Abstract
The Vibrio vulnificus putAP genes encoding a proline dehydrogenase and a proline permease are transcribed in the same direction. Proline dehydrogenase activity and the level of putA transcript were determined to reach a maximum in exponential phase and were then repressed when growth slowed down. Northern blotting and primer extension analyses revealed that transcription of putAP genes results in two different transcripts, transcript A (putA transcript) and transcript AP (putAP transcript). Expression of putAP genes was directed by two promoters, promoter PputA and promoter PputAP. A crp null mutation decreased proline dehydrogenase activity and the level of the put transcripts, indicating that transcription of putAP is under the positive control of cyclic AMP receptor protein. Proline dehydrogenase and the level of both put transcripts were increased by proline but repressed by glutamate. In contrast, the level of transcript A, not transcript AP, increased when proline dehydrogenase was induced by NaCl. Since PputA activity, not PputAP activity, was increased by NaCl, it is apparent that transcript A and transcript AP are transcribed through PputA and PputAP, respectively. Cells challenged with NaCl and various hyperosmotic stresses accumulated higher levels of glutamate than control cells, indicating that glutamate is a compatible solute in V. vulnificus.
Change in the external osmolarity is one of the most common environmental stresses that bacteria routinely encounter (10). The main strategy that members of the family Enterobacteriaceae use to adapt to high osmolarity is the accumulation of organic solutes, known as compatible solutes or osmoprotectants (6, 10, 12, 14). It has been well demonstrated that the osmolytes do not interfere with macromolecules and allow the maintenance of vital cellular functions. Hence, the osmolytes can be accumulated to high levels to prevent the loss of water caused by high external osmolarity and consequently contribute to the maintenance of the outwardly directed turgor pressure required for growth. Although a variety of novel organic osmolytes as osmoprotectants have been identified, glycine betaine, ectoine, proline, glutamate, and trehalose are probably the most widely used compatible solutes in bacteria (6, 10).
Vibrio vulnificus is an opportunistic gram-negative bacterial pathogen that commonly contaminates raw oysters and is the causative agent of food-borne diseases, such as gastroenteritis and life-threatening septicemia. Mortality from septicemia is very high (>50%), and death may occur within 1 to 2 days after the first signs of illness (2, 20). Like many other food-borne pathogenic bacteria, V. vulnificus occurs in various environments having different osmotic strengths; it naturally inhabits coastal seawaters, contaminates shellfish, survives current control practices (such as adding salt or sugar to suppress its growth), and colonizes in the human body. These facts indicate that V. vulnificus has to cope with ever-changing osmolarity in its growth environments. However, only a few studies have addressed the molecular mechanisms whereby the bacterium can survive in hyperosmotic environments (19).
In a previous report, a mutant that was sensitive to high osmolarity was screened from a library of mutants constructed by a random transposon mutagenesis (19). We identified and cloned putAP genes encoding a proline dehydrogenase and a proline permease of V. vulnificus by a transposon-tagging method (19). Functions of putAP genes were assessed by the construction of mutants, and the gene products of putAP appeared to contribute to the osmotic tolerance of V. vulnificus. However, until now, no definitive analysis of the regulatory mechanisms by which the bacterium can modulate the expression of the putAP genes has been reported. Neither the promoter(s) of the putAP genes nor trans-acting protein(s) regulating putAP expression has been identified previously. Furthermore, the question of whether the putAP genes are transcribed as a single operon or two separate transcriptional units has not been addressed previously.
Therefore, in an effort to elucidate the regulatory mechanisms of putAP expression at a molecular level, we examined the influence of the growth phase on proline dehydrogenase synthesis. We demonstrated that the transcription of the putAP genes is dependent on the growth phase, initiated at two different sites, and results in two different transcripts. The effects of a crp null mutation on proline dehydrogenase activity and the cellular level of the put transcripts were also examined. Transcription of the putAP operon has been analyzed under different conditions, and we showed that putAP transcription is induced in the presence of proline and hyperosmotic stresses but repressed in the presence of glutamate. Finally, the possible role of glutamate as a compatible solute in V. vulnificus was also examined.
MATERIALS AND METHODS
Strains, plasmids, and culture media.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli strains used for plasmid DNA replication or conjugational transfer of plasmids were grown in Luria-Bertani (LB) broth or on LB broth containing 1.5% (wt/vol) agar. Unless noted otherwise, V. vulnificus strains were grown in LB medium supplemented with 2.0% (wt/vol) NaCl (LBS). When appropriate, antibiotics were added to media at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| V. vulnificus | ||
| ATCC 29307 | Clinical isolate; virulent | Laboratory collection |
| KC74 | ATCC 29307 with crp::nptI; Kmr | 15 |
| HJK001 | ATCC 29307 with putA::nptI; Kmr | 19 |
| HJK002 | ATCC 29307 with putP::nptI; Kmr | |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir; Kmr; host for π-requiring plasmids; conjugal donor | Laboratory collection |
| Plasmids | ||
| pRK415 | IncP ori; broad-host-range vector; oriT of RP4; Tcr | 17 |
| pHJK002 | 8.3-kb SalI fragment containing putAP cloned into pUC18; Apr | 19 |
| pKC0004 | pRK415 with crp; Tcr | 15 |
Apr, ampicillin resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant.
General genetic methods.
Procedures for the isolation of plasmid DNA and genomic DNA and transformation were performed as described by Sambrook and Russell (32). Restriction and DNA-modifying enzymes were used as recommended by the manufacturer (New England Biolabs, Beverly, Mass.). DNA fragments were purified from agarose gels using the Geneclean II kit (Bio 101, Inc., Vista, Calif.). Primary DNA cloning and manipulation were conducted in E. coli DH5α, and restriction mapping was performed to confirm that transformants contained the appropriate plasmids. PCR amplification of DNA was performed using a GeneAmp PCR system 2400 (Perkin-Elmer, Norwalk, Conn.) by standard protocols.
Measurement of cell growth and proline dehydrogenase activity.
Cultures of V. vulnificus strains were grown at 30°C with aeration. Samples (5 ml) were removed at regular intervals for determination of cell densities, proline dehydrogenase activity, put transcript levels, and cellular protein concentrations. Growth was monitored by measuring the optical density at 600 nm (OD600) of the culture. Cultures with an OD600 of >0.8 were diluted prior to measurement. The proline dehydrogenase activity was determined as previously described (19). One unit of enzyme activity was defined by the method of Ostrovsky et al. (29). Protein concentrations were determined by the method of Bradford (5), with bovine serum albumin as the standard. Averages and standard errors of the mean (SEM) were calculated from at least three independent determinations.
RNA purification and analysis of the put transcripts.
Total cellular RNAs from V. vulnificus strains were isolated using a Trizol reagent kit in accordance with the manufacturer's protocol (GIBCO-BRL, Gaithersburg, Md). RNA was treated with RNase-free DNase I (Sigma, St. Louis, Mo.) to remove contaminating genomic DNA.
A series of reactions was performed according to standard procedures (32) with 20 μg of total RNA for either Northern dot blot analysis or Northern blot analysis. RNA was transferred to a nylon membrane and hybridized as previously described (9, 15). Two DNA probes, PUTAP and PUTPP, were labeled with [α-32P]dCTP using the Prime-a-gene labeling system (Promega, Madison, Wis.) and used for hybridization (9, 16). The PUTAP probe was prepared by labeling the 920-bp DNA fragment in the putA gene and amplified by PCR using primers PUTA004 and PUTA005 (Table 2). A 1.1-kb DNA fragment, containing the coding region of putP, was amplified by PCR using primers PUTP004 and PUTP005 (Table 2) and then labeled for the PUTPP probe. The blots were visualized and quantified using a phosphorimaging analyzer (BAS1500 model; Fuji Photo Film Co., Ltd, Tokyo, Japan) and the Image Gauge (version 3.12) program.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Oligonucleotide sequence (5′→3′)a | Locationb | Use |
|---|---|---|---|
| PUTA004 | CATGAATTCCTCCTTATGCGTTCTCAGTC | putA | Amplification of putA gene |
| PUTA005 | ACTCTAGATTCGGGGATGAGATTGAGGAAA | putA | Amplification of putA gene |
| PUTA0012 | GAGATCAATGCCCATAGCTTATCGAGC | putA | Identification of transcription start sites |
| PUTP004 | GTGGATCCAAGATTACTCTAGCTCAACCA | putP | Amplification of putP gene |
| PUTP005 | GCGCTGCAGTCTAATGAGGACTATCTAATGA | putP | Amplification of putP gene |
Regions of oligonucleotides not complementary to the corresponding genes are underlined.
Location to where the nucleotides are hybridized.
Primer extension analysis.
Primer extension experiments were performed with SuperScript II RNase H− reverse transcriptase (GIBCO-BRL) by the method of Sambrook and Russell (32). A 27-base oligonucleotide (PUTA0012 [Table 2]) complementary to the open reading frame of putA was used as the primer. The primer was end labeled with [γ-32P]ATP using T4 polynucleotide kinase. The cDNA products were purified and resolved on a sequencing gel along sequencing ladders generated with the same primer used for primer extension. The nucleotide sequence of pHJK002 (Table 1) was determined by the dideoxy-chain termination method with Top DNA polymerase (Bioneer, Seoul, Korea) in accordance with the manufacturer's protocols. The bands were visualized and quantified as described above for Northern analysis.
Effects of proline and glutamate and hyperosmotic stresses on the expression of putAP.
To examine the effects of proline and glutamate on the expression of putAP, cultures of V. vulnificus ATCC 29307 were grown in M9 broth (32) supplemented with various levels of proline or glutamate. Similarly, when the effects of various osmolytes on the expression of putAP were analyzed, M9-N broth in which NaCl was omitted but 200 mM proline was added was used. The inocula were from late-exponential-phase cultures in M9 broth. The cultures were adjusted to an initial pH of 7.0 with 100 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid [TES]. The put transcript, total protein, and glutamate levels and proline dehydrogenase activity of the cultures removed at an OD600 of 0.8 were analyzed and compared.
Glutamate determinations.
Cellular levels of glutamate were determined essentially as described previously (3). Cells recovered from 5 ml of culture broth were mixed with 5 ml of boiling distilled water and vortexed vigorously to extract glutamate. The cells were spun down, and glutamate in the supernatant was quantified spectrophotometrically by the method of Witt (35). The means of the results from at least three independent experiments were used.
RESULTS
Organization and sequence relatedness of the V. vulnificus putAP genes.
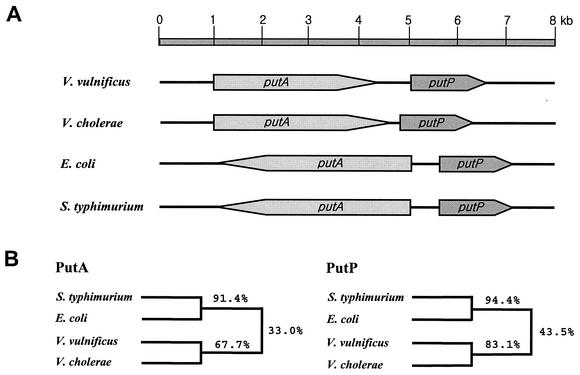
A 8.3-kb DNA fragment from V. vulnificus ATCC 29307, which carries the putAP genes, was cloned in pHJK002 (19) (Table 1). The nucleotide sequence (GenBank accession number AF454004) revealed that the two coding regions consisting of putA and putP are transcribed in the same direction (Fig. 1A). Alignment of the translated putAP sequences to those of PutAP of E. coli and Salmonella enterica serovar Typhimurium revealed about 38% amino acid identity (Fig. 1B) (http//www.ebi.ac.uk/clustalw). However, compared with the PutA proteins of E. coli and S. enterica serovar Typhimurium, the V. vulnificus PutA protein was missing approximately 70 and 200 amino acid residues at the N- and C-terminal regions, respectively. Moreover, the putAP genes of E. coli and S. enterica serovar Typhimurium, which are well characterized at the molecular level, are transcribed divergently. Further searches for amino acid sequences similar to those of V. vulnificus PutAP identified two uncharacterized coding regions from the Vibrio cholerae genome sequence database with higher levels of identity (Fig. 1B). These two coding regions, the presumed V. cholerae putAP genes, are organized in the same orientation as in V. vulnificus putAP (Fig. 1B). The difference in genetic organization indicates that the regulatory mechanisms for the expression of V. vulnificus putAP genes would be different from those for putAP genes in E. coli and S. enterica serovar Typhimurium.
FIG. 1.
Genetic organization of the putAP operons of different microorganisms and amino acid sequence relatedness of their gene products. The microorganisms were V. vulnificus, V. cholerae, E. coli, and S. enterica serotype Typhimurium. (A) The coding regions of putAP genes (shaded boxes) and chromosomal DNA (thick lines) are shown. The direction of transcription is indicated by the arrows. (B) Dendrograms showing relatedness of PutA and PutP were derived using the CLUSTALW alignment program (http//www.ebi.ac.uk/clustalw) and based on the amino acid sequences in the GenBank database (National Center for Biotechnology Information database).
Expression of the V. vulnificus putA gene as a function of growth.
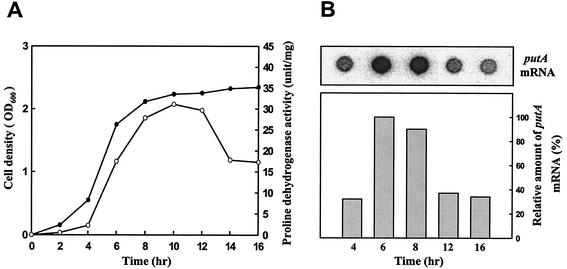
A number of the genes that contribute to stress tolerance in bacteria are induced at the onset of stationary phase. In order to examine whether the expression of putA is influenced by growth phase, the proline dehydrogenase activity of V. vulnificus ATCC 29307 was analyzed at various growth stages (Fig. 2A). Proline dehydrogenase activity appeared at the beginning of growth and increased as the growth rate increased in exponential phase. Proline dehydrogenase activity then decreased when growth slowed down at stationary phase. Growth phase regulation of proline dehydrogenase production could be manifest at either the transcriptional or posttranscriptional level. To distinguish between these two possibilities, the levels of putA mRNA were monitored during growth. The same amount of total RNA was isolated from ATCC 29307 cells at different stages of growth. The results indicated that the putA mRNA levels decreased as the culture entered stationary phase and suggested that growth phase-dependent production of proline dehydrogenase is regulated at the level of transcription (Fig. 2B).
FIG. 2.
Expression of putA as a function of growth. Samples of the culture of V. vulnificus ATCC 29307 in LBS were removed at the time points indicated. Cell density (•) and proline dehydrogenase activity (○) (A) and amount of putA mRNA (B) in samples are shown. The relative amounts of putA transcript of each dot are presented, with the amount of putA transcript at 6 h set at 100%.
Northern analysis of the put transcripts.
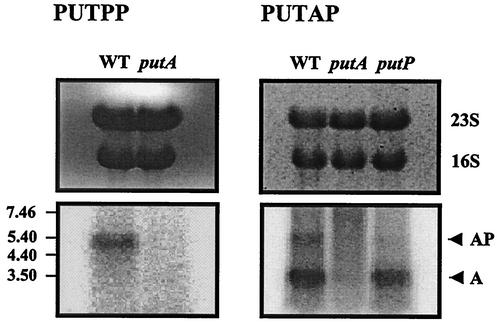
To further characterize the expression of putAP, Northern analysis was performed. When total RNA was isolated from V. vulnificus ATCC 29307 cells grown to log phase in LBS and hybridized with the PUTPP DNA probe, only a single, approximately 5.4-kb transcript, was detected (Fig. 3). On the basis of the DNA sequence of putP, it was anticipated that the putP mRNA would be approximately 1.5 kb long. Cotranscription of putA and putP was predicted to produce a 5.4-kb transcript. Therefore, it appeared that the single mRNA coded for both PutA and PutP. As a further test of this possibility, Northern blot analysis was performed again using PUTAP as a DNA probe. PUTAP also hybridized to a 5.4-kb RNA. In contrast to the result with PUTPP, however, another band of 3.5 kb hybridized with the PUTAP probe (Fig. 3). Since the putA mRNA that is approximately 3.5 kb long was expected on the basis of the DNA sequence of putA, the 3.5-kb band may represent the putA gene transcript. It is noteworthy, however, that the level of transcript AP was lower than that of transcript A in the total RNA we used.
FIG. 3.
Northern blot analysis of the put transcripts. Total RNAs were isolated from cultures of the wild-type (WT) or putA or putP mutants grown to log phase in LBS. The RNAs were separated (top) and hybridized to a 32P DNA probe corresponding to the internal coding regions of putA (PUTAP) or putP (PUTPP) (bottom). The positions of molecular size markers and of hybridization (in kilobases) are shown to the left of the gel. The positions of put transcript A and transcript AP are shown to the right of the gel.
These results supported the possibility that transcription of the putAP genes results in two different put transcripts. However, it was possible, though unlikely, that the two bands resulted from nonspecific hybridization with PUT DNA probes. To examine this possibility, total RNAs isolated from the V. vulnificus isogenic mutants, in which either putA or putP was disrupted by the insertion of nptI (19, 28), were analyzed by Northern blotting. When total RNA was isolated from the putA mutant (HJK001 [Table 1]), the bands corresponding to the two put transcripts were not detected in the Northern blot, irrespective of the DNA probe (Fig. 3). This result demonstrated that the two RNA bands resulted from specific hybridization of PUT DNA probes to the put mRNA rather than from nonspecific hybridization. These data indicate that transcription of the putAP genes results in two different species of put transcripts, transcript AP (a polycistronic putAP transcript) and transcript A (a shorter putA transcript).
Transcript A could be the result of a 3′-end decay of the longer transcript, transcript AP. To further examine this possibility, Northern analysis of total RNA isolated from the isogenic putP mutant (HJK002) (Table 1) was performed. When hybridized with the PUTAP DNA probe, transcript AP was not detected from the total RNA, indicating that transcript AP is not produced in HJK002 (Fig. 3). However, the shorter transcript, transcript A, was still detected, and there was no significant difference in band intensity of the transcript A between the wild-type strain and HJK002. Because transcript A was detected in RNA that lacked transcript AP, it was apparent that transcript A is not the result of a degradation of the longer transcript, transcript AP.
Primer extension analysis of the put transcripts.
Several different explanations were still possible for production of these different types of put transcripts. One is that the destiny of the transcripts is determined after initiation of the transcription through a promoter. That is, transcription of put is regulated by attenuation to result in the shorter transcript A and/or by antitermination (read-through) to result in transcript AP. The other possibility is that the two types of transcripts are determined at the transcription initiation step by two different promoters.
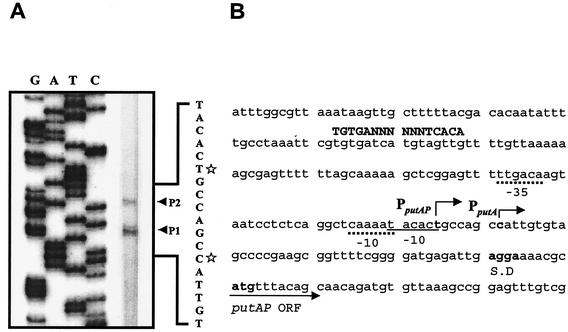
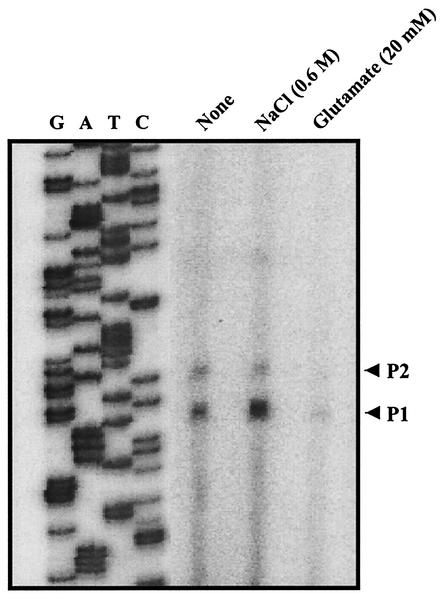
To examine these two possibilities, the transcription start sites of putAP were mapped by primer extension analysis. Total RNA was prepared from V. vulnificus ATCC 29307 grown to log phase in LBS. The primer extension analysis revealed two different transcription start sites, P1 and P2 (Fig. 4A). P1 was located 48 bp upstream of the translational initiation codon of PutA, and P2 was 6 bp from P1 (Fig. 4B). Although other explanations are possible, these results supported the possibility that two different promoters were used for transcription of the two put transcripts. We were unable in several attempts to identify any other transcription start sites by primer extension analysis using different sets of primers (data not shown).
FIG. 4.
Primer extension analysis of put transcripts and sequence of putAP upstream region. (A) The transcription start sites were determined by primer extension of the RNA derived from V. vulnificus ATCC 29307 grown to log phase in LBS. Lanes G, A, T, and C represent the nucleotide sequencing ladders of pHJK002. The transcription start sites for P1 (C) and P2 (T) are indicated by white stars. (B) Transcription start sites are indicated by bent arrows. Possible promoters (−10 and −35) are shown underlined with continuous and broken lines for the promoters PputA and PputAP, respectively. Conserved nucleotide sequences for CRP binding (4) are indicated by capital letters. The ATG translation initiation codon and putative ribosome binding site (AGGA) are indicated in boldface type. ORF, open reading frame.
The putative promoters upstream of the transcription start sites were named PputA (consisting of P1) and PputAP (consisting of P2). Based on the intensity of the bands of the reverse transcripts, the activity of PputAP was lower than that of PputA. The level of transcript A was higher than that of transcript AP in the total RNAs as determined by Northern blotting (Fig. 3). This observation suggests that transcription from the stronger promoter, PputA, results in transcript A and that transcription from the weaker promoter, PputAP, results in transcript AP.
Effect of crp mutation on proline dehydrogenase activity.
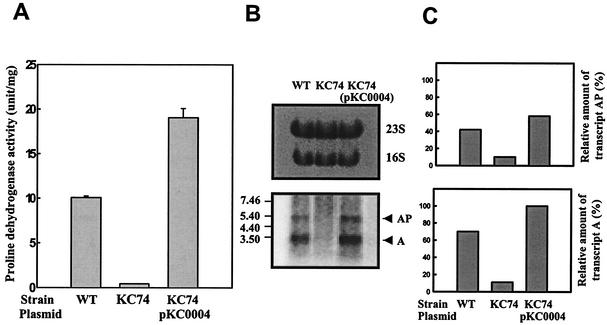
Proline dehydrogenase activity of an isogenic crp mutant of V. vulnificus, which was constructed by allelic exchange (15), was compared with that of the parental wild type. Cultures of ATCC 29307 and the crp mutant KC74 were grown in LBS to log phase (OD600 of 0.6), and the proline dehydrogenase activity of each culture was determined (Fig. 5A). While proline dehydrogenase activity was present at about 10 U per mg of cellular protein in the wild-type strain, KC74 appeared to produce much less proline dehydrogenase. The residual level of proline dehydrogenase activity in KC74 corresponded to approximately 1/10 that in the wild-type strain (Fig. 5A). The reduced production of proline dehydrogenase due to the disruption of crp suggested that cyclic AMP receptor protein (CRP) acts as a positive regulator in the production of proline dehydrogenase, at least in log-phase cultures.
FIG. 5.
Dependence of proline dehydrogenase production of V. vulnificus on crp. Proline dehydrogenase activities (A) and relative amounts of the put transcripts (B and C) were determined for the wild-type (WT) strain and the isogenic crp mutant, KC74, as indicated. Complementation of the crp mutation by functional crp (pKC0004) is also presented. Samples removed from each culture grown to log phase in LBS were analyzed for proline dehydrogenase activity and put transcripts. Means ± SEM (error bars) are shown. PUTAP was used as a DNA probe, and the Northern blot is presented as described in the legend to Fig. 3. The relative amounts of the put transcripts in each band are expressed relative to the amount of the transcript A of KC74(pKC0004).
To rule out the possibility that the decreased proline dehydrogenase activity was due to polar effects of the crp insertion on downstream genes, we examined whether reintroduction of crp on a plasmid could complement the mutation. To do this, plasmid pKC0004 that had been constructed by subcloning crp into pRK415 (15, 17), was transferred into V. vulnificus KC74 by conjugation. The proline dehydrogenase activity expressed by KC74(pKC0004) was about twofold higher than that of ATCC 29307 (Fig. 5A). Therefore, the decreased proline dehydrogenase activity of KC74 resulted from the inactivation of crp rather than any polar effects on any genes downstream of crp.
In order to characterize the role of CRP in more detail, the levels of put transcript in the wild-type strain and KC74 were compared by Northern blot analysis (Fig. 5B and C). When PUTAP was used as the DNA probe, the put transcripts were readily detected in the wild-type strain and in KC74(pKC0004). Transcript A and transcript AP were barely detected in KC74, indicating that CRP exerts its effects on the production of proline dehydrogenase at the level of transcription from both put promoters. Furthermore, the levels of both put transcripts in KC74(pKC0004) recovered, and the pattern of the recovery was similar to that observed when the proline dehydrogenase activities were compared directly. Taking these results together led us to conclude that expression of putAP in V. vulnificus is under the positive control of CRP and that CRP affects the level of proline dehydrogenase production by activating both put promoters.
Effects of proline and glutamate on the expression of putAP.
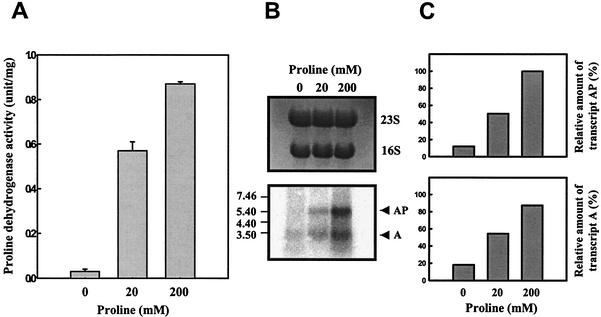
To examine whether expression of putAP is modulated by proline, proline dehydrogenase activities of cultures in M9 broth (32) supplemented with different levels of proline were analyzed. When the amount of proline was increased to 200 mM, the level of proline dehydrogenase activity reached higher levels than that observed in the absence of proline (Fig. 6A). A Northern blot analysis using a PUTAP DNA probe revealed that the addition of proline resulted in increased levels of transcript A and transcript AP (Fig. 6B and C). It was apparent from these results that the expression of putAP is subject to induction by proline in V. vulnificus.
FIG. 6.
Induction of expression of V. vulnificus putAP genes by proline. Samples were removed from the culture of ATCC 29307 grown to log phase in M9 medium supplemented with various levels of proline as indicated. Samples were analyzed for determination of proline dehydrogenase activity (A) and put transcripts (B and C). Means ± SEM (error bars) are shown. PUTAP was used as a DNA probe, and the Northern blot is presented as described in the legend to Fig. 3. The relative amounts of the put transcripts in each band are expressed relative to the amount of transcript AP as expressed in the presence of 200 mM proline.
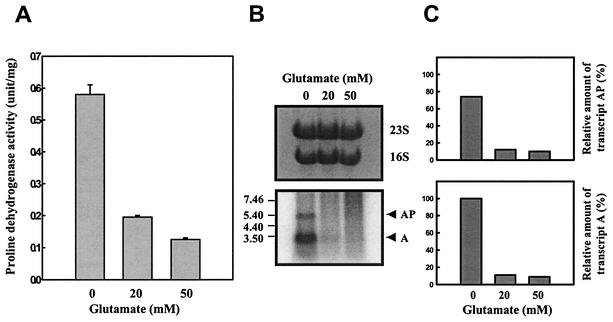
To further investigate regulation of putAP expression, the effect of adding glutamate on the production of proline dehydrogenase was examined. To do this, cultures of V. vulnificus were grown in M9 broth supplemented with 200 mM proline alone or with 200 mM proline and glutamate together. As shown in Fig. 7A, the level of proline dehydrogenase activity induced in the presence of proline alone was repressed by the addition of glutamate. It was possible that the decrease of proline dehydrogenase activity was due to inhibition of the enzyme activity itself by increasing glutamate. However, the cellular level of put transcripts determined by a Northern blot analysis using a PUTAP DNA probe was lower in response to the presence of glutamate (Fig. 7B and C). This reduction of put transcripts brought about by glutamate indicates that the expression of the V. vulnificus putAP genes is repressed by glutamate.
FIG. 7.
Repression of expression of the V. vulnificus putAP operon by glutamate. Samples were removed from the culture of ATCC 29307 grown to log phase in M9 medium supplemented with 200 mM proline and various levels of glutamate as indicated. Samples were analyzed for determination of proline dehydrogenase activity (A) and put transcripts (B and C). Means ± SEM (error bars) are shown. PUTAP was used as a DNA probe, and the Northern blot is presented as described in the legend to Fig. 3. The relative amounts of the put transcripts in each band are expressed relative to the amount of transcript A as expressed in the absence of glutamate.
Effects of hyperosmotic stress on proline dehydrogenase activity and cellular level of glutamate.
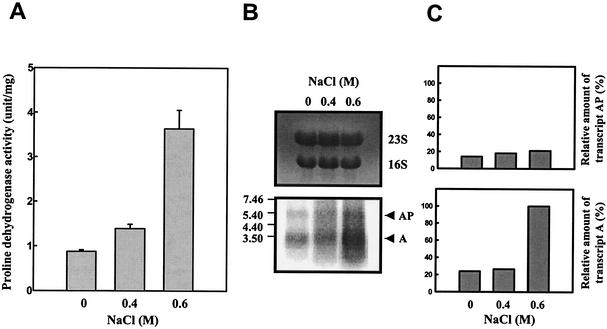
It has been proposed that since inactivation of putA renders V. vulnificus more sensitive to hyperosmotic stress, proline dehydrogenase contributes to the osmotic tolerance of the bacterium (19). As a further test of this hypothesis, we examined whether expression of the put genes is modulated in response to osmotic stress. When cultures in M9-N medium with proline were grown with increasing levels of NaCl, the levels of proline dehydrogenase activity increased (Fig. 8A). Northern blot analysis using a PUTAP DNA probe demonstrated that the level of transcript A also increased in response to excess NaCl. However, no significant increase in the level of transcript AP was observed in the culture grown with NaCl, indicating that the elevated level of transcript A is responsible for the increased proline dehydrogenase activity (Fig. 8B and C).
FIG. 8.
Induction of expression of the V. vulnificus putAP operon by NaCl. Samples were removed from the culture of ATCC 29307 grown to log phase in M9-N medium supplemented with 200 mM proline and various levels of NaCl as indicated. Samples were analyzed for determination of proline dehydrogenase activity (A) and put transcripts (B and C). Means ± SEM (error bars) are shown. PUTAP was used as a DNA probe, and the Northern blot is presented as described in the legend to Fig. 3. The relative amounts of the put transcripts in each band are expressed relative to the amount of transcript A as expressed in the presence of 0.6 M NaCl.
The activities of the put promoters were compared by primer extension of the total RNAs isolated from cultures of ATCC 29307 grown with or without NaCl (Fig. 9). The activity of promoter PputA increased in response to NaCl, but the activity of PputAP was not significantly affected. This indicates that the higher level of proline dehydrogenase activity observed in the presence of NaCl is mainly due to the increased activity of the PputA promoter. These results combined together agreed with our previous hypothesis that transcription of transcript A is directed by promoter PputA and that transcription of transcript AP is directed by promoter PputAP. In contrast, the activities of both PputA and PputAP were reduced when the RNA isolated from the cells grown in the presence of glutamate was analyzed by primer extension (Fig. 9). This observation indicates that the activities of both Pput promoters were repressed by glutamate. This result was not unexpected, since both transcript A and transcript AP were lower in the cells grown with glutamate as determined by Northern blot analysis (Fig. 7B and C).
FIG. 9.
Effects of NaCl and glutamate on the activities of put promoters. Promoter activities were determined by primer extension of the RNA derived from the culture of ATCC 29307 grown to log phase in M9-N (None) and in M9-N supplemented with NaCl (0.6 M) or glutamate (20 mM) as indicated. Lanes G, A, T, and C contain the nucleotide sequencing ladders of pHJK002. The transcription start sites for PputA (P1) and PputAP (P2) are indicated by arrows.
Since proline dehydrogenase converts proline into glutamate, it is reasonable to assume that glutamate, rather than proline, plays a role in counteracting osmotic stress. To test this possibility, the accumulation of glutamate was measured in cells grown with 0.6 M NaCl. Compared with cells grown in M9-N medium alone, cells of V. vulnificus in M9-N medium supplemented with NaCl accumulated greater amounts of glutamate (Table 3). To examine whether this increased level of proline dehydrogenase activity and glutamate accumulation occurred as a result of increasing osmotic stress or NaCl itself, the effects of other osmolytes on proline dehydrogenase activity and on the accumulation of glutamate were analyzed. As shown in Table 3, the cells accumulated excess glutamate and produced higher levels of proline dehydrogenase when they were osmotically stressed, regardless of the type of osmolytes. Since the number and impermeability of the dissolved particles are different, osmolarity of the osmolytes in culture broth would be not same. However, it was obvious that glutamate accumulated in excess and proline dehydrogenase activity increased when cells were exposed to increased osmolarity. These results indicate that the increased level of proline dehydrogenase activity and glutamate accumulation results from increased osmotic strengths in culture media and that glutamate, rather than proline, is a possible compatible solute counteracting hyperosmotic stress in V. vulnificus.
TABLE 3.
Glutamate accumulation and proline dehydrogenase activity in response to osmotic stress
| Osmolytea | Amount of glutamate (nmol mg of protein−1) | Proline dehydrogenase activity (U mg of protein−1) |
|---|---|---|
| None (control) | 253.85 | 1.37 |
| NaCl | 575.35 | 5.85 |
| KCl | 559.45 | 5.74 |
| Sucrose | 485.45 | 4.14 |
NaCl, KCl, and sucrose were present at a final concentration of 0.6 M.
In summary, we have found that transcription of the V. vulnificus putAP genes is initiated by two different promoters, PputA and PputAP. The level of putAP expression is growth phase dependent and is under the control of CRP. Transcription of the putAP genes is induced by proline and hyperosmotic stress and repressed by glutamate. Accumulation of glutamate in response to hyperosmolarity indicates that glutamate plays a physiologically significant role as a compatible solute for osmoprotective purposes in V. vulnificus.
DISCUSSION
Proline is catabolized via the gene products of putA and putP (23, 25). The putP gene encodes a proline permease with a high affinity for proline (22, 31). Proline is then converted into glutamate by the action of two enzymes, proline dehydrogenase and ▵1-pyrroline-5-carboxylate dehydrogenase. In enteric bacteria, putA encodes the PutA protein, which catalyzes both steps of proline utilization (23, 25). Although the functions of putAP gene products are quite conserved among the different microorganisms, the genetic organization and regulatory mechanisms that control the expression of the genes are highly divergent (8, 18). In Agrobacterium tumefaciens and Rhodobacter capsulatus, putA is organized as a monocistronic transcriptional unit, and putP is not located adjacent to putA (8, 18). In E. coli, S. enterica serovar Typhimurium, and Pseudomonas putida, the putA and putP genes are transcribed divergently (23, 34). In these bacteria, the PutA protein is a bifunctional enzyme; in addition to its enzymatic activities, PutA functions as a proline-responsive repressor of the putA and putP genes (1, 22, 24, 26, 30). Besides this autoregulation by PutA, the put genes are catabolite repressed, requiring cyclic AMP and CRP (7). In Klebsiella aerogenes, the expression of proline utilization genes is positively regulated by the Nac (nitrogen assimilation control) protein that is expressed under conditions of general nitrogen limitation (21).
As observed in E. coli and S. enterica serovar Typhimurium, the V. vulnificus putA gene belongs to the put operon together with the putP gene, which encodes the major proline permease (19). The putA gene encodes a proline dehydrogenase, revealing that the function of the gene product of putA is well conserved as observed in other enteric bacteria (19). However, the amino acid sequence homology of V. vulnificus PutA with those of E. coli and S. enterica serovar Typhimurium is quite lower than that observed between PutA proteins from E. coli and S. enterica serovar Typhimurium (Fig. 1). Furthermore, the organization of the putAP genes of V. vulnificus is different from that of other enteric bacteria, such that the transcription orientations of both genes are the same direction rather than divergent. This information that its sequence and genetic organization differ from those of putAP of other enteric bacteria suggests that the putAP genes are probably not expressed and modulated in the same way as observed in other enteric bacteria. To determine whether V. vulnificus putAP genes are autoregulated by PutA, we assessed the expression of putA by constructing a put-lux fusion reporter. When introduced with the reporter, the levels of luminescence in the wild-type and putA mutant cells were comparable (data not shown). This result suggests that the V. vulnificus putAP operon is not autoregulated by the PutA protein.
Recently, the putAP operon from P. aeruginosa was identified (27). The features of the P. aeruginosa putAP operon are remarkably similar to those of the putAP operon of V. vulnificus. Transcription of the P. aeruginosa putAP genes is in the same direction and results in two transcripts, one for putA and the other for putAP. Moreover, it is apparent that the P. aeruginosa putAP genes are not autoregulated by the PutA protein. Expression of the P. aeruginosa putAP genes are activated by a transcriptional activator, PruR, a product of the pruR gene located upstream of putA. Transcription of the putAP genes from a promoter appeared to be attenuated by a stem-loop terminator structure located 10 bp downstream of putA, and only a small portion of the transcription resulted in the putAP transcript. However, an extensive sequence analysis was not able to identify a pruR gene (or homologs of pruR) in the upstream region of V. vulnificus putAP operon (data not shown). Searching for any secondary structures of RNA or sequences conserved for attenuation or antitermination in the putA-putP intergenic region of V. vulnificus was not successful (DNASIS version 2.6, Hitachi Software Engineering Co., Ltd., Tokyo, Japan).
Northern blot and primer extension analyses were performed to determine whether expression of the putAP operon results in two different types of put transcripts. In the meanwhile, it was obvious that transcription of V. vulnificus putAP is directed by two different promoters. The two put promoters appear to be independent of one another, as PputA is induced but PputAP is not by the presence of increasing osmotic stress (NaCl). Furthermore, the observation that the relative amount of each transcript is changed in distinct ways in different growth conditions suggests that the shorter transcript, transcript A, is probably not produced simply by the 3′-end degradation of the longer transcript, transcript AP. Taken together, these results suggested that transcription initiating from PputA results in transcript A and that transcription initiating from PputAP results in transcript AP. However, additional work, such as construction of mutants which lack either PputA or PputAP, is needed to establish the specific mechanisms which result in the two different types of put transcripts. Nevertheless, the differential utilization of the two promoters, requiring distinct regulation, may permit precise adjustment of put expression in response to environmental signals.
We have previously shown that the V. vulnificus strain with a null mutation in the putA gene exhibits impaired growth with proline as the sole carbon and nitrogen source and that its survival is impaired under hyperosmotic stress (19). These pleiotropic effects of putA mutation can be explained if proline dehydrogenase converts proline into glutamate for a dual purpose, for dissimilation and/or assimilation of glutamate and for accumulation of glutamate as an osmoprotectant. Many bacteria accumulate glutamate in response to osmotic stress (6, 11). The availability of compatible solutes in the environment determines whether synthesis or uptake will predominate in the cell (6). The endogenous glutamate that is accumulated in response to osmotic stress is synthesized by glutamate dehydrogenase in enteric bacteria (11). Our data revealed that expression of proline dehydrogenase and accumulation of glutamate in cells increased in response to hyperosmotic stress. These results suggest that the increased level of proline dehydrogenase plays a significant, if not the sole, role for the increased accumulation of glutamate in response to increasing osmotic stress in V. vulnificus. However, when exogenous glutamate is abundant, it has also been observed that the putA mutant HJK001 can take up and utilize glutamate as the sole carbon and nitrogen source and accumulate excess glutamate in response to hyperosmotic stress (data not shown).
In the present study, it was found that expression of the V. vulnificus putAP operon is induced in response to hyperosmotic stress. It has been reported previously that many of the stress-induced genes in enteric bacteria are induced at the onset of stationary phase and are regulated by RpoS (σ38), an alternative sigma factor (13, 33). However, the proline dehydrogenase activity appeared at the beginning of growth and reached a maximum in late exponential phase. Consistent with this, neither the cellular level of the put transcripts nor the proline dehydrogenase activity was affected by the mutation of rpoS (data not shown). On the basis of homology to consensus sequences of the E. coli σ70 promoter, the putative put promoter sequences consisting of −10 and −35 segments were identified (Fig. 4B). The assigned sequences for −10 regions (CAAAAT or TACACT) scored a 67% homology to the −10 consensus sequences (TATAAT) of the E. coli σ70 promoter. Although no sequence of the −35 region assigned with respect to PputA revealed a reasonable homology to −35 consensus sequences, the PputAP −35 region (TTGACA) is apparently identical to the consensus sequence. The homology of put promoters with the consensus sequences of the E. coli σ70 promoter suggests that put promoters are most probably recognized by the V. vulnificus homolog of the σ70 RNA polymerase. In addition to these sequences, a location for a possible CRP binding site was identified by consensus sequences (TGTGAN6-8TCACA [4]). However, it has not yet been established whether CRP activates put promoters directly by binding to the upstream sequences of putAP.
Acknowledgments
This study was supported in part by a grant to S.H.C. from Korea Health 21 research and development project of the Ministry of Health & Welfare (01-PJ1-PG1-01CH14-0001) of the Republic of Korea.
REFERENCES
- 1.Allen, S. W., A. Senti-Willis, and S. R. Maloy. 1993. DNA sequence of the putA gene from Salmonella typhimurium: a bifunctional membrane-associated dehydrogenase that binds DNA. Nucleic Acids Res. 21:1676.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blake, P. A., R. E. Weaver, and D. G. Hollis. 1980. Diseases of humans (other than cholera) caused by vibrios. Annu. Rev. Microbiol. 34:341-367. [DOI] [PubMed] [Google Scholar]
- 3.Botsford, J. L., M. Alvarez, R. Hernandez, and R. Nichols. 1994. Accumulation of glutamate by Salmonella typhimurium in response to osmotic stress. Appl. Environ. Microbiol. 60:2568-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bremer, E., and R. Kramer. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria, p. 79-98. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 7.Chen, L. M., and S. Maloy. 1991. Regulation of proline utilization in enteric bacteria: cloning and characterization of the Klebsiella put control region. J. Bacteriol. 173:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, K., and S. C. Winans. 1996. The putA gene of Agrobacterium tumefaciens is transcriptionally activated in response to proline by an Lrp-like protein and is not autoregulated. Mol. Microbiol. 22:1025-1033. [DOI] [PubMed] [Google Scholar]
- 9.Choi, H. K., N. Y. Park, D. Kim, H. J. Chung, S. Ryu, and S. H. Choi. 2002. Promoter analysis and regulatory characteristics of vvhBA encoding cytolytic hemolysin of Vibrio vulnificus. J. Biol. Chem. 277:47292-47299. [DOI] [PubMed] [Google Scholar]
- 10.Csonka, L. N. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Csonka, L. N., and A. D. Hanson. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569-581. [DOI] [PubMed] [Google Scholar]
- 12.Galinski, E. A. 1995. Osmoadaptation in bacteria. Adv. Microb. Physiol. 37:273-328. [PubMed] [Google Scholar]
- 13.Hengge-Aronis, R. 1996. Regulation of gene expression during entry into stationary phase, p. 1497-1512. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 14.Imhoff, J. F. 1986. Osmoregulation and compatible solutes in eubacteria. FEMS Microbiol. Rev. 39:57-66. [Google Scholar]
- 15.Jeong, H. S., K. C. Jeong, H. K. Choi, K. J. Park, K. H. Lee, J. H. Rhee, and S. H. Choi. 2001. Differential expression of Vibrio vulnificus elastase gene in a growth phase-dependent manner by two different types of promoters. J. Biol. Chem. 276:13875-13880. [DOI] [PubMed] [Google Scholar]
- 16.Jeong, K. C., H. S. Jeong, S. E. Lee, S. S. Chung, J. H. Rhee, A. M. Starks, G. M. Escudero, P. A. Gulig, and S. H. Choi. 2000. Construction and phenotypic evaluation of a Vibrio vulnificus vvpE mutant for elastolytic protease. Infect. Immun. 68:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 18.Keuntje, B., B. Masepohl, and W. Klipp. 1995. Expression of the putA gene encoding proline dehydrogenase from Rhodobacter capsulatus is independent of NtrC regulation but requires an Lrp-like activator protein. J. Bacteriol. 177:6432-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, H. J., J. H. Lee, J. E. Rhee, H. S. Jeong, H. K. Choi, H. J. Chung, S. Ryu, and S. H. Choi. 2002. Identification and functional analysis of the putAP genes encoding Vibrio vulnificus proline dehydrogenase and proline permease. J. Microbiol. Biotechnol. 12:318-326. [Google Scholar]
- 20.Klontz, K. C., S. Lieb, M. Schreiber, H. T. Janowski, L. M. Baldy, and R. A. Gunn. 1988. Syndromes of Vibrio vulnificus infections. Clinical and epidemiologic features in Florida cases, 1981-1987. Ann. Intern. Med. 109:318-323. [DOI] [PubMed] [Google Scholar]
- 21.Macaluso, A., E. A. Best, and R. A. Bender. 1990. Role of the nac gene product in nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J. Bacteriol. 172:7249-7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maloy, S., and J. Roth. 1983. Regulation of proline utilization in Salmonella typhimurium: characterization of put::Mud(Ap lac) operon fusion. J. Bacteriol. 154:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maloy, S. R. 1987. The proline utilization operon, p. 1513-1519. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 24.Maloy, S. R., and V. Stewart. 1993. Autogenous regulation of gene expression. J. Bacteriol. 175:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menzel, R., and J. Roth. 1992. Purification of the putA gene product: a bifunctional membrane-bound protein from Salmonella typhimurium responsible for the two-step oxidation of proline to glutamate. J. Biol. Chem. 256:9755-9761. [PubMed] [Google Scholar]
- 26.Muro-Pastor, A. M., P. Ostrovsky, and S. Maloy. 1997. Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J. Bacteriol. 179:2788-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakada, Y., T. Nishijyo, and Y. Itoh. 2002. Divergent structure and regulatory mechanism of proline catabolic system: characterization of the putAP proline catabolic operon of Pseudomonas aeruginosa PAO1 and its regulation by PruR, an AraC/XylS family protein. J. Bacteriol. 184:5633-5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 29.Ostrovsky, P., K. O'Brien, and S. Maloy. 1991. Regulation of proline utilization in Salmonella typhimurium: a membrane-associated dehydrogenase binds DNA in vitro. J. Bacteriol. 173:211-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrovsky, P., and S. Maloy. 1993. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc. Natl. Acad. Sci. 90:4295-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratzkin, B., and J. Roth. 1978. Cluster of genes controlling proline degradation in Salmonella typhimurium. J. Bacteriol. 133:744-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Tanaka, K., Y. Takayanagi, N. Fujita, A. Ishihama, and H. Takahashi. 1993. Heterogeneity of the principal σ factor in Escherichia coli: the rpoS gene product, σ38, is a second principal σ factor of RNA polymerase in stationary-phase Escherichia coli. Proc. Natl. Acad. Sci. 90:3511-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilchez, S., L. Molina, C. Ramos, and J. L. Ramos. 2000. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 182: 91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witt, I. 1974. Determination with glutamate dehydrogenase and 3-acetyl pyridine analogue of NAD (APAD), p. 1713. In U. Berger (ed.), Methods of enzymatic analysis, vol. 4. Academic Press, New York, N.Y.