Abstract
Holin proteins are phage-induced integral membrane proteins which regulate the access of lytic enzymes to host cell peptidoglycan at the time of release of progeny viruses by host cell lysis. We describe the identification of the membrane-containing phage PRD1 holin gene (gene XXXV). The PRD1 holin protein (P35, 12.8 kDa) acts similarly to its functional counterpart from phage lambda (gene S), and the defect in PRD1 gene XXXV can be corrected by the presence of gene S of lambda. Several nonsense, missense, and insertion mutations in PRD1 gene XXXV were analyzed. These studies support the overall conclusion that the charged amino acids at the protein C terminus are involved in the timing of host cell lysis.
Holins are small integral membrane proteins required for host cell lysis by most double-stranded DNA (dsDNA) and dsRNA bacteriophages. Holin proteins act by permeabilizing the host cell cytoplasmic membrane, allowing the phage endolysin to reach its peptidoglycan target and determine the timing of host cell lysis (51). Mutant holin alleles causing premature or delayed lysis have been isolated in the bacteriophage λ and T4 systems (27, 39-41). The holin proteins characterized so far share a number of typical properties. Typically, holins have at least one putative α-helical transmembrane region and a highly charged hydrophilic C-terminal domain. Many holin genes have a dual-start motif that permits the synthesis of two products of different lengths: the effector and inhibitor forms of the holin protein (21, 51, 54). The membrane permeabilization function seems to be nonspecific, as every heterologous holin-endolysin pair tested has proven to be lytically competent (51).
Bacteriophage PRD1 is a broad-host-range phage that infects gram-negative bacterial species such as Escherichia coli and Salmonella enterica carrying a receptor-encoding IncP multidrug resistance plasmid (38). It is a member of the Tectiviridae family, a group of icosahedral bacteriophages with a linear dsDNA genome and an internal membrane component (2, 38). The virion consists of a protective protein coat that surrounds the protein-rich lipid membrane, which in turn encloses the phage genome (1, 13). The phage membrane is composed of approximately half lipid and half phage-encoded membrane-associated protein and has been proposed to function in phage DNA entry (1, 7, 16, 19, 20).
As for typical lytic dsDNA phages, progeny virions are liberated from PRD1-infected cells via disruption of the host cell. Gene XV and complementation group A, which are involved in this process, have been identified by analysis of phage nonsense mutants (32), suggesting that a two-component, holin-endolysin system also operates in phage PRD1. The product of gene XV, protein P15, is a soluble β-1,4-N-acetylmuramidase that effectively degrades the peptidoglycan moiety of the gram-negative bacterial cell wall, causing lysis (14). Earlier attempts to isolate PRD1 nonsense mutants yielded a total of six group A amber mutants that were deficient in lytic ability but not particle assembly. These mutants formed plaques with halos, and the lysis defect could be reversed by the addition of chloroform to the infected cells (32). As the mutants were all leaky and showed enhanced plating on many of the complementation strains, the corresponding protein could not be identified (32, 35). Similarly, amber mutants deficient in putative holin function have been isolated in the closely related tectivirus PR4, but the corresponding protein was not definitively identified (49). A small membrane protein, P24, was proposed as the best candidate.
For phages without a membrane component, the identification of holin genes by sequence analysis is relatively straightforward, as usually only a small number of holin gene candidates have been observed. In the case of membrane-containing viruses, this approach may not be equally productive. The PRD1 genome contains 11 open reading frames (ORFs) without known functions, most of which are predicted to contain at least one transmembrane helix (7, 19). Based on primary sequence analysis, PRD1 holin was assigned to ORF m. Accordingly, when transmembrane channel-forming proteins were classified into families, the identity of the PRD1 holin m family was established (42). However, it has been shown that PRD1 ORF m corresponds to gene XVIII, encoding protein P18 (3, 19). Protein P18 is needed for infectivity and functions in phage DNA entry (1, 20, 32), making it improbable that P18 would be involved in membrane permeabilization. Cells infected with gene XVIII amber mutants are not defective for lysis.
As the original class A mutants deficient in holin function (32) were no longer viable and thus were no longer available for analysis, a new attempt to isolate and characterize PRD1 holin mutants was carried out.
MATERIALS AND METHODS
Bacteria, plasmids, and phages.
The bacterial strains, phages, and plasmids used in this study are listed in Tables 1 and 2. PRD1 insertional mutants 13272, 13273, 13284, 13290, 13300, and 13315 were obtained from Harri Savilahti, Institute of Biotechnology, University of Helsinki, Helsinki, Finland. The insertional mutants were generated by Mu in vitro transposition technology in order to identify PRD1 genomic regions tolerant to transposition insertion and to recognize genes not obligatory for virus propagation. The exact sites of Mu insertion into ORF t in the PRD1 genome were revealed by sequencing (50). Cells were grown in Luria-Bertani (LB) medium (43), and when appropriate, ampicillin (150 μg/ml), chloramphenicol (25 μg/ml), kanamycin (25 μg/ml), and/or tetracycline (20 μg/ml) was added. Cell growth and lysis profiles were monitored by measuring culture turbidity with a Klett-Summerson colorimeter. Infections of S. enterica cells were performed with a multiplicity of infection (MOI) of 6 to 10. In the case of E. coli hosts, considerably higher MOIs of 20 to 120 were used, due to the less efficient binding of PRD1 particles to these cells (28).
TABLE 1.
Bacterial strains and bacteriophages used in this study
| Strain or phage | Genotype and/or relevant properties | Reference |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | dcm hsdS(rB− mB−) gal(λcIts857 ind1 Sam7 nin5 lac UV5-T7 gene 1); host strain for protein production from pET32a expression vector | 48 |
| HMS174 | Host for gene cloning and complementation analysis | 15 |
| S. enterica serovar Typhimurium LT2 strains | ||
| DS88 | ΔH2, H1-i::Tn10 (Tcs) (pLM2); standard nonsuppressor host for PRD1 | 6 |
| DB7154(pLM2) | DB7100 leuA414(Am) hisC527(Am) supD10; suppressor host | 52 |
| DB7156(pLM2) | DB7100 leuA414(Am) hisC527(Am) supF30 suppressor host | 52 |
| PSA(pLM2) | supE; suppressor host | 34 |
| Bacteriophages | ||
| PRD1 | ||
| wt | wt | 38 |
| H1 | nta 13224 G→T; Asp81(GAT)→Tyr(TAT) in ORF t | This study |
| H3 | nt 13214 G→T; Trp77(TGG)→Cys(TGT) in ORF t | This study |
| H4 | nt 13214 G→T; Trp77(TGG)→Cys(TGT) in ORF t | This study |
| H9 | nt 12976 G→A in ORF t RBS | This study |
| sus711 | nt 13183 G→A; Trp67(TGG)→Am(TAG) in ORF t | This study |
| sus712 | nt 13183 G→A; Trp67(TGG)→Am(TAG) in ORF t | This study |
| sus714 | nt 13183 G→A; Trp67(TGG)→Am(TAG) in ORF t | This study |
| sus715 | nt 13006 G→A; Trp8(TGG)→Am(TAG) in ORF t | This study |
| sus716 | nt 13006 G→A; Trp8(TGG)→Am(TAG) in ORF t | This study |
| sus718 | nt 13006 G→A; Trp8(TGG)→Am(TAG) in ORF t | This study |
| 13272 | Insertion of mini-Mu transposon into PRD1 genome replaces residues 99-117 of gp t with amino acid sequence EAAHEKRESVSR | 50 |
| 13273 | Insertion of mini-Mu transposon creates a new stop codon following nt 13273; gp t is truncated to 98 aab | 50 |
| 13284 | Insertion of mini-Mu transposon into PRD1 genome replaces residues 102-117 of gp t with amino acid sequence EAAHEKRESVSR | 50 |
| 13290 | Insertion of mini-Mu transposon into PRD1 genome replaces residues 105-117 of gp t with amino acid sequence EAAHEKRESVSR | 50 |
| 13300 | Insertion of mini-Mu transposon creates a new stop codon following nt 13300; gp t is truncated to 107 aa | 50 |
| 13315 | Insertion of mini-Mu transposon creates a new stop codon following nt 13315; gp t is truncated to 112 aa | 50 |
Nucleotides (nt) refer to positions in the recently updated PRD1 genomic sequence (GenBank accession no. M69077).
aa, amino acids.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Replicon | Selective marker | Reference |
|---|---|---|---|---|
| pET32a | High-level expression vector | pBR322 | Apr | 30 |
| pJB15 | Encodes PRD1 receptor | IncPα | Tcr | 24 |
| pLM2 | Encodes PRD1 receptor | IncPα | Kmr | 34 |
| pLysS | Produces small amount of phage T7 lysozyme that reduces the basal expression of genes cloned under the T7 promoter of pET32a vector | p15a | Cmr | 47 |
| pPR26 | PSU18 + nta 12984-13337 from PRD1 genome (ORF t) | p15a | Cmr | This study |
| pPR60 | PET32a + late promoter pR′ and gene S105 from lambda genome | pBR322 | Apr | This study |
| pPR61 | PET32a + nt 12984-13337 from PRD1 genome (ORF t) | pBR322 | Apr | This study |
| pPR62 | PSU18 + nt 4907-5294 from PRD1 sus715 genome (gene XXXI and ORF d) | p15a | Cmr | This study |
| pPR63 | PSU18 + nt 9427-9681 from PRD1 sus715 genome (ORF h) | p15a | Cmr | This study |
| pPR64 | PSU18 + nt 8332-8460 from PRD1 sus715 genome (ORF i) | p15a | Cmr | This study |
| pPR65 | PSU18 + nt 10044-10166 from PRD1 sus715 genome (ORF l) | p15a | Cmr | This study |
| pPR66 | PSU18 + nt 11090-11200 from PRD1 sus715 genome (ORF q) | p15a | Cmr | This study |
| pPR67 | PSU18 + nt 2415-3587 from PRD1 sus715 genome (ORF a, gene XV, ORF b) | p15a | Cmr | This study |
| pPR68 | PSU18 + nt 10617-10823 from PRD1 sus715 genome (ORF o) | p15a | Cmr | This study |
| pPR69 | PSU18 + nt 12984-13888 from PRD1 sus715 genome (ORFs t, u, v) | p15a | Cmr | This study |
| pPR70 | PSU18 + nt 12984-13337 from PRD1 sus715 genome (ORF t) | p15a | Cmr | This study |
| pPR71 | PSU18 + nt 12984-13337 from PRD1 sus712 genome (ORF t) | p15a | Cmr | This study |
| pPR72 | PSU19 + nt 12984-13337 from PRD1 sus711 genome (ORF t) | p15a | Cmr | This study |
| pPR73 | PSU19 + nt 12984-13337 from PRD1 sus714 genome (ORF t) | p15a | Cmr | This study |
| pPR74 | PSU19 + nt 12984-13337 from PRD1 sus716 genome (ORF t) | p15a | Cmr | This study |
| pPR75 | PSU18 + nt 12984-13337 from PRD1 sus718 genome (ORF t) | p15a | Cmr | This study |
| pPR76 | PSU18 + nt 12535-13692 from PRD1 H1 genome (ORF t + flanking sequences) | p15a | Cmr | This study |
| pPR77 | PSU18 + nt 12984-13337 from PRD1 H3 genome (ORF t) | p15a | Cmr | This study |
| pPR78 | PSU18 + nt 12535-13692 from PRD1 H3 genome (ORF t+ flanking sequences) | p15a | Cmr | This study |
| pPR79 | PSU19 + nt 12984-13337 from PRD1 H4 genome (ORF t) | p15a | Cmr | This study |
| pPR80 | PSU19 + nt 12535-13692 from PRD1 H4 genome (ORF t+ flanking sequences) | p15a | Cmr | This study |
| pPR81 | PSU18 + nt 12984-13337 from PRD1 H9 genome (ORF t) | p15a | Cmr | This study |
| pPR82 | PSU18 + nt 12535-13692 from PRD1 H9 genome (ORF t+ flanking sequences) | p15a | Cmr | This study |
| pS105 | Contains lambda late promoter pR′ and the lysis cassette, genes S105, R, Rz, and Rzl | pBR322 | Apr | 45 |
| pSU18/19b | Low-copy-number cloning vector | p15a | Cmr | 9 |
| pQ | Lambda gene Q cloned in pZS∗24 vector, containing the lac/ara hybrid promoter inducible by IPTG and arabinose | pSC101* | Kmr | 23 |
Nucleotides (nt) refer to positions in the recently updated PRD1 genomic sequence (GenBank accession no. M69077).
The only difference between vectors pSU18 and pSU19 is the opposite orientation of the polycloning site. Genes cloned into vector pSU18 can be expressed from the lac promoter. In the cloning of PRD1 ORFs, pSU19 was employed in those cases in which no correct constructs were obtained with pSU18.
For the production of wild-type (wt) and sus712 and sus715 mutant particles, DS88 cells were infected at an MOI of 6. Incubation at 37°C was continued until lysis occurred or, in the case of sus712 and sus715 mutants, for 2 h. Cells infected with mutant phages were collected by centrifugation (Sorvall GS3 rotor, 2,440 × g, 8 min, 4°C), resuspended in 20 mM K-phosphate buffer (pH 7.2) (1/100 of the original volume), and disrupted by a passage through a 0.375-in. French pressure cell (400 lb/in2). The original culture volume was restored with the same buffer, and the lysate was cleared by centrifugation (Sorvall GS3 rotor, 10,800 × g, 20 min, 4°C). Viruses were precipitated from lysates with polyethylene glycol 6000, purified in 5 to 20% rate zonal sucrose gradients followed by 20 to 70% sucrose equilibrium gradients, and collected by differential centrifugation as previously described (6).
DNA manipulations.
Plasmids pPR26 and pPR62 to pPR82 were constructed by amplifying the PRD1 genomic regions specified in Table 2 by PCR and inserting them into a low-copy-number plasmid, pSU18/19. In the case of pPR61, wt ORF t was transferred from pPR26 into the BamHI-HindIII site of pET32a for high-level protein expression. Plasmid pPR60 was constructed by transferring a 1,685-bp EcoRV-HindIII fragment containing the bacteriophage lambda late promoter pR′ and gene S105 from pS105 (45) into the pET32a vector. All cloning procedures were performed by using standard molecular techniques (43).
Isolation and mapping of phage mutants.
For the isolation of holin mutants, wt PRD1 phages were plated on DS88 host cells in soft LB agar supplemented with ethidium bromide (15 μg/ml). After overnight incubation at 37°C, the soft agar was removed from the plates and transferred to 500-ml flasks. Three milliliters of LB medium/plate was added, and incubation was continued for 2 h at 37°C. Debris was removed by centrifugation (Sorvall SS34 rotor, 7,600 × g, 20 min, 4°C), and the supernatant was collected. In order to enrich for holin mutant phages, DS88 cells were infected with the resulting phage stock as follows. An overnight culture of DS88 cells was diluted 50-fold into 50 ml of LB broth and incubated at 37°C until the cell density was approximately 109 CFU/ml (170 Klett units). Cells were infected at an MOI of 0.5, and incubation was continued for 2 h with aeration. The culture was transferred to ice and incubated for 1 h to allow complete lysis of cells infected with wt PRD1. The remaining (nonlysed) cells were collected by centrifugation (Sorvall GSA rotor, 4,060 × g, 10 min, 4°C), resuspended in 10 ml of 20 mM K-phosphate buffer (pH 7.2), and transferred into 14-ml polypropylene tubes (Falcon). One milliliter of chloroform (CHCl3) was added, and the suspension was vortexed vigorously for 1 min. The aqueous and organic phases were allowed to separate, and the aqueous phase was collected and cleared by centrifugation (Sorvall SS34 rotor, 14,500 × g, 20 min, 4°C). The amount of infectious virions in the resulting specimen was determined and used in the following infections. After the enrichment procedure was repeated five times, mutant viruses were isolated either based on their unusual plaque morphology or, in the case of amber mutants, by selecting them on suppressor and nonsuppressor host lawns as described previously (32). Each mutant was purified by three consecutive rounds of single-plaque isolations before further analysis.
The mapping of mutations was carried out by sequencing. Several ORFs (putative holin genes [Table 2]) from the PRD1 amber mutant sus715 genome were cloned into pSU18/19 vectors and used as templates for sequencing reactions to identify the TAG amber codon. In addition, fragments containing nucleotides 12535 to 13692 from the genomes of PRD1 mutants H1, H3, H4, and H9 and nucleotides 12984 to 13337 from the genomes of PRD1 amber mutants sus711, sus712, sus714, sus716, and sus718 were cloned and sequenced. All sequencing reactions were performed with an automated sequencer (Perkin-Elmer ABI Prism 377XL) at the DNA Synthesis and Sequencing Laboratory, Institute of Biotechnology, University of Helsinki.
Electron microscopy.
For thin-section electron microscopy, S. enterica DS88 cells were grown to a density of 109 CFU/ml and infected with H1, H9, sus712, or sus715 phage stock at an MOI of 10. Samples were taken at 100 min (H1) or 180 min (H9, sus712, and sus715) postinfection (p.i.) and fixed with 3% (vol/vol) glutaraldehyde in 20 mM K-phosphate buffer (pH 7.2). After a 20-min incubation at room temperature, cells were washed twice and prepared for transmission electron microscopy as described previously (4). Micrographs were taken with a JEOL 1200 EX electron microscope operating at 60 kV (Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki).
Analytical methods.
Protein concentrations were determined by Coomassie brilliant blue staining with bovine serum albumin as a standard (12). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described previously (37). The nucleotide sequence of ORF t was analyzed for helical transmembrane domains by using the prediction programs TMPRED (26) (http://www.ch.embnet.org) and TMHMM version 2.0 (29, 46) (http://www.cbs.dtu.dk).
RESULTS
Bacteriophage PRD1 uses a two-component lysis system that is dependent on the host energy state.
The addition of energy poisons like cyanide or dinitrophenol to infected cells triggers premature lysis in phage lambda-infected cells, as well as in other phage infections that employ a holin-dependent lysis system. A prerequisite for premature lysis is that an adequate amount of phage endolysin has accumulated in the cytosol (17, 51). In the case of thermally induced λ cIts lysogens, an acute lysis event can be induced by cyanide addition at 30 min p.i., while normally lysis starts approximately 40 to 50 min p.i. (51, 53).
A holin-endolysin system has been proposed to operate in bacteriophage PRD1 infections (19, 32). We tested the dependence of the PRD1 lysis system on the host cell energy state by treating infected Salmonella cells with cyanide (KCN). In order to detect the presence of PRD1 endolysin (protein P15) in infected cells, corresponding samples were also treated with chloroform. The results are shown in Fig. 1. Cells infected with wt PRD1 enter the lysis phase at approximately 45 to 50 min p.i. Premature lysis could be triggered by cyanide addition at 35 min p.i., and when added just before the start of normal lysis, the lysis of cyanide-induced infected cells ensued instantaneously. The addition of cyanide at 25 min p.i. did not cause a decrease in culture turbidity even though the cells could be induced to lyse by chloroform treatment at the same time point (Fig. 1b). These results indicate that a two-component lysis system, dependent on the energy state of the host cell, is functioning in PRD1-infected cells.
FIG. 1.
(a) Effect of cyanide addition to PRD1-infected and noninfected DS88 cells. At various time points, 5 mM KCN was added to the cultures. Symbols: •, DS88 cells, no KCN added; ▾, DS88 cells, KCN added at 35 min p.i.; ○, PRD1-infected cells, no KCN added; ▿, PRD1-infected cells, KCN added at 25 min p.i.; ▪, PRD1-infected cells, KCN added at 35 min p.i.; □, PRD1-infected cells, KCN added 45 min p.i. (b) Chloroform treatment of PRD1-infected DS88 cells at different times after infection. •, no CHCl3 added; ○, CHCl3 added at 5 min p.i.; ▾, CHCl3 added at 15 min p.i.; ▿, CHCl3 added at 25 min p.i.; ▪, CHCl3 added at 35 min p.i.; □, CHCl3 added at 45 min p.i.
Isolation of PRD1 holin mutants. (i) Isolation of lysis-deficient mutants.
The established procedure to isolate phage mutants with holin defects is to infect cells at a low MOI and collect the cells that fail to undergo lysis (53). Membranes of these cells are then chemically permeabilized by chloroform, allowing the phage endolysin to escape from the cytosol and reach the host cell peptidoglycan layer. As a consequence, cells infected with holin-defective phages lyse. As holins are not expected to interfere with particle formation, liberated holin mutant phages should be infectious and can be enriched by repeating the procedure.
PRD1 virions contain an internal membrane component, and as a result, CHCl3 addition can be expected to have some effect on phage viability. Treatment with 10% CHCl3 caused an ∼85% reduction in the titer of wt PRD1 stock (from 1.8 × 1012 PFU/ml to 3.0 × 1011 and 2.4 × 1011 PFU/ml after 30 and 60 s of treatment, respectively). The remaining 15% infectivity was considered sufficient for mutant isolation. After the enrichment procedure was repeated twice, plaques with unusual morphology appeared, and after five cycles, their proportion had increased to approximately 10% of all plaques. Two different types of unusual plaque morphologies were detected: smaller-than-wt plaques (diameter of about 1 mm) and wt-sized plaques (diameter of about 4 mm), both surrounded with turbid halos. Five plaques of each morphology group were isolated (H1 to H10), and four of the isolates (H1, H3, H4, and H9) then were chosen for further studies. Of these, H1 and H9 produced small plaques and H3 and H4 produced wt-sized plaques. For isolation of amber mutants, 900 plaques were picked from amber suppressor host lawns and plated on nonsuppressor host lawns. Among these, six amber mutants were recovered (sus711, sus712, sus714, sus715, sus716, and sus718).
(ii) Isolated amber mutants have defective holin function.
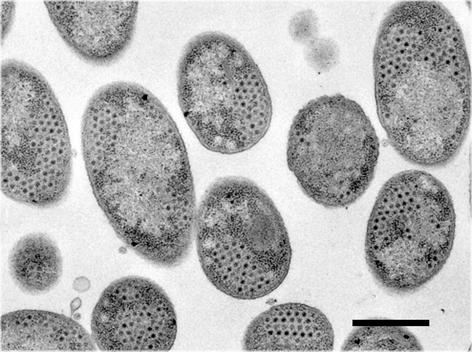
When propagated on nonsuppressor DS88 cells, amber mutants sus711, sus712, sus714, sus715, sus716, and sus718 did not result in lysis even if incubation was continued for 360 min p.i. Instead, the turbidity of the mutant-infected cell cultures increased similarly to that of the noninfected control culture. Thin-section electron microscopy showed that the cytoplasm of mutant-infected DS88 cells contained a large number of DNA-filled PRD1 particles (Fig. 2), confirming that particle assembly was not affected. In all cases, CHCl3 addition to mutant-infected cells at 40 min p.i. caused immediate lysis. To confirm the absence of holin function, we tested the ability of phage λ holin (S105 protein) to complement the lysis defect in sus712 and sus715mutants. As shown for sus715 in Fig. 3a, induction of S105 production from pPR60 led to lysis of mutant-infected cells. Similar results were obtained with sus712, indicating that the missing activity in sus712 and sus715 mutant infections can be restored by λ S105 protein and that PRD1 endolysin can cross membranes permeabilized by λ S105.
FIG. 2.
Electron micrograph of thin-sectioned DS88 cells infected with the sus715 mutant and harvested at 180 min p.i. The cells are filled with DNA-containing PRD1 particles that have accumulated in the cytosol during the prolonged infection cycle. Similar results were obtained with thin-sectioned DS88 cells infected with sus712, H1, or H9 mutant phages. Bar, 500 nm.
FIG. 3.
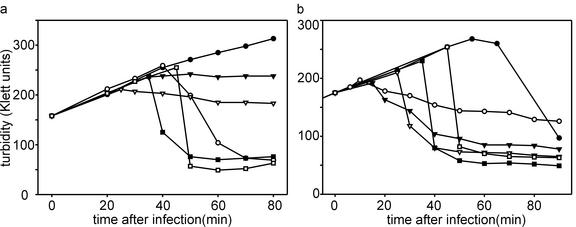
(a) Complementation of the PRD1 sus715 mutant with bacteriophage lambda gene S105. E. coli HMS174 cells carrying pPR60 encoding the λ late promoter and holin gene S105 were infected with wt PRD1 and sus715 mutant phages at an MOI of 20. At 120 min after infection, S105 protein production was initiated by inducing λ Q protein production from plasmid pQ with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 0.4% l-arabinose as described previously (23). The time of induction is marked by an arrow. Symbols: •, HMS174(pPR60)(pQ)(pJB15) control cells, no induction; ○, HMS174(pPR60)(pQ)(pJB15) cells, induction at 120 min p.i.; ▾, HMS174(pPR60)(pQ)(pJB15) cells infected with wt PRD1, no induction; ▿, cells infected with sus715 mutant, no induction; ▪, cells infected with sus715 mutant, induction at 120 min p.i. (b) Growth of E. coli cells expressing protein P35. Symbols: •, BL21(DE3)(pPR61) cells, no induction; ○, BL21(DE3)(pPR61) cells induced with 1 mM IPTG; ▾, BL21(DE3)(pLysS)(pPR61) cells, no induction; ▿, BL21(DE3)(pLysS)(pPR61) cells induced with 1 mM IPTG. (c) Complementation of sus715 and H9 mutants with PRD1 gene XXXV. BL21(DE3)(pPR61)(pLM2) cells containing gene XXXV cloned in vector pET32a were infected with wt or mutant PRD1 at an MOI of 120. When the growth of wt-infected cell stopped at 100 min p.i., P35 production was induced by adding 1 mM IPTG (arrow). Symbols: •, BL21(DE3)(pPR61)(pLM2) cells infected with wt PRD1, no induction; ○, cells infected with sus715 mutant, no induction; ▿, cells infected with H9 mutant, no induction; ▾, cells infected with sus715, induction at 100 min p.i; ▪, cells infected with H9 mutant, induction at 100 min p.i.
To examine the protein composition of mutant virions, sus712 and sus715 particles were purified from infected nonsuppressor DS88 host cells as described in Materials and Methods. When the mutant particles were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, no loss of or altered protein mobility was detected compared to wt particles. The specific infectivities of the purified sus712 and sus715 particles on suppressor hosts (1.6 × 1010 and 2.0 × 1010 PFU/μg of protein, respectively) were similar to that of wt PRD1 (1.9 × 1010 PFU/μg of protein). Based on these results, we conclude that the amber mutations sus712 and sus715 do not affect particle assembly or infectivity but are targeted to a nonstructural protein(s) functioning in membrane permeabilization.
PRD1 gene XXXV encodes holin protein P35. (i) Identification of ORF t as the PRD1 holin gene.
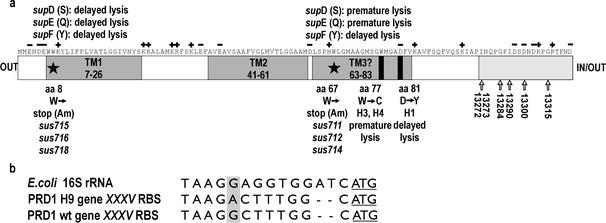
Similar to the case for previously isolated complementation group A mutants (32, 35), the sus711 to sus718 mutants were leaky and could not be mapped by complementation analysis. As most of the PRD1 genes can now be assigned to a known function (5, 19), the number of potential holin genes could be restricted to ∼10. The small number of candidate genes allowed an alternative approach, mapping by gene sequencing. The holin gene candidates from the sus715 amber mutant were cloned into pSU18/19 and sequenced. The amber codon TAG was found in only one of the sequenced ORFs, ORF t. This result was verified by cloning and sequencing ORF t from all of the other isolated mutants. In the case of nonamber mutants with altered plaque morphology, a fragment that contained an additional 449 bp upstream and 355 bp downstream of ORF t was sequenced to reveal possible mutations in nearby noncoding regions. As shown in Table 1 and Fig. 4, all mutants contained single base changes that were observed either in the coding region of ORF t or in its putative ribosome binding site (RBS) (7). Two different mutations were observed among the amber mutants. In sus711, sus712, and sus714, a transition from G to A changed the codon TGG, encoding Trp67, to the amber stop codon TAG. In mutants sus715, sus716, and sus718, a similar TGG→TGA transition occurred at codon 8. In the H1 mutant, codon 81 GAT (Asp) was changed to TAT (Tyr), while in mutants H3 and H4, codon 77 TGG (Trp) was replaced by TGT (Cys). In the case of the H9 mutant, the coding sequence of ORF t was identical to that of wt PRD1, and in addition, a G-to-A change at position 12976 that reduces the complementarity of the ORF t RBS to the host 16S rRNA was detected (Fig. 4b).
FIG. 4.
(a) Amino acid sequence and locations of predicted α-helical transmembrane domains in P35. Positions of charged amino acids in the wt sequence are indicated by + and −. Potential membrane-spanning helices TM1 to TM3 detected by the TMPRED prediction program are highlighted in grey. The end positions of amber fragments in sus712 and sus715 mutants (66 or 7 amino acids, respectively) are indicated by stars, and lysis phenotypes observed in suppressor hosts (with different amino acids inserted) are shown at the top. Amino acid substitutions detected in missense mutants H1, H3, and H4 at positions 81 and 77, respectively, are marked by rectangles. The insertion sites of mini-Mu transposons in the 13272, 13273, 13284, 13290, 13300, and 13315 mutants are indicated by arrows (50). aa, amino acid. (b) Shine-Dalgarno sequences of gene XXXV in genomes of wt PRD1 and the H9 mutant. For comparison, the nucleotide sequence complementary to E. coli 16S rRNA is shown. A G→A change in the H9 RBS is highlighted in grey. Translational start codons (ATG) are underlined.
We propose, based on the results presented here, that ORF t corresponds to the complementation group A identified in earlier studies (32) and is associated with holin activity. According to PRD1 nomenclature, ORFs that have been confirmed to encode protein products are designated with Roman numerals (5, 7, 32). Thus, we assign ORF t to gene XXXV encoding protein P35.
(ii) Protein P35 overproduction is lethal to E. coli.
For overexpression purposes, gene XXXV was inserted downstream of the T7 promoter of pET32a, resulting in the construct pPR61. When the production of P35 was induced in BL21(DE3)(pPR61) cells, culture turbidity ceased to increase at approximately 30 min after induction (Fig. 3b). The determination of viable cell counts showed that by 2 h following induction, the amount of plasmid-containing cells decreased from 1.2 × 108 to 7.3 × 104 CFU/ml (0.06% of plasmid-containing cells remained viable).
We also tested the effects of the coexpression of PRD1 protein P35 and bacteriophage T7 endolysin. Phage T7 endolysin is a natural inhibitor of T7 RNA polymerase (36) that can allow genes with relatively toxic products to be established in the same cell under the control of T7 promoter (in this case gene XXXV in pPR61 plasmid). T7 endolysin was expressed from plasmid pLysS, which provides only low levels of the protein. This is due to the fact that in pLysS, endolysin gene 3.5 is orientated in the opposite direction as the tet promoter (47). In the absence of membrane permeabilization, the T7 endolysin has little effect on cell integrity and growth. Instead, 60 min after induction, cultures of BL21(DE3)(pLysS)(pPR61) cells coexpressing PRD1 protein P35 and T7 endolysin showed signs of lysis (Fig. 3b). No decrease in culture turbidity was detected when BL21(DE3)(pLysS)(pET32a) control cells were induced. The lysis of induced BL21(DE3)(pLysS)(pPR61) cells indicates that phage T7 lysozyme can cross the host cytoplasmic membrane via the putative membrane lesion caused by P35.
Despite the lack of complementation with gene XXXV in plating assays, the lysis defect exhibited by sus712 and sus715 mutant-infected BL21(DE3)(pPR61)(pLM2) cells could be corrected by inducing P35 expression after infection, as is shown for sus715 in Fig. 3c.
Alterations in protein P35 cause variation in the rate of lysis.
The potential effects of nucleotide substitutions on gene XXXV function were probed by infecting DS88 cells with H1, H3, and H9 missense mutant particles. Mutant phages sus712 and sus715 were analyzed with three different amber suppressor hosts, resulting in a variety of amino acid substitutions at codons 8 and 67 (Table 1; Fig. 4). In addition, six PRD1 insertion mutants (13272, 13273, 13284, 13290, 13300, and 13315) (50) generated with mini-Mu in vitro transposition technology were analyzed. These mutations cause truncations and/or small insertions in the C-terminal part of protein P35, some of which affect the total charge of the terminus (Table 1; Fig. 4). Similar to the case for H1 and H9 mutants, the insertion mutants produced smaller-than-wt plaques on standard DS88 host cells.
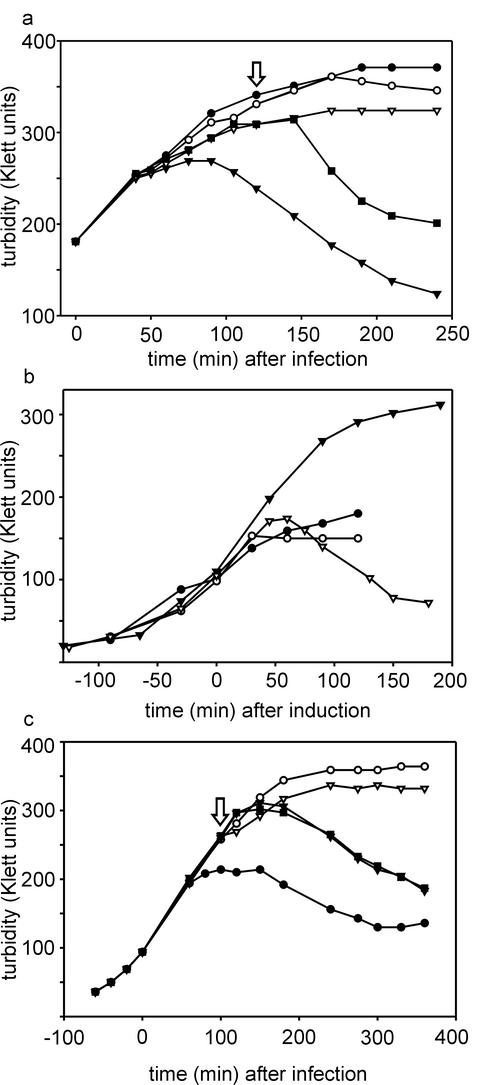
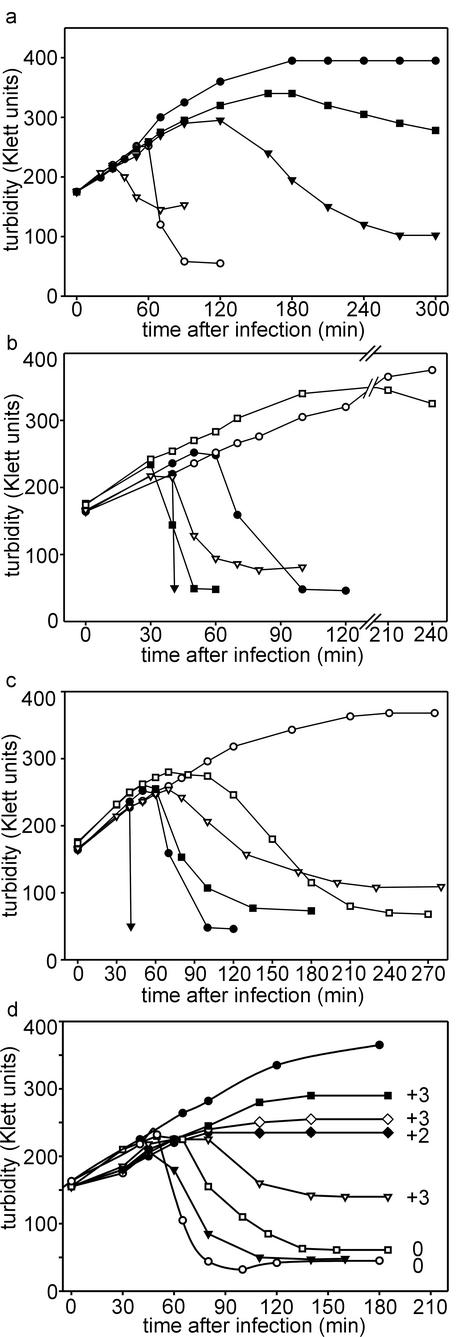
When the lysis curves of mutant-infected cells were determined, two different lysis phenotypes were detected (Fig. 5). Premature lysis was seen in H3 mutant infections as well as in sus712-infected DB7154(pLM2) and PSA(pLM2) suppressor hosts. In all other cases lysis was delayed.
FIG. 5.
(a) Lysis profiles of wt PRD1- and H1, H3, and H9 mutant-infected DS88 cells. Symbols: •, DS88 cells with no added phage; ○, PRD1 wt-infected cells; ▾, H1 mutant-infected cells; ▿, H3 mutant-infected cells; ▪, H9 mutant-infected cells. (b) Lysis profiles of PRD1 sus712 mutant with nonsuppressor and different suppressor hosts. Symbols: ○, DS88 nonsuppressor host; ▾, DS88 host, chloroform added at 40 min p.i.; ▿, PSA(pLM2) supE host; ▪, DB7154(pLM2) supD host; □, DB7156(pLM2) supF host; •, control DS88 cells infected with wt PRD1. (c) Lysis profiles of PRD1 sus715 mutant with nonsuppressor and suppressor hosts. Symbols: ○, DS88 nonsuppressor host; ▾, DS88 host, chloroform added at 40 min p.i.; ▿, PSA(pLM2) supE host; ▪, DB7154(pLM2) supD host; □, DB7156(pLM2) supF host; •, control DS88 cells infected with wt PRD1. (d) Lysis profiles of PRD1 insertion mutants 13272 (▪), 13273 (⧫), 13284 (▿), 13290 (◊), 13300 (□), and 13315 (▾) with DS88 host. •, noninfected control cells; ○, cells infected with wt PRD1. Numbers following the lysis curves indicate the change in the total charge of the P35 C-terminal domain caused by insertions and truncations compared to wt P35.
The titers of lysates obtained from H3 and H4 infections (2.6 × 1010 and 2.2 × 1010 PFU/ml, respectively) associated with the premature lysis phenotype were approximately 1 log unit lower than those of wt or H1 and H9 mutant lysates (2.5 × 1011, 6.4 × 1011, and 2.6 × 1011 PFU/ml, respectively). This shows that approximately 90% of PRD1 progeny particles are assembled in the last 10 to 15 min before lysis, which is in good agreement with results from earlier electron microscopy studies that approximated the time needed to produce visible DNA-containing virus particles as 40 min (3, 31, 33).
DISCUSSION
In this study, the bacteriophage PRD1 holin gene was identified and cloned, and mutant viruses deficient in holin function were isolated. It was demonstrated that the expression of gene XXXV and its protein product, P35, were essential for the membrane permeabilization step in host cell lysis. Many characteristics typical of holins are associated with PRD1 protein P35, a small (12.8-kDa) protein with two or three putative α-helical transmembrane domains and a hydrophilic C terminus. P35 expression is lethal to E. coli cells but does not cause lysis unless a peptidoglycan-degrading enzyme is coexpressed with it. In addition to PRD1 endolysin (1,4-β-N-acetylmuramidase protein P15), P35 is also compatible with bacteriophage T7 lysozyme (gene 3.5 amidase) (25). In the absence of P35 activity, the growth of PRD1-infected cells continues for several hours after infection, but immediate lysis can be induced with chloroform.
The lysis of induced BL21(DE3)(pLysS)(pPR61) cells caused by the combined synthesis of T7 endolysin and PRD1 protein P35 is not as immediate as the lysis of Salmonella cells infected with wt PRD1. This is probably due to the low levels of T7 lysozyme provided by plasmid pLysS. However, the amount of T7 endolysin produced seems to be sufficient to cause the difference in the P35 holin triggering times detected between induced BL21(DE3)(pPR61) cells (holin function detected at ∼30 min after induction) and BL21(DE3)(pLysS)(pPR61) cells (detected at ∼60 min after induction) (Fig. 3b). Higher levels of T7 endolysin provided by plasmids pLysE and pLysH are known to both increase the lag time and reduce the maximum level of expression of target recombinant genes cloned under the T7 promoter due to the inhibition of T7 RNA polymerase by T7 endolysin. Unlike the holin genes of lambdoid phages, which encode both the effector and inhibitor forms of holin protein (8, 11, 51), PRD1 gene XXXV lacks the dual start motif. In addition, its position in the PRD1 genome differs from those of most other phage holin genes, which tend to be located in the immediate vicinity of the endolysin gene. In lambdoid phages, both the holin and endolysin genes are included in a lysis cassette that is under the control of a late promoter (51, 53). In bacteriophage PRD1, the endolysin gene XV and holin gene XXXV are located at the opposite ends of the linear 15-kb genome. Gene XV is transcribed from an early promoter, PE131, while the holin gene XXXV is located in the late operon OL3, which contains the genes encoding viral structural proteins (18, 19, 44). The latter parallels the T7 phage, where the endolysin and holin production are temporally separated (25). In PRD1, the OL3 genes are expressed from a double promoter, and the operon contains four potential transcription terminator hairpin loops (18). The transcription terminators regulate the relative levels of proteins produced from OL3 so that the major capsid protein is favored over all other gene products of the operon. As all four hairpin loops are located upstream of gene XXXV, we hypothesize that a relatively small amount of protein P35 is produced during infection. A simple putative mechanism for the regulation of PRD1 lysis initiation might involve slow accumulation of P35 to a critical level, triggering the subsequent lysis event. This theory also is supported by the fact that efficient lysis cannot be induced by cyanide addition until ∼35 min p.i., indicating the time point by which a sufficient amount of holin protein has accumulated in the cells.
The majority of holins can be divided in two classes according to the number of predicted α-helical transmembrane domains (54). Class I holins are 95 to 130 residues long and have three putative transmembrane domains. Class II holins are shorter (65 to 95 residues) and contain only two potential membrane-spanning α-helices (51, 54). The hydrophilic C-terminal domain has been located in cytosol in the prototype holins of classes I and II, i.e., the S proteins of λ and phage 21, respectively (22, 51). When PRD1 protein P35 was analyzed for α-helical transmembrane domains with the prediction programs TMHMM and TMPRED, two different models were obtained. Both programs detected membrane-spanning domains designated TM1 and TM2 (Fig. 4). In addition, the TMPRED program predicted the presence of a third transmembrane helix at residues 63 to 83 (TM3 in Fig. 4). Both programs assign TM1 an out-in topology and assign TM2 an in-out topology. Based on the cytosolic location of the C-terminal domains of holin proteins and the size of P35 (117 amino acids), the class I holin model (three membrane-spanning α-helices, periplasmic N terminus, and cytoplasmic C terminus) seems more likely. However, a thorough biochemical and genetic analysis of P35 will be necessary to define its membrane topology.
The C-terminal domains of holins are highly hydrophilic and contain clusters of charged residues, with a positive overall net charge (10). By constructing nonsense mutations of phage λ gene S, Bläsi and coworkers showed that as long as at least one basic residue is retained at the C terminus, truncated forms of S protein remained lysis proficient (10). The C-terminal domain of λ S was proposed to serve as a reservoir of charge involved in the regulation of initiation of lysis. The timing of lysis initiation loosely correlates with the number of positively charged residues at the C terminus, with an increasing positive charge correlating to a delay in the onset of lysis (10). The lysis curves of PRD1 mutants containing small insertions and truncations in the C-terminal part of P35 indicate the presence of a similar regulatory system in PRD1 (Fig. 5d). The positive charge increase in the C-terminal part of P35 was associated with a delay in the onset of lysis. Two of the mutants, 13300 and 13315, that encode truncated forms of P35 (107 and 112 residues, respectively) but retain the wt net charge in the C terminus display wt lysis initiation. In addition, in the H1 mutant, the replacement of Asp 81 with Tyr leads to an increase in the net charge of +1 for the C-terminal domain of protein P35. The lysis initiation in the H1 mutant was delayed, supporting a proposed role in regulatory function for the C terminus of protein P35.
The delay in lysis initiation detected in the H9 mutant infections is likely caused by decreased P35 production. The G-to-A transition detected in the Shine-Dalgarno sequence of gene XXXV makes it less complementary to the host 16S rRNA sequence and therefore probably hinders ribosome binding. In support of this, lysis initiation in H9 mutant-infected BL21(DE3)(pPR61)(pLM2) cells was accelerated by induction of P35 production from pPR61 (Fig. 3c).
A delay in lysis initiation was observed for the sus715 mutant, in which the amber mutation in tryptophan codon 8 was suppressed by insertion of serine, glutamine, or tyrosine residues, respectively, in the supD, supE, and supF hosts. Based on secondary-structure predictions, Trp 8 is located in the beginning of TM1 (residues 7 to 26). Replacement of a large hydrophobic residue with a hydrophilic one might impede the insertion of TM1 into the membrane and thus cause the observed lysis phenotype.
Here, we have assigned PRD1 holin function to P35, the protein product of gene XXXV. The PRD1 lysis system resembles those described for lambdoid phages, and the PRD1 holin defect can be overcome by the presence of lambda S protein. These observations support the idea that holins are nonspecific membrane gates that under appropriate conditions make the plasma membrane permeable to peptidoglycan-digesting enzymes, leading to cell lysis. Intriguing questions remain concerning the regulation of lysis initiation and the mechanisms of membrane permeabilization by holin proteins. We obtained initial information along these lines by studying altered PRD1 holins, and we are applying electrochemical methods to describe, in more detail, functional alterations of the membrane permeability caused by PRD1 holin and its derivatives.
Acknowledgments
We are grateful to Ry Young and Ing-Nang Wang at Texas A&M University for providing plasmids pS105 and pQ and to Harri Savilahti at the Institute of Biotechnology, University of Helsinki, for PRD1 insertion mutants 13272, 13273, 13284, 13290, 13300, and 13315. We warmly thank Sari Korhonen for her skillful technical assistance.
This work was supported by Finnish Academy research grants 164298 and 172621 (Finnish Centre of Excellence Programme [2000 to 2005]) and by EC project CEBIOLA (ICAI-CT-2000-70027).
REFERENCES
- 1.Bamford, D., and L. Mindich. 1982. Structure of the lipid-containing bacteriophage PRD1: disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J. Virol. 44:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamford, D. H., and H.-W. Ackermann. 2000. Family Tectiviridae, p. 111-116. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 3.Bamford, D. H., J. Caldentey, and J. K. H. Bamford. 1995. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281-319. [DOI] [PubMed] [Google Scholar]
- 4.Bamford, D. H., and L. Mindich. 1980. Electron microscopy of cells infected with nonsense mutants of bacteriophage phi6. Virology 710:222-228. [DOI] [PubMed] [Google Scholar]
- 5.Bamford, J. K., J. J. Cockburn, J. Diprose, J. M. Grimes, G. Sutton, D. I. Stuart, and D. H. Bamford. 2002. Diffraction quality crystals of PRD1, a 66-MDa dsDNA virus with an internal membrane. J. Struct. Biol. 139:103-112. [DOI] [PubMed] [Google Scholar]
- 6.Bamford, J. K. H., and D. H. Bamford. 1990. Capsomer proteins of bacteriophage PRD1, a bacterial virus with a membrane. Virology 177:445-451. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, J. K. H., A.-L. Hänninen, T. M. Pakula, P. M. Ojala, N. Kalkkinen, M. Frilander, and D. H. Bamford. 1991. Genome organization of membrane-containing bacteriophage PRD1. Virology 183:658-676. [DOI] [PubMed] [Google Scholar]
- 8.Barenboim, M., C.-Y. Chang, F. dib Hajj, and R. Young. 1999. Characterization of the dual start motif of a class II holin gene. Mol. Microbiol. 32:715-727. [DOI] [PubMed] [Google Scholar]
- 9.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 10.Bläsi, U., P. Fraisl, C.-Y. Chang, N. Zhang, and R. Young. 1999. The C-terminal sequence of the lambda holin constitutes a cytoplasmic regulatory domain. J. Bacteriol. 181:2922-2929.10217787 [Google Scholar]
- 11.Bläsi, U., and R. Young. 1996. Two beginnings for a single purpose: the dual start holins in the regulation of phage lysis. Mol. Microbiol. 21:675-682. [DOI] [PubMed] [Google Scholar]
- 12.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 13.Butcher, S. J., D. H. Bamford, and S. D. Fuller. 1995. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 14:6078-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldentey, J., A.-L. Hänninen, and D. H. Bamford. 1994. Gene XV of bacteriophage PRD1 encodes a lytic enzyme with muramidase activity. J. Eur. Biochem. 225:341-346. [DOI] [PubMed] [Google Scholar]
- 15.Campbell, J. L., C. C. Richardson, and F. W. Studier. 1978. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc. Natl. Acad. Sci. USA 75:2276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis, T. N., E. D. Muller, and J. E. Cronan, Jr. 1982. The virion of the lipid-containing bacteriophage PR4. Virology. 120:287-306. [DOI] [PubMed] [Google Scholar]
- 17.Garrett, J., and R. Young. 1982. Lethal action of the bacteriophage lambda S gene. J. Virol. 44:886-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grahn, A. M., J. K. H. Bamford, M. C. O'Neill, and D. H. Bamford. 1994. Functional organization of the bacteriophage PRD1 genome. J. Bacteriol. 176:3062-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grahn, A. M., S. J. Butcher, J. K. H. Bamford, and D. H. Bamford. PRD1—dissecting the genome, structure and entry. In R. Calendar (ed.), The bacteriophages, in press. Oxford Press, Oxford, United Kingdom.
- 20.Grahn, A. M., R. Daugelavicius, and D. H. Bamford. 2002. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 46:1199-1209. [DOI] [PubMed] [Google Scholar]
- 21.Graschopf, A., and U. Bläsi. 1999. Molecular function of the dual-start motif in the lambda S holin. Mol. Microbiol. 33:569-582. [DOI] [PubMed] [Google Scholar]
- 22.Gründling, A., U. Bläsi, and R. Young. 2000. Genetic and biochemical evidence for three transmembrane domains in the class I holin, lambda S. J. Biol. Chem. 275:769-776. [DOI] [PubMed] [Google Scholar]
- 23.Gründling, A., M. D. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. USA 98:9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hänninen, A.-L., D. H. Bamford, and J. K. H. Bamford. 1997. Probing the phage PRD1 specific proteins during infection by monoclonal and polyclonal antibodies. Virology 227:198-206. [DOI] [PubMed] [Google Scholar]
- 25.Hausmann, R. 1988. The T7 group, p. 259-290. In R. Calendar (ed.), The bacteriophages. Plenum Press, New York, N.Y.
- 26.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler. 374:166. [Google Scholar]
- 27.Johnson-Boaz, R., C.-Y. Chang, and R. Young. 1994. A dominant mutation in the bacteriophage lambda S gene causes premature lysis and an absolute defective plating phenotype. Mol. Microbiol. 13:495-504. [DOI] [PubMed] [Google Scholar]
- 28.Kotilainen, M. M., A. M. Grahn, J. K. H. Bamford, and D. H. Bamford. 1993. Binding of an Escherichia coli double-stranded DNA virus PRD1 to a receptor coded by an IncP-type plasmid. J. Bacteriol. 175:3089-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 30.LaVallie, E. R., E. A. DiBlasio, S. Kovacic, K. L. Grant, P. F. Schendel, and J. M. McCoy. 1993. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Bio/Technology 11:187-193. [DOI] [PubMed] [Google Scholar]
- 31.Lundström, K. H., D. H. Bamford, E. T. Palva, and K. Lounatmaa. 1979. Lipid-containing bacteriophage PR4: structure and life cycle. J. Gen. Virol. 43:538-592. [DOI] [PubMed] [Google Scholar]
- 32.Mindich, L., D. Bamford, C. Goldthwaite, M. Laverty, and G. Mackenzie. 1982. Isolation of nonsense mutants of lipid-containing bacteriophage PRD1. J. Virol. 44:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mindich, L., D. Bamford, T. McGraw, and G. Mackenzie. 1982. Assembly of bacteriophage PRD1: particle formation with wild-type and mutant viruses. J. Virol. 44:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mindich, L., J. Cohen, and M. Weisburd. 1976. Isolation of nonsense suppressor mutants in Pseudomonas. J. Bacteriol. 126:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mindich, L., and T. McGraw. 1983. Molecular cloning of bacteriophage PRD1 genomic fragments. Mol. Gen. Genet. 190:233-236. [DOI] [PubMed] [Google Scholar]
- 36.Moffatt, B. A., and F. W. Studier. 1987. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell 49:221-227. [DOI] [PubMed] [Google Scholar]
- 37.Olkkonen, V. M., and D. H. Bamford. 1989. Quantitation of the adsorption and penetration stages of bacteriophage phi6 infection. Virology 171:229-238. [DOI] [PubMed] [Google Scholar]
- 38.Olsen, R. H., J. Siak, and R. H. Gray. 1974. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J. Virol. 14:689-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raab, R., G. Neal, J. Garrett, R. Grimaila, R. Fusselman, and R. Young. 1986. Mutational analysis of bacteriophage lambda lysis gene S. J. Bacteriol. 167:1035-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramanculov, E., and R. Young. 2001. Functional analysis of the phage T4 holin in a lambda context. Mol. Gen. Genomics 265:345-353. [DOI] [PubMed] [Google Scholar]
- 41.Ramanculov, E., and R. Young. 2001. Genetic analysis of the T4 holin: timing and topology. Gene 265:25-36. [DOI] [PubMed] [Google Scholar]
- 42.Saier, M. H., Jr. 2000. Families of proteins forming transmembrane channels. J. Membr. Biol. 175:165-180. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Savilahti, H., and D. H. Bamford. 1987. The complete nucleotide sequence of the left very early region of Escherichia coli bacteriophage PRD1 coding for the terminal protein and DNA polymerase. Gene 57:121-130. [DOI] [PubMed] [Google Scholar]
- 45.Smith, D. L., D. K. Struck, J. M. Scholtz, and R. Young. 1998. Purification and biochemical characterization of the lambda holin. J. Bacteriol. 180:2531-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnhammer, E. L. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 47.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 48.Studier, F. W., and B. A. Moffat. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 49.Vanden Boom, T., and J. E. J. Cronan. 1990. Nonsense mutants defining seven new genes of the lipid-containing bacteriophage PR4. Virology 177:11-12. [DOI] [PubMed] [Google Scholar]
- 50.Vilen, H., J.-M. Aalto, A. Kassinen, L. Paulin, and H. Savilahti. 2003. A direct transposon insertion tool for modification and functional analysis of virus genomes. J. Virol. 77:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 52.Winston, F., D. Botstein, and J. H. Miller. 1979. Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 137:433-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, R., and U. Bläsi. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17:191-205. [DOI] [PubMed] [Google Scholar]