Abstract
In Bradyrhizobium japonicum, a gene named nnrR was identified which encodes a protein with high similarity to FNR/CRP-type transcriptional regulators. Mutant strains carrying an nnrR null mutation were unable to grow anaerobically in the presence of nitrate or nitrite, and they lacked both nitrate and nitrite reductase activities. Anaerobic activation of an nnrR′-′lacZ fusion required FixLJ and FixK2. In turn, N oxide-mediated induction of nir and nor genes encoding nitrite and nitric oxide reductase, respectively, depended on NnrR. Thus, NnrR expands the FixLJ-FixK2 regulatory cascade by an additional control level which integrates the N oxide signal required for maximal induction of the denitrification genes.
Denitrification is an alternative form of respiration in which bacteria sequentially reduce nitrate (NO3−) or nitrite (NO2−) to nitrogen gas (N2) via the intermediates nitric oxide (NO) and nitrous oxide (N2O), when oxygen becomes limiting. Reduction of nitrogen oxides is coupled to energy conservation and permits cell growth under microoxic or anoxic conditions. Denitrification is initiated by reduction of nitrate to nitrite catalyzed by the respiratory (dissimilatory) nitrate reductase. Yet this reaction is not unique to denitrification, because it also occurs in dissimilatory and assimilatory ammonification, both of which result in the formation of ammonia. Thus, the defining reaction in denitrification is the reduction of nitrite to the first gaseous intermediate, NO. For reviews see references 6, 28, and 42.
Bradyrhizobium japonicum is a soil bacterium with the capability of reducing NO3− simultaneously to NH4+ and N2 when cultured anaerobically with nitrate as the terminal electron acceptor and sole source of nitrogen (34). The recently published genome sequence (20; also see http://www.kazusa.or.jp/rhizobase) indicated that B. japonicum lacks genes encoding a membrane-bound nitrate reductase. As with Pseudomonas sp. strain G-179 (8), the first step of denitrification in B. japonicum depends on the napEDABC genes, which specify a periplasmic nitrate reductase (M. J. Delgado, E. J. Bedmar, and P. Müller, personal communication). Subsequent denitrification reactions are catalyzed by the products of nirK (37), norCBQD (24), and nosRZDYFLX (E. J. Bedmar, unpublished data), which encode reductases for nitrite, nitric oxide, and nitrous oxide, respectively.
Microaerobic induction of transcription from the nir, nor, and nos promoter regions depends on the fixLJ and fixK2 genes, whose products form the FixLJ-FixK2 regulatory cascade (2, 24, 25, 37, and E. J. Bedmar, unpublished). Hence, B. japonicum fixLJ and fixK2 mutants are unable to grow anaerobically with nitrate as the terminal electron acceptor.
FixLJ is a two-component regulatory system consisting of the heme-based sensor kinase FixL and the FixJ response regulator (32 and references therein). In B. japonicum, the only known gene that apparently is directly controlled by FixJ is fixK2. Its product, FixK2, is a transcriptional activator for a large group of genes involved in anaerobic or microaerobic metabolism (14, 25). FixK2 belongs to the bacterial family of FNR/CRP-type transcriptional regulators. FNR (fumarate and nitrate reductase regulator) of Escherichia coli has four domains: (i) the N-terminal redox-sensing domain where three cysteine residues together with a centrally located cysteine coordinate binding of a redox-responsive [4Fe-4S]2+ cluster; (ii) the central β-roll domain that interacts with RNA polymerase; (iii) a long α helix involved in protein dimer formation; and (iv) the C-terminal helix-turn-helix DNA binding motif (for reviews see references 16, 21, and 33). Oxygen-responsive FNR homologs have been identified in a number of other bacteria, e.g., FnrA of Pseudomonas stutzeri (9), ANR of Pseudomonas aeruginosa (40), FnrL of Rhodobacter sphaeroides (41), FnrP of Paracoccus denitrificans (35), and FixK1 of B. japonicum (3). They all contain the N-terminal cysteine-rich motif. By contrast, FixK2 of B. japonicum and its homologs lack that cysteine motif, and there is no evidence that members of this protein class are redox responsive (14). Another category of FNR-like activator proteins comprises R. sphaeroides NnrR (22, 31), P. denitrificans NNR (36), P. aeruginosa DNR (4), and P. stutzeri DnrD (39). These regulators also lack the cysteine signature, and they control expression of nir and nor genes in response to NO or a chemically related species. The precise sensing mechanism is still unknown. The consensus DNA binding site for E. coli FNR is TTGAT-N4-ATCAA (FNR box), which usually is located around position −41.5 relative to the transcription start of FNR-activated promoters (30). A similar motif is found at a comparable position in many target promoters of the FNR-like proteins described above (42). Accordingly, C-terminal amino acid residues predicted to make contact with DNA (Glu209, Ser212, Arg213) (23) are conserved in FNR homologs.
Here we report on the identification and functional characterization of the B. japonicum nnrR gene whose product (NnrR) mediates control of denitrification genes in response to N oxide. We provide evidence that nnrR itself is a target gene for the FixLJ-FixK2 regulatory cascade, thereby extending it by an additional control level.
Identification and mutational analysis of B. japonicum nnrR.
This gene was originally identified as orf236 in the course of analyzing a gene cluster which encodes a heme uptake system (26; position numbers 1807 to 2517 on the complementary strand of the sequence were deposited under GenBank accession no. AJ311165). The nnrR gene is 710 bp in length, has a G+C content of 68.5 mol%, and encodes a protein of 236 amino acids with a calculated molecular mass of 25,870 Da. The B. japonicum NnrR protein shares significant similarity with NnrR of R. sphaeroides strains (47 to 49% identity) (AAB69132, AAC44402, AAD27624) (22, 31) and with an uncharacterized, predicted protein from Rhodopseudomonas palustris (74% identity) (ZP_00008933). The N terminus of B. japonicum NnrR lacks the cysteine motif that is characteristic for redox-responsive FNR-like proteins. Near the C terminus it contains a predicted helix-turn-helix motif likely to be involved in DNA binding (F177PISRQDIAQMTGTTLHTVSRILSGWEQQGLV208; MotifScan; http://www.expasy.org/prosite/).
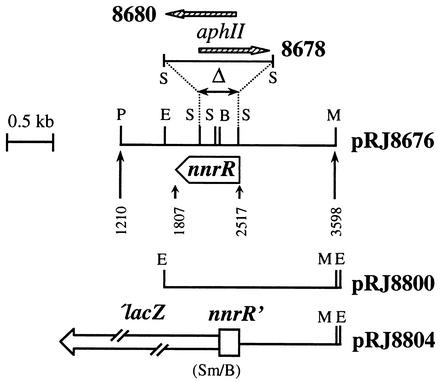
A 2.39-kb PvuII/MluI fragment harboring nnrR was subcloned in plasmid pRJ8676 (Fig. 1). For functional analysis, two nnrR-internal SalI fragments comprising 425 bp were deleted and replaced by a kanamycin resistance gene cassette (aphII) on a 1.22-kb SalI fragment from pBSL86 (Fig. 1) (1). Conventional marker exchange mutagenesis (17, 18) resulted in mutant strains 8678 and 8680 (Fig. 1).
FIG. 1.
Physical map of the B. japonicum nnrR locus present on plasmid pRJ8676 and genetic constructions relevant to this work. Numbers below vertical arrows refer to nucleotide positions as given in the GenBank database (accession no. AJ311165). The genotype of deletion-insertion mutants 8678 and 8680 is indicated above the physical map, with the orientation of the inserted aphII resistance gene cassette emphasized by horizontal arrows and the deleted DNA region indicated by the horizontal double-headed arrow. Below are the insert of plasmid pRJ8800 (cloned in the broad-host-range vector pRK290 [12]), which was used in complementation experiments, and the structure of the translational nnrR′-′lacZ fusion on the chromosomally integrated plasmid pRJ8804. Note that the EcoRI site at the right end of the plasmid inserts originates from the vector pUCBM20 (Boehringer Mannheim, Mannheim, Germany) that was used for construction of pRJ8676. B, BsaBI; E, EcoRI; M, MluI; P, PvuII; S, SalI; Sm, SmaI. Restriction sites in parentheses were destroyed during the cloning procedures.
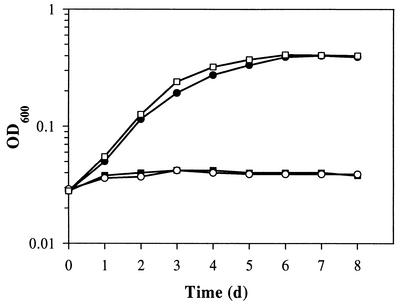
In contrast to the wild type, cells of B. japonicum 8678 did not grow when cultured anaerobically in yeast extract-mannitol (YEM) medium (10) supplemented with 10 mM KNO3 (Fig. 2). The same results were obtained with mutant 8680 or when nitrate was replaced by KNO2 (100 μM) as the final electron acceptor (data not shown). Complementation of mutant strain 8678 with plasmid pRJ8800 containing the wild-type nnrR gene (Fig. 1) restored growth under these conditions (Fig. 2). For the determination of nitrate and nitrite reductase activities, cells were first grown aerobically in YEM medium, collected by centrifugation, washed twice with YEM, and then incubated anaerobically for 96 h in the same medium supplemented with 10 mM KNO3 in completely filled, rubber-stoppered serum bottles. Enzyme activities were assayed as described previously (11). Unlike wild-type cells, nnrR mutant cells lacked both nitrate and nitrite reductase activity. Both nnrR mutants were also used in soybean plant infection tests (15, 17) and were found to have a wild-type phenotype with regard to nodulation and nitrogen fixation (acetylene reduction) (data not shown).
FIG. 2.
Anaerobic growth of wild-type B. japonicum (•), nnrR mutant strain 8678 (▪), strain 8678 harboring the vector pRK290 (control) (○), and the nnrR-containing plasmid pRJ8800 (□). Cells were inoculated into YEM medium plus 10 mM KNO3, and anaerobic growth was followed by monitoring the optical density at 600 nm (OD600).
Role of nnrR in the regulation of nirK and norCBQD genes.
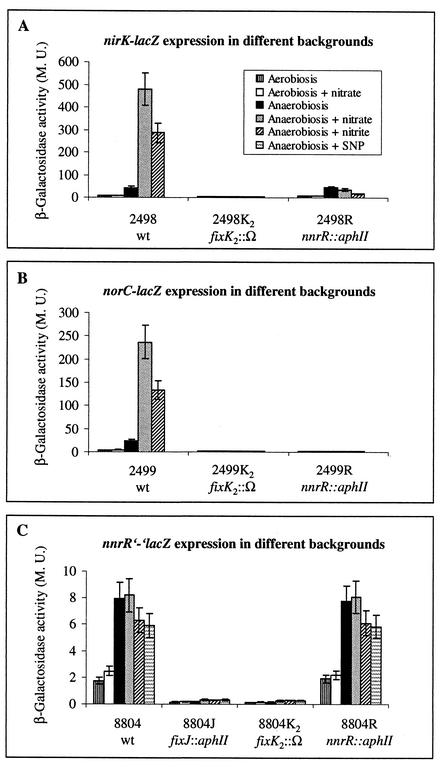
Chromosomally integrated, transcriptional lacZ fusions to the promoters of nirK and norCBQD were used in regulatory studies. For construction of the nirK-lacZ fusion, an 878-bp EcoRI/PstI fragment from pNIRLZ (37) was cloned into the pSUP202-based (29) lacZ fusion vector pSUP3535 (H. M. Fischer, unpublished data), resulting in plasmid pRJ2498. The norC-lacZ fusion was constructed by cloning an EcoRI fragment of about 4.3 kb from pNORLZ (24) into vector pSUP3535, yielding plasmid pRJ2499. The fusion plasmids pRJ2498 and pRJ2499 were integrated by homologous recombination into the chromosome of wild-type B. japonicum, fixK2 mutant 9043 (25), and nnrR mutant 8678 (this work), resulting in strains 2498, 2498K2, 2498R, 2499, 2499K2, and 2499R, respectively. Expression of the lacZ reporter gene was determined in β-galactosidase activity assays with cells grown under different conditions (Fig. 3A and B).
FIG. 3.
β-Galactosidase activities resulting from expression of an nirK-lacZ (A), norC-lacZ (B), or nnrR′-′lacZ (C) fusion chromosomally integrated in different B. japonicum backgrounds. The relevant genotype of host strains is indicated below strain designations (for details see the text). Cells were first grown aerobically in YEM medium, collected by centrifugation, and then incubated aerobically or anaerobically (filled rubber-stoppered tubes) for 96 h in the same medium supplemented with 10 mM KNO3, 10 mM KNO2 (periodically added to 100 μM in regular 24-h intervals), or 10 μM SNP. The legend shown in panel A is also valid for panels B and C. Values are means ± standard errors from at least two independent experiments with two cultures assayed in duplicate. β-Galactosidase activity is expressed in Miller units (M. U.). wt, wild type.
Only background expression levels were observed with both fusions in aerobically grown cells, regardless of the presence or absence of nitrate (10 mM KNO3). Maximal induction was observed in wild-type cells that were incubated anaerobically in the presence of nitrate. Nitrate (or an N oxide derived from it) is crucial for maximal induction, because expression levels were only about 10% of the maximal values when no nitrate was added to the anaerobic cultures. Supplementation with nitrite (repeated addition of KNO2 to 100 μM in regular 24-h intervals) (Fig. 3A and B) or 10 μM sodium nitroprusside (SNP), an NO+-generating agent (reference 7 and data not shown), instead of nitrate resulted in half-maximal expression of the reporter fusions. No significant expression was observed in the fixK2 mutant background under all conditions tested, confirming our previous finding that FixK2 is absolutely required for activation of both the nirK and the norCBQD promoter (Fig. 3A, strain 2498K2, and B, strain 2499K2) (24, 37). Induction of the norCBQD promoter was completely abolished in the absence of a functional nnrR gene (Fig. 3B, strain 2499R). By contrast, anaerobic induction of the nirK promoter was retained in the nnrR mutant background (Fig. 3A, strain 2498R), implying that the nirK and the norCBQD promoter exhibit slight differences with regard to their dependence on FixK2 in the absence of NnrR.
Regulation of nnrR expression.
To evaluate a potential regulatory link between FixLJ-FixK2 and NnrR, we studied expression of nnrR in different backgrounds. For this purpose, a translational fusion of nnrR (87th codon) to lacZ was constructed by cloning a 1,356-bp EcoRI-BsaBI fragment from pRJ8676 into pSUP482 (H. M. Fischer, unpublished) to yield plasmid pRJ8804 (Fig. 1). This plasmid was integrated by homologous recombination into the chromosome of wild-type B. japonicum, fixJ mutant 7360 (2), fixK2 mutant 9043 (25), and nnrR mutant 8678 (this work), resulting in strains 8804, 8804J, 8804K2, and 8804R, respectively. In strain 8804R, the nnrR::aphII null mutation was retained after integration of the nnrR′-′lacZ fusion. Cells of these strains were grown under different conditions, and nnrR expression was determined in β-galactosidase activity assays (Fig. 3C).
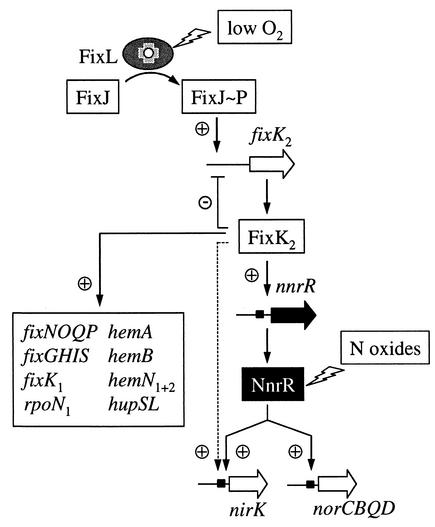
Expression of nnrR′-′lacZ was quite low (<10 Miller units) under all growth conditions. In anaerobically incubated cells, expression was significantly enhanced three- to fourfold compared to that for aerobically cultivated cells, regardless of the absence or presence of KNO3, KNO2, or SNP. The same expression pattern was observed in the nnrR mutant background, indicating that nnrR does not control its own expression. By contrast, only very low nnrR′-′lacZ expression was detected in cells of strains 8804J and 8804K2, regardless of the incubation conditions. These findings are compatible with a model that places NnrR in the FixLJ-FixK2 cascade downstream of FixK2 (Fig. 4). The presence of a putative, albeit rather poorly conserved, FixK2 box upstream of nnrR further supports this model (TTGCG-N4-CGCAA53, with the underlined nucleotides matching the consensus FNR/FixK box [the nucleotide position number refers to the translational start of nnrR]) (14, 42).
FIG. 4.
Location and function of NnrR in the FixLJ-FixK2 regulatory cascade of B. japonicum. Details are explained in the text. Note that even though the model suggests a direct hierarchical organization of FixLJ, FixK2, NnrR, and denitrification genes, additional control levels situated in between cannot be excluded. Similarly, N oxide signaling to NnrR may be direct or indirect. The dashed arrow refers to anaerobic activation of nirK (but not of norCBQD) by FixK2, which is retained in an nnrR mutant background. Filled boxes symbolize putative binding sites for FixK2 and NnrR which are similar to that of the consensus FNR box (TTGAT-N4-ATCAA).
With NnrR we have added to the FixLJ-FixK2 cascade an additional control level which integrates the N oxide signal, presumably either NO2− or NO or both, that is critical for maximal induction of the B. japonicum denitrification genes. Our data indicate that N oxide control of B. japonicum denitrification genes occurs at the nnrR posttranscriptional level, yet the identity of the signaling molecule and the sensing mechanism remain unknown. Nitric oxide (NO) has been suggested or shown to act as a signal molecule for NnrR of R. sphaeroides (22, 31), NNR of P. denitrificans (19, 36), DNR of P. aeruginosa (5), and DnrD of P. stutzeri (38). It seems likely that NnrR and its homologues can switch from an inactive to an active form in response to NO.
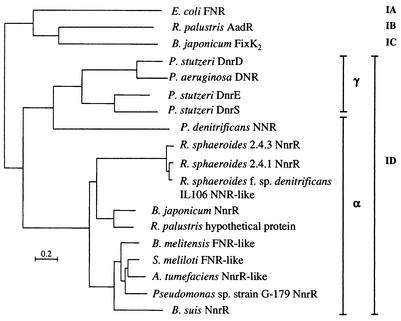
NnrR-like proteins constitute a distinct cluster within the ID subgroup of the FNR family of transcriptional regulators.
Based on overall amino acid sequence similarity and distinct structural features, the family of FNR-like proteins has been divided into four subgroups, with E. coli FNR (IA), R. palustris AadR (IB), rhizobial FixK proteins (IC), and P. aeruginosa DNR (ID) as representative reference proteins of each group (14, 16, 39). We determined the phylogenetic relationship of 15 members of subgroup ID (including B. japonicum NnrR) and compared it with those of the other subgroups of the protein family (Fig. 5). The proteins of subgroup ID can be further divided into two subclusters that largely follow the classification of the proteobacterial subgroups of the respective bacterial species. One subcluster is formed by DNR-like proteins from γ-proteobacterial Pseudomonas species, while the other branch comprises NnrR-like proteins from various α-proteobacterial species. The NNR protein of the α-proteobacterium P. denitrificans represents an exception in that it clusters with the γ-proteobacterial DNR-like proteins. The position of the NnrR-like protein of Pseudomonas sp. strain G-179 within the group of α-proteobacterial homologues is not surprising, because this species is most likely a rhizobial strain as deduced from 16S rRNA analyses (8). Zumft and coworkers noted characteristic distinctions between DNR- and NnrR-like proteins which may eventually lead to their classification in different subgroups (39, 43): (i) in contrast to the EXXSR motif in DNR-like proteins, the recognition helix for DNA binding of NnrR-like regulators includes a conserved HXXSR motif; (ii) DNR- and NnrR-like proteins seem to correlate with the regulation of cytochrome cd1- and Cu-containing nitrite reductase enzymes, respectively. The NnrR protein of B. japonicum perfectly matches these criteria.
FIG. 5.
Phylogenetic tree of 15 FNR-like transcriptional activator proteins belonging to subgroup ID (14, 16, 39). For comparison, one representative reference protein of each subgroup IA, IB, and IC is included in the tree. Amino acid sequences were aligned with the program T-COFFEE (27; http://www.ch.embnet.org/software/TCoffee.html), and phylogenetic analyses were performed with the phylogeny inference package PHYLIP (13; http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html). The scale bar represents a distance of 0.2 substitutions per position. Accession numbers of individual protein sequences are the following: E. coli FNR, AAC74416; R. palustris AAdR, Q01980; B. japonicum FixK2, CAA06287; P. stutzeri DnrD, CAC14591; P. aeruginosa DNR, BAA08744; P. stutzeri DnrE, CAB40906; P. stutzeri DnrS, CAB40908; P. denitrificans NNR, AAA69977; R. sphaeroides 2.4.3 NnrR, AAC44402; R. sphaeroides 2.4.1 NnrR, AAB69132; R. sphaeroides f. sp. denitrificans IL-106 NNR-like, AAD27624; B. japonicum NnrR, CAC38738; R. palustris NnrR-like, ZP_00008933; Brucella suis NnrR, AAN33482; Brucella melitensis FNR-like, NP_541964; Pseudomonas sp. strain G-179 NnrR, AAB96771; Sinorhizobium meliloti FNR-like, NP_435925; Agrobacterium tumefaciens NnrR-like, NP_534858.
Acknowledgments
We thank Zöhre Ucurum for excellent technical assistance. We are grateful to Stéphane Vuilleumier for helpful advice in the phylogenetic analysis of NnrR.
This work was supported by a grant from the Swiss Federal Institute of Technology, Zürich, and by grant BMC2002-04126-C03-02 from the Dirección General de Investigación and Junta de Andalucía PAI/CVI-275.
REFERENCES
- 1.Alexeyev, M. F. 1995. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques 18:52-54. [PubMed] [Google Scholar]
- 2.Anthamatten, D., and H. Hennecke. 1991. The regulatory status of the fixL-like and fixJ-like genes in Bradyrhizobium japonicum may be different from that in Rhizobium meliloti. Mol. Gen. Genet. 225:38-48. [DOI] [PubMed] [Google Scholar]
- 3.Anthamatten, D., B. Scherb, and H. Hennecke. 1992. Characterization of a fixLJ-regulated Bradyrhizobium japonicum gene sharing similarity with the Escherichia coli fnr and Rhizobium meliloti fixK genes. J. Bacteriol. 174:2111-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai, H., Y. Igarashi, and T. Kodama. 1995. Expression of the nir and nor genes for denitrification of Pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett. 371:73-76. [DOI] [PubMed] [Google Scholar]
- 5.Arai, H., T. Kodama, and Y. Igarashi. 1999. Effect of nitrogen oxides on expression of the nir and nor genes for denitrification in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 170:19-24. [DOI] [PubMed] [Google Scholar]
- 6.Baker, S. C., S. J. Ferguson, B. Ludwig, M. D. Page, O. M. Richter, and R. J. van Spanning. 1998. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 62:1046-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bates, J. N., M. T. Baker, R. Guerra, Jr., and D. G. Harrison. 1991. Nitric oxide generation from nitroprusside by vascular tissue: evidence that reduction of the nitroprusside anion and cyanide loss are required. Biochem. Pharmacol. 42(Suppl.):S157-S165. [DOI] [PubMed] [Google Scholar]
- 8.Bedzyk, L., T. Wang, and R. W. Ye. 1999. The periplasmic nitrate reductase in Pseudomonas sp. strain G-179 catalyzes the first step of denitrification. J. Bacteriol. 181:2802-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuypers, H., and W. G. Zumft. 1993. Anaerobic control of denitrification in Pseudomonas stutzeri escapes mutagenesis of an fnr-like gene. J. Bacteriol. 175:7236-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, R. M., and C. A. Appleby. 1972. Anaerobic-nitrate, symbiotic and aerobic growth of Rhizobium japonicum: effects on cytochrome P450, other haemoproteins, nitrate and nitrite reductases. Biochim. Biophys. Acta 275:347-354. [DOI] [PubMed] [Google Scholar]
- 11.Delgado, M. J., J. Olivares, and E. J. Bedmar. 1989. Nitrate reductase activity of free-living and symbiotic uptake hydrogenase-positive and uptake hydrogenase-negative strains of Bradyrhizobium japonicum. Arch. Microbiol. 151:166-170. [Google Scholar]
- 12.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1989. PHYLIP—phylogeny inference package. Cladistics 5:164-166. [Google Scholar]
- 14.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Göttfert, M., S. Hitz, and H. Hennecke. 1990. Identification of nodS and nodU, two inducible genes inserted between the Bradyrhizobium japonicum nodYABC and nodIJ genes. Mol. Plant-Microbe Interact. 3:308-316. [DOI] [PubMed] [Google Scholar]
- 16.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol 44:1-34. [DOI] [PubMed] [Google Scholar]
- 17.Hahn, M., and H. Hennecke. 1984. Localized mutagenesis in Rhizobium japonicum. Mol. Gen. Genet. 193:46-52. [Google Scholar]
- 18.Hahn, M., L. Meyer, D. Studer, B. Regensburger, and H. Hennecke. 1984. Insertion and deletion mutations within the nif region of Rhizobium japonicum. Plant Mol. Biol. 3:159-168. [DOI] [PubMed] [Google Scholar]
- 19.Hutchings, M. I., N. Shearer, S. Wastell, R. J. van Spanning, and S. Spiro. 2000. Heterologous NNR-mediated nitric oxide signaling in Escherichia coli. J. Bacteriol. 182:6434-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 21.Kiley, P. J., and H. Beinert. 1999. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 22.Kwiatkowski, A. V., and J. P. Shapleigh. 1996. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Biol. Chem. 271:24382-24388. [DOI] [PubMed] [Google Scholar]
- 23.Lazazzera, B. A., D. M. Bates, and P. J. Kiley. 1993. The activity of the Escherichia coli transcription factor Fnr is regulated by a change in oligomeric state. Genes Dev. 7:1993-2005. [DOI] [PubMed] [Google Scholar]
- 24.Mesa, S., L. Velasco, M. E. Manzanera, M. J. Delgado, and E. J. Bedmar. 2002. Characterization of the norCBQD genes, encoding nitric oxide reductase, in the nitrogen fixing bacterium Bradyrhizobium japonicum. Microbiology 148:3553-3560. [DOI] [PubMed] [Google Scholar]
- 25.Nellen-Anthamatten, D., P. Rossi, O. Preisig, I. Kullik, M. Babst, H. M. Fischer, and H. Hennecke. 1998. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J. Bacteriol. 180:5251-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nienaber, A., H. Hennecke, and H. M. Fischer. 2001. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41:787-800. [DOI] [PubMed] [Google Scholar]
- 27.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, D. J., and N. J. Watmough. 1999. Inorganic nitrogen metabolism in bacteria. Curr. Opin. Chem. Biol. 3:207-219. [DOI] [PubMed] [Google Scholar]
- 29.Simon, R., U. Priefer, and A. Pühler. 1983. Vector plasmids for in vivo and in vitro manipulation of gram-negative bacteria, p. 98-106. In A. Pühler (ed.), Molecular genetics of the bacteria-plant interaction. Springer Verlag, Heidelberg, Germany.
- 30.Spiro, S. 1994. The FNR family of transcriptional regulators. Antonie van Leeuwenhoek 66:23-36. [DOI] [PubMed] [Google Scholar]
- 31.Tosques, I. E., J. Shi, and J. P. Shapleigh. 1996. Cloning and characterization of nnrR, whose product is required for the expression of proteins involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 178:4958-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuckerman, J. R., G. Gonzalez, and M. A. Gilles-Gonzalez. 2001. Complexation precedes phosphorylation for two-component regulatory system FixL/FixJ of Sinorhizobium meliloti. J. Mol. Biol. 308:449-455. [DOI] [PubMed] [Google Scholar]
- 33.Unden, G., S. Achebach, G. Holighaus, H. G. Tran, B. Wackwitz, and Y. Zeuner. 2002. Control of FNR function of Escherichia coli by O2 and reducing conditions. J. Mol. Microbiol. Biotechnol. 4:263-268. [PubMed] [Google Scholar]
- 34.Vairinhos, F., W. Wallace, and D. J. D. Nicholas. 1989. Simultaneous assimilation and denitrification of nitrate by Bradyrhizobium japonicum. J. Gen. Microbiol. 135:189-193. [Google Scholar]
- 35.van Spanning, R. J., A. P. De Boer, W. N. Reijnders, H. V. Westerhoff, A. H. Stouthamer, and O. J. Van Der. 1997. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol. Microbiol. 23:893-907. [DOI] [PubMed] [Google Scholar]
- 36.van Spanning, R. J., E. Houben, W. N. Reijnders, S. Spiro, H. V. Westerhoff, and N. Saunders. 1999. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J. Bacteriol. 181:4129-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velasco, L., S. Mesa, M. J. Delgado, and E. J. Bedmar. 2001. Characterization of the nirK gene encoding the respiratory, Cu-containing nitrite reductase of Bradyrhizobium japonicum. Biochim. Biophys. Acta 1521:130-134. [DOI] [PubMed] [Google Scholar]
- 38.Vollack, K. U., and W. G. Zumft. 2001. Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 183:2516-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollack, K.-U., E. Härtig, H. Körner, and W. G. Zumft. 1999. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol. Microbiol. 31:1681-1694. [DOI] [PubMed] [Google Scholar]
- 40.Ye, R. W., D. Haas, J. O. Ka, V. Krishnapillai, A. Zimmermann, C. Baird, and J. M. Tiedje. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 177:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeilstra-Ryalls, J. H., and S. Kaplan. 1995. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J. Bacteriol. 177:6422-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zumft, W. G. 2002. Nitric oxide signaling and NO dependent transcriptional control in bacterial denitrification by members of the FNR-CRP regulator family. J. Mol. Microbiol. Biotechnol. 4:277-286. [PubMed] [Google Scholar]