ACC |
| 907R | Reverse | CCG TCA ATT CCT TTR AGT |
| 1098Ra | Reverse | GGG TCT CGC TCG TTS CC |
| 1391R | Reverse | GAC GGG CGG TCT GTR CA |
R = A or G, M = A or C, S = C or G, K = G or T.
Sequences of other cultivated methanogens were obtained from the Ribosomal Database Project (20) or GenBank. The sequences with the following accession numbers, and that of Nankai-1, were manually aligned in the Genetic Data Environment (30): M. marisnigri (type strain, JR1), M59134, M. bourgensis (type strain, MS2), AF095269; M. thermophilicus (type strain, CR1), M59129, M. marisnigri (strain CoCam), AF028693; Methanogenium marinum (type strain, AK1), AF531178; Methanogenium cariaci (type strain, JR1), M59130; and Methanococcoides burtonii (type strain, DSM 6242), X65537. A maximum-likelihood tree was constructed by using fastDNAml (23). Sequence positions whose alignment was not clear were masked for the construction of the tree (24). The tree generated was then edited in Tree Tool, version 1.0 (http://rdp.cme.msu.edu/download/programs/TreeTool), a phylogenetic data editor and tree formatter. A distance matrix was generated with the PHYLIP nucleic acid sequence distance matrix program, version 3.5c (11; http://evolution.genetics.washington.edu/phylip.html). The Jukes-Cantor model was used to correct observed sequence differences to get a more accurate estimate of the evolutionary distance between the sequences (16). Bootstrapping was done with 100 randomly generated subsets by the fastDNAml_boot program (23).
Spectroscopic DNA-DNA hybridization.
Interstrain DNA hybridization values were determined by the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH Identification Service (Braunschweig, Germany) as briefly summarized here. Genomic DNA was isolated from cell pellets of Nankai-1 and M. marisnigri JR1 by chromatography on hydroxyapatite by the procedure of Cashion et al. (5). DNA-DNA hybridization was carried out as described by De Ley et al. (9), with the modifications described by Huss et al. (13) and Escara and Hutton (10), by using a model 2600 spectrophotometer equipped with a model 2527-R thermoprogrammer and plotter (Gilford Instrument Laboratories, Inc., Oberlin, Ohio). Renaturation rates were computed with the TRANSFER.BAS program (14).
Nucleotide sequence accession number.
The 16S rDNA of strain Nankai-1 was sequenced, and the sequence was deposited in the GenBank database under accession number AF531178.
RESULTS
Sediment sample.
The sediment sample was sealed in a canning jar under inert gas at the collection site and shipped at 4°C to the laboratory within 2 days of collection. Upon arrival, the sample was immediately transferred into an anaerobic jar, flushed with inert gas to ensure anaerobic conditions, and placed at 2°C until processing 3 days later. The sample was opened in a glove bag (Coy Products, Grass Lake, Mich.) containing an atmosphere of 5% hydrogen, 5% carbon dioxide, and 90% nitrogen. The outer surface layer of the core was removed with sterilized tools, and the pared core was then pared again in a similar fashion.
Although sulfate is present at high concentrations in seawater (approximately 28 mM), it is absent in marine sediments more than a few meters below the surface because of the activity of sulfate-reducing bacteria (7). During the drilling process, seawater was used as drilling fluid and intrusion of drilling fluid into the recovered core would be evident by the presence of sulfate. Core samples from the 247-m depth were devoid of sulfate (Waseda et al., Int. Symp. Methane Hydrates Resour. Future), indicating that samples were not contaminated by drilling fluids and were representative of deep subsurface sediments.
Enrichment of methanogens.
Enrichment was performed at the Idaho National Engineering and Environmental Laboratory (28). Approximately 10 g from the final inner pared core sample was placed in a serum vial containing 90 ml of MSH medium (22) without KCl. The vial was stoppered and shaken vigorously for 10 s. H2 was added (2 atm), cultures were incubated statically at 21°C for 2 months, and methane was detected in the headspace. This enrichment culture was maintained by periodic transfer into MSH medium or MSH medium with Trypticase peptones and yeast extract reduced to 1 g of each per liter and with either 1 atm of H2 or 80 mM formate added as the catabolic substrate. Strain Nankai-1 was isolated from this culture.
Isolation.
Epifluorescence microscopy with an O2 filter set (Zeiss, Oberkochen, Germany) revealed a large number of nonfluorescent microbes (presumably nonmethanogens) and a smaller number of methanogens in the enrichment culture grown in MSH medium with 80 mM formate. To increase the relative numbers of methanogens, the culture was transferred to MSH media with a reduced amount of yeast extract and Trypticase peptones (1 g of each). After growth, the culture was serially diluted and transferred to roll tube media for isolation. All roll tube cultures were incubated at room temperature. A colony appeared after 1 month and was transferred to fresh MSH media. This culture was again diluted and inoculated into two sets of roll tube media, one incubated at 21°C and the other incubated at 37°C. Similar numbers of colonies appeared in each set of roll tube media of the same dilution, indicating that the dominant methanogen could grow at 37°C, as well as at 21°C. A colony was picked from the 21°C cultures, and the roll tube isolation procedure was repeated at 37°C to ensure purity. No contaminants appeared after visual microscopic observation or in culture medium without the methanogenic substrate. This culture was named strain Nankai-1 and deposited in the Oregon Collection of Methanogens (Portland State University, Portland, Oreg.) as OCM 780, in the Subsurface Microbial Culture Collection (Portland State University) as SMCC 780W, and in the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH as DSMZ 15122.
Morphology.
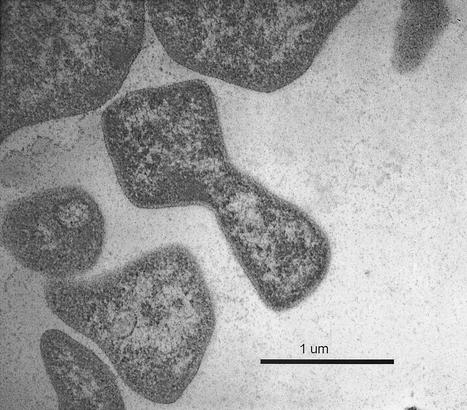
Strain Nankai-1 was an epifluorescent, irregularly shaped coccus with an average cell diameter of 0.8 to 2 μm. Gram staining results were negative, although thin-section electron micrographs show no outer membrane (Fig. 1). We were unable to detect motility during observations of wet mounts, although flagella were detected by electron microscopy. The typical arrangement of cells was diploid or in clusters when observed under a light microscope. Nankai-1 appeared as smooth, light yellow, transparent colonies with entire edges when grown on agar media. The colony diameter was less than 1 mm after 2 months of incubation at 37°C.
FIG. 1.
Electron micrograph of strain (str.) Nankai-1. This thin-section micrograph shows the irregular shape and lack of a thick cell wall.
Electron microscopy.
Figure 1 is a thin-section electron micrograph of strain Nankai-1 that shows a typical coccoid methanogen with irregular shapes and the absence of a cell wall other than an S-layer. Negatively stained electron micrographs revealed flagella in strain Nankai-1 and in M. marisnigri JR1. For each of these two strains, some cells had no visible flagella and others had a single flagellum (data not shown).
Phylogenetic analysis.
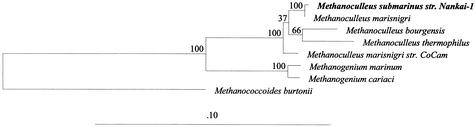
Basic Local Alignment Search Tool results for the 16S rDNA sequence of strain Nankai-1 indicated the greatest similarity to that of M. marisnigri JR1 from the Black Sea (99.1% sequence similarity). The sequence of M. marisnigri CoCam was 98.1% similar. A maximum-likelihood phylogenetic tree (Fig. 2) shows that Nankai-1 clusters with other M. marisnigri sequences. The direct comparison of the aligned sequences indicated that the sequence of strain Nankai-1 was most similar to that of M. marisnigri JR1, with 99.78% sequence identity. DNA hybridization indicated that the genomic sequence similarity of these two strains was 49%.
FIG. 2.
Phylogenetic tree based on 16S rDNA sequences. The scale bar represent 0.1 nucleotide substitution per sequence position. This tree was constructed by the maximum-likelihood method of phylogenetic analysis.
Growth requirements.
Strain Nankai-1 grew with hydrogen (2 atm) or formate (80 mM) as an electron donor for reduction of carbon dioxide. Cells did not catabolize acetate (40 mM), trimethylamine (20 mM), or methanol (20 mM), either alone or in combination with formate. The concentrations of these potential catabolic substrates did not appear to be inhibitory because when cultures were incubated with those concentrations plus 5 mM formate, methane was formed in approximately the amount expected from the 5 mM formate. Thus, it appeared that the cells were not inhibited from using the formate although the other substrate was not catabolized.
Cultures did not grow with formate as a catabolic substrate in MSH mineral medium. Strain Nankai-1 grew in this medium when acetate (5 mM) was added as an anabolic substrate. Yeast extract alone (1 g liter−1) could also satisfy the nutritional requirements of strain Nankai-1, perhaps because yeast extract contains acetate. Trypticase peptones or a vitamin solution (2) did not support growth. The fastest growth occurred when both yeast extract (1 g liter−1) and Trypticase peptones (1 g liter−1) were present.
Effects of temperature, pH, and salinity on growth rate.
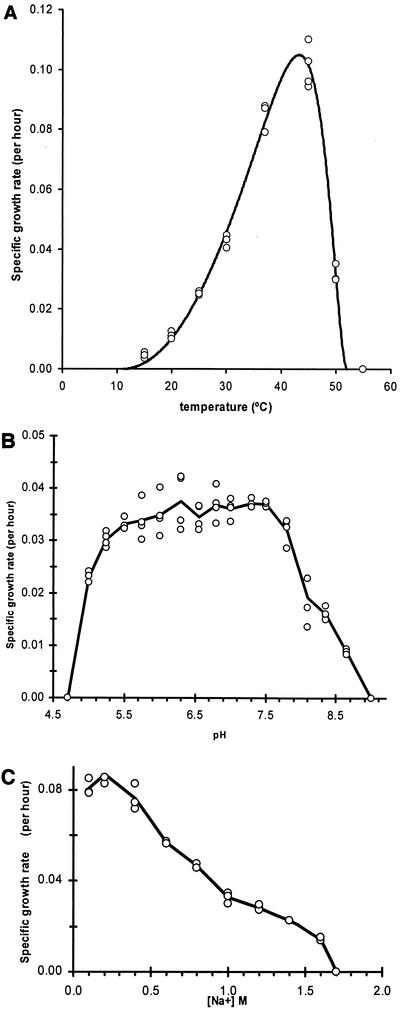
Strain Nankai-1 grew faster at 45°C than at any other temperature tested (Fig. 3A). The square-root equation (26) indicated that the optimal temperature was 43°C, the minimum temperature was 11°C, and the maximum temperature was 52°C. Strain Nankai-1 grew fastest at pH values between 6.0 and 7.5, with similar growth rates throughout this range (Fig. 3B). Strain Nankai-1 grew fastest at Na+ concentrations between 0.1 and 0.4 M. Growth was observed in up to 1.6 M Na+, but no growth was observed in medium with an Na+ concentration of 1.7 M (Fig. 3C).
FIG. 3.
Effects of environmental factors on the growth rate of strain Nankai-1. (A) Specific growth rate at various temperatures. Data points (circles) were fitted with the square-root equation (solid curve). (B) Specific growth rate at various pH values. (C) Specific growth rate in media with various salinities.
DISCUSSION
Several lines of evidence support the hypothesis that the source and habitat of strain Nankai-1 are at 247 m below the sea floor in the Nankai Trough. Sulfate measurements and other controls indicated that the sample from which strain Nankai-1 was isolated was not contaminated by the drilling mud or overlying sediments. The physiological requirements of strain Nankai-1 are consistent with its being adapted to growth in that environment. Strain Nankai-1 grew at marine salinity, at temperatures consistent with the source of the sample, and at pH values that are likely to occur in situ. Organic marine sediments at depths such as this typically accumulate acetate and have pH values well below 7 (36). Further, the inability of Nankai-1 to grow at temperatures below 10°C suggests that it was not a contaminant from the upper sediment layers, which are cooler.
The optimum growth temperature noted for strain Nankai-1 was approximately 30°C higher than the in situ temperature. That this strain is optimally adapted to such high temperatures is consistent with other findings from these sediments. Reed et al. (28) found that thermophiles and hyperthermophiles were the closest cultured representatives for more than 60% of the clones that were obtained from a nearby borehole in the forearc basin of the Nankai Trough. Experimental incubations of these sediments with acetate or hydrogen as electron donors indicated the presence of methanogens that were able to make methane at temperatures of at least 35°C (28). Strain Nankai-1 grew slowly and produced methane at temperatures near the estimated in situ temperature (15 to 16°C) of its source.
The presence of strain Nankai-1 in these sediments supports the supposition that biological methanogenesis occurs in environments where methane hydrates form. The high partial pressures of methane that occur in these environments make methanogenesis thermodynamically less favorably there, but a study (6) of the relationship between threshold H2 concentration and methane partial pressure suggests that methanogens can carry out methanogenesis with only a small amount of energy coupled to ATP synthesis.
The phylogenetic analysis of 16S rDNA sequences indicates that strain CoCam, previously classified as M. marisnigri (15), should probably be classified outside this species if the species is to remain monophyletic. Figure 2 shows that strain CoCam may be less closely related to M. marisnigri than are the type strains of M. bourgensis and M. thermophilicus. Strain Nankai-1 was certainly more closely related to the type strain (strain JR1) of M. marisnigri (bootstrap value of 100%), yet strain Nankai-1 and M. marisnigri JR1 had a DNA reassociation value of only 49%.
Although the 16S rDNA sequences of strain Nankai-1 and M. marisnigri JR1 were 99.1% similar, the DNA reassociation value was so low (49%) as to indicate that strain Nankai-1 should be classified as a separate species. There are other examples of microbes whose 16S rDNA sequences are nearly identical but whose DNA reassociation values are below 70% (33). In such cases, taxonomy should be guided by the reassociation values (33, 35). In addition to phylogenetic distance, this separation is supported by two phenotypic distinctions: strain Nankai-1 did not require peptones for growth, and its maximum temperature was much higher (54°C). M. marisnigri JR1 grows well at 45°C but does not grow at 50°C (19). The low sequence similarity of the DNAs of strain Nankai-1 and M. marisnigri JR1 and the important phenotypic differences meet the recommendations for placement of an organism into a new species (35). Therefore, we propose a new species, M. submarinus, with strain Nankai-1 as the type strain.
Methanoculleus submarinus.
sub.ma.rin′us (M.L. preposition sub, under; M.L. adj. marinus marine, of the sea; submarinus, from under the sea).
Irregular cocci 0.8 to 2 μm in average diameter, occurring singly and sometimes in pairs. Nonmotile but possesses flagella. Fimbriae are not present. H2 plus CO2 or formate serves as a catabolic substrate, with CH4 as the product. Acetate is the sole organic nutrient required for cell carbon. Fastest growth was at a specific growth rate of about 2.4 day−1; fastest growth occurred at 45°C, with a salinity of 0.1 to 0.4 M Na+ and a pH of 4.8 to 7.7. Isolated from deep marine sediments where methane hydrates occur. Type strain: Nankai-1 (= OCM 780 = SMCC 780W = DSMZ 15122).
Acknowledgments
We thank Karl Rusterholtz, Martin Sobieraj, Jane Boone, and Amy Banta for help in the analysis of the 16S rDNA sequences and Henry Aldrich (University of Florida) for preparing the electron micrographs. We thank T. Uchida (Japanese Petroleum Exploration Company) for providing access to the core samples.
This work was supported by a grant from the U.S. Department of Energy's Subsurface Science program to the Subsurface Microbial Culture Collection (DOE prime contract DE-FG02-96ER62210 through purchase order G09965 from Florida State University), by a subcontract from Lockheed Martin Idaho Tech/Bechtel (K-99-180960), and by the National Science Foundation's LExEn program (prime contract OCE-0085607 via a subcontract from the University of California at Irvine [L00OCE0085607]). The Idaho National Engineering and Environmental Laboratory was funded through contract DE-AC07-76IDO1570. Research done there was supported by the U.S. Department of Energy's Fossil Energy Office and the Idaho National Engineering and Environmental Laboratory's Directed Research and Development program.
REFERENCES
- 1.Appenzeller, T. 1991. Fire and ice under the deep sea floor. Science 252:1790-1792. [DOI] [PubMed] [Google Scholar]
- 2.Balch, W. E., L. J. Magrum, G. E. Fox, R. S. Wolfe, and C. R. Woese. 1977. An ancient divergence among the bacteria. J. Mol. Evol. 9:305-311. [DOI] [PubMed] [Google Scholar]
- 3.Bidle, K. A., M. Kastner, and D. H. Bartlett. 1999. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP site 892B). FEMS Microbiol. Lett. 177:101.. [DOI] [PubMed] [Google Scholar]
- 4.Blunier, T. 2000. Frozen methane escapes from the sea floor. Science 288:68-69. [Google Scholar]
- 5.Cashion, P., M. A. Holder-Franklin, J. McCully, and M. Franklin. 1977. A rapid method for base ratio determination of bacterial DNA. Anal. Biochem. 81:461-466. [DOI] [PubMed] [Google Scholar]
- 6.Chong, S. C., Y. Liu, M. Cummins, D. L. Valentine, and D. R. Boone. 2002. Methanogenium marinum sp. nov., a H2-using methanogen from Skan Bay, Alaska, and kinetics of H2 utilization. Antonie van Leeuwenhoek 81:263-270. [DOI] [PubMed] [Google Scholar]
- 7.Claypool, G. E., and K. A. Kvenvolden. 1983. Methane and other hydrocarbon gases in marine sediments. Annu. Rev. Earth Planet. Sci. 11:299-327. [Google Scholar]
- 8.Cragg, B. A., R. J. Parkes, J. C. Fry, A. J. Weightman, P. A. Rochelle, and J. R. Maxwell. 1996. Bacterial populations and processes in sediments containing gas hydrates (ODP leg 146: Cascadia margin). Earth Planet. Sci. Lett. 139:497-507. [Google Scholar]
- 9.De Ley, J., H. Cattoir, and A. Reynaerts. 1970. The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 10.Escara, J. F., and J. R. Hutton. 1980. Thermal stability and renaturation of DNA in dimethylsulphoxide solutions: acceleration of renaturation rate. Biopolymers 19:1315-1327. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 12.Hungate, R. E. 1969. A roll tube method for cultivation of strict anaerobes, p. 117-132. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology, vol. 3B. Academic Press, Inc., New York, N.Y.
- 13.Huss, V. A. R., H. Festl, and K. H. Schleifer. 1983. Studies on the spectrophotometric determination of DNA hybridization from renaturation rate. Syst. Appl. Microbiol. 4:184-192. [DOI] [PubMed] [Google Scholar]
- 14.Jahnke, K.-D. 1992. Basic computer program for evaluation of spectroscopic DNA renaturation data from a Gilford System 2600 spectrometer on a PC/XT/AT type personal computer. J. Microbiol. Methods 15:61-73. [Google Scholar]
- 15.Joulian, C., B. M. Ollivier, B. K. C. Patel, and P. A. Roger. 1998. Phenotypic and phylogenetic characterization of dominant culturable methanogens isolated from ricefield soils. FEMS Microbiol. Ecol. 25:135-145. [Google Scholar]
- 16.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y.
- 17.Lanoil, B. D., R. Sassen, M. T. LaDuc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maestrojuán, G. M., and D. R. Boone. 1991. Characterization of Methanosarcina barkeri MST and 227. Methanosarcina mazei S-6T and Methanosarcina vacuolata Z-761T. Int. J. Syst. Bacteriol. 41:267-274. [Google Scholar]
- 19.Maestrojuán, G. M., D. R. Boone, L. Xun, R. A. Mah, and L. Zhang. 1990. Transfer of Methanogenium bourgense, Methanogenium marisnigri, Methanogenium olentangyi, and Methanogenium thermophilicum to the genus Methanoculleus gen. nov., emendation of Methanoculleus marisnigri, and description of new strains of Methanoculleus bourgense and Methanoculleus marisnigri. Int. J. Syst. Bacteriol. 40:117-122. [Google Scholar]
- 20.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. G. M., T. M. Schimidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 22.Ni, S., and D. R. Boone. 1991. Isolation and characterization of a dimethyl sulfide-degrading methanogen, Methanolobus siciliae HI350, from an oil well, characterization of M. siciliae T4/MT, and emendation of M. siciliae. Int. J. Syst. Bacteriol. 41:410-416. [DOI] [PubMed] [Google Scholar]
- 23.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 24.Page, R. D. M., and E. C. Holmes. 1998. Molecular evolution: a phylogenetic approach. Blackwell Science Ltd., Cambridge, England.
- 25.Powell, G. E. 1983. Interpreting gas kinetics of batch culture. Biotechnol. Lett. 5:437-440. [Google Scholar]
- 26.Ratkowsky, D. A., R. K. Lowry, T. A. McMeekin, A. N. Stokes, and R. E. Chandler. 1983. Model for bacterial culture growth rate throughout the entire biokinetic temperature range. J. Bacteriol. 154:1222-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratkowsky, D. A., J. Olley, T. A. McMeekin, and A. Ball. 1982. Relationship between temperature and growth rate of bacterial cultures. J. Bacteriol. 149:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed, D. W., Y. Fujita, M. E. Delwiche, D. B. Blackwelder, P. P. Sheridan, T. Uchida, and F. S. Colwell. 2002. Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol. 68:3759-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan, E. D. 1998. Clathrate hydrates of natural gases, second ed. Marcel Dekker, Inc., New York, N.Y.
- 30.Smith, S. W., R. Overbeek, C. R. Woese, W. Gilbert, and P. M. Gillevet. 1994. The genetic data environment: an expandable GUI for multiple sequence analysis. Comput. Appl. Biosci. 10:671-675. [DOI] [PubMed] [Google Scholar]
- 31.Sowers, K. R., and K. M. Noll. 1995. Techniques for anaerobic growth, p. 15-47. In F. T. Robb, K. R. Sowers, H. J. Schreier, S. DasSarma, and E. M. Fleischmann (ed.), Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, N.Y.
- 32.Springer, E., M. S. Sachs, C. R. Woese, and D. R. Boone. 1995. Partial gene sequences for the A subunit of methyl-coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol. 45:554-559. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 34.Valentine, D. L., and D. R. Boone. 2000. Diversity of methanogens, p. 291-302. In J. Seckbach (ed.), Microbial diversity. Kluwer Press, Dordrecht, The Netherlands.
- 35.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 36.Wellsbury, P., K. Goodman, T. Barht, B. A. Cragg, S. P. Barnes, and R. J. Parkes. 1997. Deep marine biosphere fuelled by increasing organic matter availability during burial and heating. Nature (London) 388:573-576. [Google Scholar]
- 37.Zwietering, M. H., I. Jongenburger, F. M. Rombouts, and K. van't Riet. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]