Abstract
In the dystrophin-mutant mdx mouse, an animal model for Duchenne muscular dystrophy (DMD), damaged skeletal muscles are efficiently regenerated and thus the animals thrive. The phenotypic differences between DMD patients and the mdx mice suggest the existence of factors that modulate the muscle wasting in the mdx mice. To identify these factors, we searched for mRNAs affected by the mdx mutation by using cDNA microarrays with newly established skeletal muscle cell lines from mdx and normal mice. We found that in the mdx muscle cell line, 12 genes, including l-arginine:glycine amidinotransferase and thymosin β4, are up-regulated, whereas 7 genes, including selenoprotein P and a novel regeneration-associated muscle protease (RAMP), are down-regulated. Northern blot analysis and in situ hybridization revealed that RAMP mRNA is predominantly expressed in normal skeletal muscle and brain, and its production is enhanced in the regenerating area of injured skeletal muscle in mice. RAMP expression was much lower in individual muscle cell lines derived from biopsies of six DMD patients compared to a normal muscle cell line. These results suggest that RAMP may play a role in the regeneration of skeletal muscle and that its down-regulation could be involved in the progression of DMD in humans.
Point mutations or out-of-frame deletions in the dystrophin gene on the X-chromosome are known to cause Duchenne muscular dystrophy (DMD).1 This disease occurs with a frequency of 1 of 3500 newborn males, which makes it the most common lethal myopathy. Dystrophin is a large membrane-associated protein that plays an important role in linking the intracellular cytoskeletal actin filaments to the sarcolemmal membrane.2 In addition, it forms a multicomponent complex denoted as dystrophin-associated protein complex, which contains dystroglycans, sarcoglycans, syntrophins, and nitric oxide synthase.3,4 Thus, dystrophin not only mechanically protects the sarcolemma from muscle contraction-induced tension,5 it also affects intracellular signaling pathways, particularly in the Ca2+-dependent enzymatic cascade.6
The mdx mutant mouse strain carries a nonsense mutation at position 3185 of the murine dystrophin gene.7,8 However, despite the lack of subsarcolemmal dystrophin protein in these mice, their skeletal muscle degeneration is less severe than it is in DMD patients as after an initial period of skeletal muscle necrosis at 3 to 4 weeks of age, regenerative activity in the mdx mice gradually compensates for the muscle damage in the hindlimb.9 As a result, the adult mdx mice show little functional disability. In contrast, in DMD patients, there is an imbalance between muscle degeneration and repair that leads to the loss of muscle fibers and increased fibrosis.10 Consistent with these observations is that recent DNA microarray analyses revealed that mRNAs encoding proteins related to the muscle regeneration process are more abundantly expressed in the skeletal muscle of mdx mice than in the skeletal muscle of normal control mice.10–13 Examples of these muscle-regenerating proteins are insulin-like growth factor-2, transforming growth factor β, procollagens, and osteopontin. The down-regulation of myostatin mRNA in the skeletal muscle of the mdx mouse is also related to its higher regeneration capacity.10,12 Intriguingly, recent reports demonstrated that transgenic overexpression of insulin-like growth factor-1 in muscle14 or administration of anti-myostatin neutralizing antibody15 blocked the degeneration and fibrosis in the diaphragm in mdx mice, suggesting that the enhancement of muscle regenerative capacity may be a promising therapeutic approach for DMD.
Although the extensive gene profiling of DMD patient biopsies versus normal muscle samples has provided many clues about the secondary loss of or changes in DMD muscle,16,17 it is difficult to be sure that net change observed in the gene expression of DMD muscle reflects an altered genetic program in the muscle cells because the necrotic DMD muscle areas are filled with many macrophages and other inflammatory immune cells. Intact muscle biopsies from young patients also contain many blood cells. Thus, it is difficult to be sure that a net change observed in the gene expression of DMD muscle reflects an altered genetic program in the muscle cells. To overcome this problem, we first immortalized skeletal muscle cells from mdx and control mice and compared their expression of several gene sets by using cDNA microarrays. We also established muscle cell lines from biopsies taken from DMD patients, Becker muscular dystrophy (BMD) patients, and an unaffected person to investigate the behavior of the genes whose expression patterns were found to be altered in the mdx muscle cell line. In this study, we report that, relative to the control murine muscle cell line, the transcription of 12 genes in the mdx muscle cell line is up-regulated while mRNA levels of 7 other genes is down-regulated. Among the down-regulated genes was a novel gene that we found encoded a secreted protease termed regeneration-associated muscle protease (RAMP). We found that RAMP mRNA levels are also often decreased in the muscle cell lines derived from the DMD and BMD patients.
Materials and Methods
Mice
Breeding pairs of C57BL/10 ScSn-Dmdmdx (mdx) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and propagated in a standard pathogen-free animal facility in the institute. C57BL/10 (B10) mice were purchased from Nihon SLC (Hamamatsu, Japan). All animal experiments were based on institutionally approved protocols.
Primary Culture of Skeletal Muscle and Immortalization
Ex vivo culture of skeletal muscle was done according to the published protocol with a slight modification.18 In, brief, the hindlimb muscles were taken from 2-month-old B10 and mdx mice, thoroughly minced into a coarse slurry, and enzymatically dissociated with 5% trypsin (Difco, Detroit, MI) in phosphate-buffered saline (PBS) at 37°C for 30 minutes. The dissociated muscle tissues were resuspended in Dulbecco’s modified Eagle’s minimum essential medium-high glucose type (Sigma, St. Louis, MO) supplemented with 20% fetal calf serum and 0.5% penicillin-streptomycin (Sigma), triturated by using a 10-ml pipette, and passed through a 100-μm nylon mesh. The cells were cultured in Dulbecco’s modified Eagle’s medium containing 20% fetal calf serum (JRH Biosciences, Lenexa, KS), 4% Ultroser G (Biosepra, Cergy-Saint-Christophe, France) and 0.5% penicillin-streptomycin in gelatin-coated flasks for 24 hours. The nonadherent cells were then transferred to new flasks for the subculture of the primary myoblastic cells. Two days later, half of the cells were infected for 1 day with a recombinant retrovirus producing a temperature-sensitive form of the simian virus 40 large T antigen (SV40 tsT)19 and then cultured at the permissive temperature of 32.5°C until continuously growing cells appeared. The retrovirus was produced as previously described by using the PLAT-E packaging cell line20 and the pMESVts retrovirus vector19 (a kind gift from Dr. Drinkwater, University of Wisconsin Medical School). After 1 month of culture, the Ultroser G was removed from the growth medium and myoblastic subclones were isolated by limiting dilution. For myotube formation, the culture medium was changed to Dulbecco’s modified Eagle’s medium supplemented with 5% horse serum (Invitrogen, Carlsbad, CA) followed by incubation at 39.5°C for 7 days.
Immunocytochemistry
Cells grown in a 4-well culture dish (Nunc, Roskilde, Denmark) were treated with 0.1% Triton X-100 (Wako, Osaka, Japan) in PBS for 1 minute and fixed in cold methanol for 2 minutes. After rinsing with PBS, cells were blocked with 2% bovine serum albumin (Sigma) in PBS for 1 hour and incubated with rabbit anti-desmin antibody (Progen, Heidelberg, Germany) (1:100 dilution) for 1 hour. Samples were washed with PBS and incubated with anti-rabbit immunoglobulin conjugated with horseradish peroxidase (Jackson Immunoresearch Laboratories, West Grove, PA) (1:250 dilution) for 1 hour. After rinsing with PBS three times, the peroxidase activity was visualized by incubation with 0.25% diaminobenzidine (Sigma) in PBS containing 0.075% H2O2 and 3.36 mmol/L NiCl2 (Nakalai, Kyoto, Japan).
cDNA Microarray Analysis
Poly(A)+ RNAs were isolated by using the FastTrack 2.0 (Invitrogen). According to the previously published method,21,22 mouse microarrays carrying ∼4000 different cDNAs derived from a mouse fetus at 17.5 days post coitum and an adult mouse brain were hybridized with Cy3- and Cy5-labled cDNA probes prepared from the mRNAs of the C57BL/10- and mdx-derived cell lines cultured in the growth media at 32.5°C. The fluorescent signals were quantified by ScanArray 4000 (GSI Lumonics, Moorpark, CA) and the data were analyzed by QuantArray software (GSI Lumonics).
Construction of Human Microdystrophin cDNA and Transfection
Construction of the human microdystrophin cDNA containing four rod repeats and three hinges was performed based on a published report.23 By using cDNA derived from human skeletal muscle cells (Cambrex, Baltimore, MD) as the template, two polymerase chain reactions (PCRs) were independently performed with two sets of primers, namely, HD1 (5′-CTCGAGATGCTTTGGTGGGAAGAAGT-3′) and HD2(5′-TCTTTCAAGGGTATCCACAGTAATCTGCCTCTTC-3′) orHD3 (5′-ATTACTGTGGATACCCTTGAAAGACTCCAGGAAC-3′) and HD4 (5′-GCGGCCGCCTACATTGTGTCCTCTCTCAT-3′). Subsequently, a mixture of the two PCR products was reamplified with the HD1 and HD4 primers to obtain a long PCR band of 4.6 kb pairs that was subsequently cloned into a pPCR-Script Amp SK(+) vector (Stratagene, La Jolla, CA). A XhoI/NotI fragment was inserted into the reverse tet-regulated retrovirus vector pLRT-X24 (a kind gift from Dr. Hagiwara, Tokyo Medical and Dental University, Tokyo, Japan). A subclone of the mdx-derived muscle cell line was transfected with the pLRT-microdystrophin vector by FuGENE 6 (Roche, Mannheim, Germany) and cultured in growth medium containing blasticidin S (Wako) at 5 μg/ml for 3 weeks to establish stable transfectants.
Western Blot Analysis
Total cell lysates were separated by electrophoresis on a 5 to 10% sodium dodecyl sulfate-polyacrylamide gel, transferred onto a polyvinylidene difluoride membrane (Millipore, Bedford, MA), and incubated with anti-dystrophin DYS2 monoclonal antibody (1:100 dilution; Novocastra, Newcastle, UK). Immunoreacting bands were visualized by using the ECL-Plus detection reagent (Amersham Pharmacia Biotech, Piscataway, NJ).
Northern Blot Analysis and Reverse Transcriptase (RT)-PCR
Poly(A)+ RNAs (2 μg) from various tissues of adult (12 to 16 weeks old) C57BL/6 mice were separated by electrophoresis on a 1% formaldehyde-agarose gel, transferred to a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech), and hybridized with [α-32P] dCTP-labeled probes prepared from each cDNA at 42°C overnight. To verify the amount of RNA loaded in each lane, the blot was rehybridized with a β-tubulin probe. After washing under the most stringent of conditions, the membrane was subjected to autoradiography. For RT-PCR analysis, total RNAs were prepared from skeletal muscle tissues and various muscle cell lines using TRIzol (Invitrogen). Five μg of the RNA from each sample was reverse-transcribed by using the superscript preamplification system for first strand cDNA synthesis and an oligo(dT) primer (Invitrogen). Part (1/125) of the cDNA mixture was subjected to a PCR reaction using 56°C as an annealing temperature, ExTaq DNA polymerase (Takara, Tokyo, Japan), and the specific primer sets listed in Table 1.
Table 1.
Summary of the Expression Pattern and Primer Sequences of the Genes that Are Up-Regulated and Down-Regulated in Mdx Mice
| Name of gene/protein | Accession no. | Mdx/B10 cell line* | Mdx/B10 muscle† | Patient/normal human cell | Sense primer sequence (5′–3′) | Anti-sense primer sequence (5′–3′) | PCR band size (bp) |
|---|---|---|---|---|---|---|---|
| 1 SCHIP-1 | NM_01392 | ↑ | → | → | GTCTATCAGACAGAAGTTGGC | GAAGATCAGCGACGGGAGAC | 415 |
| 2 OX-2 | AF004023 | ↑ | N.T. | N.T. | |||
| 3 Arg/Gly amidinotransferase | U07971 | ↑ | ↑ | ↑ | GGAAGTGATAGTGGGCAGAGC | CAGGATGTCTCGAGGCATTGC | 293 |
| 4 MAD/MEF2C | L08895 | ↑ | N.T. | N.T. | |||
| (m) CTCGAGATGAGCCACGGGAAGAGAAC | (m) GCGGCCGCCTAGCTGGAGACGGCCATCA | (m) 476 | |||||
| 5 PC3 | M60921 | ↑ | ↑ | → | (h) ATGAGCCACGGGAAGGAAGGGAAC | (h) GGGTCAGCTCGCTGGGCAGC | (h) 304 |
| 6 mc7 | AJ278191 | ↑ | N.D. | → | GACGGCACGTGTGACGAGTG | CCTCGCTGTCTTCTTCACTC | 422 |
| 7 CGI 61 | AF151819 | ↑ | → | → | CCTCCTTAGCAAAGCTGAATG | TGCAGCATCCAAATCCAGTC | 393 |
| (m) CCGGAATTCATGTCTGACAAACCCG | (m) GCTCTAGATTACGATTCGCCAGCTTG | (m) 135 | |||||
| 8 Thymosin β4 | M34043 | ↑ | ↑ | → | (h) CAACCATGTCTCTGAGAAACCCG | (h) GATTCGCCTGCTTGCTTCTCC | (h) 135 |
| 9 EST-MNCb4008 Telomeric repeat binding | BF168890 | ↑ | → | ↑ | GGAGCAAAGAATGCATAAGC | CAGTGGGAAGAGAGGCCATG | |
| 10 Factor 2 | NM_02058 | ↑ | ↑ | → | CCTTTCCTGCCAACTCTTCCAC | GAGACTCTGGTTGGCCAGAG | 300 |
| 11 EST-MNCb1040 | AU035914 | ↑ | ↑ | N.T. | (m) ATGTGAGGCCGTTTGCGCGGA | (m) TTTTCTAATGGAATGCTTCCCC | (m) 540 |
| 12 mKIAA1039 protein | AK122424 | ↑ | ↑ | → | AGGTTTCCGAGGAGGCCTGG | CTTGTAGGATGGGGTCTCTG | |
| (m) CCGGAATTCGAGAGCCAAGGCCAAAGC | (m) GCTCTAGATTAGTTTGAATGTCATTTC | (m) 1085 | |||||
| 13 Selenoprotein P | X99807 | ↓ | ↑ | → | (h) CAATGTGGAGAAGCCTGGGG | (h) AGATGTGTGTATTTAATCG | (h) 312 |
| (m) GCGAAGGCTCTGAAAGTGG | (m) GGGGTAACTCAGAATGCAGGGTTC | (m) 908 | |||||
| 14 GARG16 Phosphatidic acid | Q64828 | ↓ | ↑ | → | (h) GACTGTGAGGAAGGATGGGC | (h) GGTCTGTGAGGACATGTTGG | (h) 330 |
| 15 Phosphatase type 2B | BC005558 NM_0320 03 | ↓ | ↓ | → | CCGCAGCCAGCGCCATGCAAA | GTAATAGATCCGGTAGAATTC | 344 |
| 16 Ectonucleotide pyrophosphatase | ↓ | → | → | GATCACAAACCAGAGGGCAG | TCTGTGGAGTTCATGGCTTC | 684 | |
| 17 Phosphodiesterase 5/amyloid beta (A4) precursor like protein 2 | NM_009691 | ↓ | → | → | GAGGCTCTTGCAGCCAATGC | CTGGAACTAGCAGGACATCAC | 337 |
| 18 Lysosomal membrane glycoprotein | J05287 | ↓ | → | → | TGTCTGCTGGCTACCATGGG | CTGCACTGCAGTCTTGAGCTG | 418 |
| 19 EST-MNCb1423 (RAMP) | XM_14918 | ↓ | ↑ | ↓ | CTGGGCAGCGCTGTGAAAATC | GTAATGGTGTCTCCCTTGAC | 366 |
The mRNA expression levels were determined by Northern blot analyses (*) or by semi-quantitative RT-PCR (†).
N.D., not detectable; N.T., not tested; M, mouse; H, human.
Muscle Injury Model and in Situ Hybridization Analysis
A crush-injury was given by puncturing the gastrocnemius muscle of 8-week-old male C57BL/10 mice with a 23-gauge needle. At different time points (5 hours to 14 days) after the injury, the gastrocnemius muscles were isolated and frozen in liquid nitrogen for RNA extraction. For in situ hybridization analysis, the tibialis anterior muscle samples from C57BL/10 and mdx mice were dissected on the day 6 after the crush injury or the injection of 100 μl of 10 μmol/L cardiotoxin (Wako), and they were frozen in isopentane precooled in liquid nitrogen. Ten-μm cryostat longitudinal sections were prepared and fixed in 4% paraformaldehyde in PBS (pH 7.4) and treated with 1 μg/ml of proteinase K (Wako) in PBS at room temperature for 7 minutes. After being acetylated with acetic anhydride in triethanolamine, the sections were hybridized with a digoxigenin-labeled anti-sense or sense RNA probe at 65°C for 18 hours and subjected to the colorimetric detection of signals as previously described.25
Establishment of Myoblastic Cell Lines from Patient Biopsy Samples
Skeletal muscle biopsy samples were provided from a normal donor (52 years of age), two BMD patients (1 to 6 years of age), and six DMD patients (1 to 12 years of age) after obtaining the informed consent of donors or their parents. The enzymatically dissociated human cells were subjected to primary culture in Dulbecco’s modified Eagle’s medium supplemented with 20% fetal calf serum and 4% Ultroser G or 5% of chicken embryo extract. Fibroblastic cells were removed by a 1-hour attachment in the tissue culture plate. Remaining myoblastic cells were subcultured for 1 to 2 weeks at 37°C, and infected with the SV40 tsT retrovirus for 2 days followed by a continuous culture at 32.5°C. The amphotropic retrovirus was produced by using the PLAT-A packaging line (SM and TK, unpublished). The above-described experimental protocols were approved by the ethical committees of the institute and associated universities.
Results
Establishment of Skeletal Muscle-Derived Cell Lines from Mdx and B10 Mice
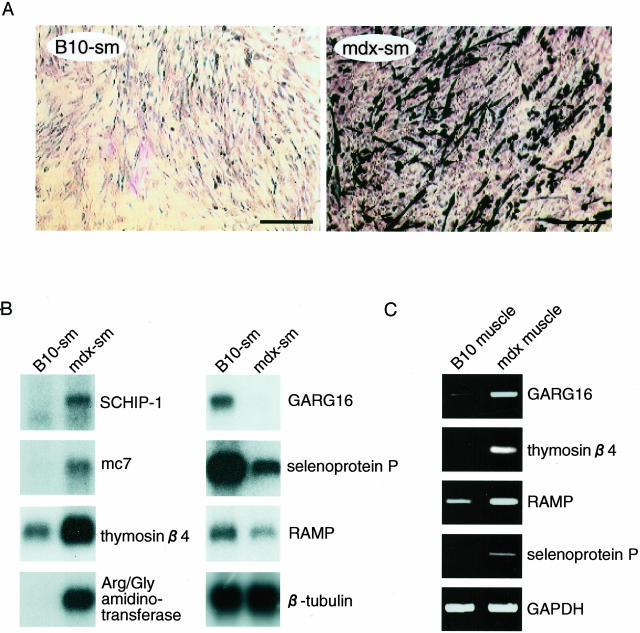
To assess whether the gene expression program of myogenic cells in skeletal muscle is affected by the mdx mutation, we immortalized the myoblastic cells isolated from the skeletal muscle of mdx and B10 mice. The immortalization was achieved by introducing SV40 tsT by retrovirus-mediated gene transfer into the primary myoblast culture. The newly established skeletal muscle cell lines derived from the mdx mice were termed mdx-sm whereas those from the B10 control mice were termed B10-sm. In the absence of viral challenge, the primary cells ceased growing after the third to fourth passage. In contrast, the mdx-sm and B10-sm cell lines have proliferated for more than a year when grown at 32.5°C. Their doubling times are comparable (data not shown). When these cells are cultured at 39.5°C, all stop proliferating and myotube formation is initiated. This indicates that the cells have been successfully immortalized in a temperature-dependent manner. In terms of their morphology, both cell lines are myoblastic in general but some of the mdx-sm cells spontaneously differentiate into myotube-like structures in the presence of horse serum even at the permissive temperature of 32.5°C. Concordantly, the frequency of desmin-positive myotubes in the mdx-sm cell line is much higher than that in the B10-sm cell line (Figure 1A). This phenotypic difference was also observed when six subclones of the mdx-sm cell line (mdx-sm1 to mdx-sm6) and two subclones of the B10-sm cell line (B10-sm1, B10-sm2) were compared. Thus, it appears that the higher differentiation capacity of mdx-sm cells is not because of the mixed cell populations in the founding lines, rather it results from the altered genetic program in the mdx myoblasts. For the following studies, we used the mdx-sm2 and B10-sm1 subclones as representative cell lines.
Figure 1.
Identification of up- and down-regulated genes in the mdx-derived skeletal muscle cell line. A: Extent of spontaneous myogenic differentiation in the B10-sm and mdx-sm myoblastic cell lines derived from B10 and mdx mice, respectively. Cells cultured in differentiation-inducing conditions were immunostained with anti-desmin antibody. B: Northern blot analysis of various mdx up- and down-regulated genes in mdx-sm and B10-sm cell lines. The integrity and amount of loaded RNAs were assessed by probing with β-tubulin cDNA. C: RT-PCR analysis of various mdx up- and down-regulated genes in the intact skeletal muscle of B10 and mdx mice. Total RNAs were converted to cDNA by random hexamers and subjected to 20 cycles of amplification with specific primer sets for each gene as indicated. DNA bands after ethidium bromide staining are shown with GAPDH, which acts as the control that ensures equal amount of template cDNA were used.
Identification of Differentially Expressed Genes in Mdx-sm Cells by cDNA Microarray Analysis
DNA tip analysis has been shown to be a powerful way to compare the overall gene expression profiles of test cells with those of control cells. We used mouse cDNA microarrays holding ∼4000 distinct mouse cDNA fragments (>0.5 kb) originating from a mouse fetus and an adult mouse brain. After hybridization with fluorescently labeled cDNAs from the mdx-sm and B10-sm cells, we selected those genes in which the ratio of mdx-sm fluorescence to that of B10-sm was either higher than 1.5 or lower than 0.62 when both fluorescent color combinations were used (data not shown). In this way, we identified 20 up-regulated and 21 down-regulated genes in the mdx-sm cells.
To confirm the differential expression of these candidate genes, we performed Northern blot analyses with mRNAs from mdx-sm and B10-sm cells. As shown in Figure 1B, mRNAs for schwannomin-interacting protein 1 (SCHIP-1),26 mc7 (dystrophin-interacting protein),27 thymosin β4 (G-actin sequestering protein),28 and l-arginine:glycine (Arg/Gly) amidinotransferase29 are more abundantly expressed in mdx-sm cells than in B10-sm cells, whereas the mRNA levels of GARG16 (glucocorticoid-attenuated response gene),30 selenoprotein P (anti-oxidant protein),31 and EST-MNCb1423 (accession number, XM 149185; designated as RAMP in this study) are lower in mdx-sm cells. In addition, eight other genes including myogenic transcription factor MEFC32 were found to be up-regulated in mdx-sm cells while four other genes were down-regulated (data not shown, Table 1). The expression patterns of these genes were similar in all of the mdx-sm subclones (data not shown).
Of the 19 differentially expressed genes, 15 encode proteins with known function and 4 are registered in the public database only as expressed sequenced tags (Table 1). When we examined the mRNA expression of these genes in the intact skeletal muscle of mdx and B10 mice by RT-PCR, five genes, including thymosin β4 and Arg/Gly amidinotransferase showed the same patterns revealed by the cell lines (Figure 1C, Table 1). In contrast, the mRNAs for GARG16, selenoprotein P, and RAMP were increased in the intact muscle of mdx mice (Figure 1C, Table 1). Although the exact reason for these discrepancies remains to be determined, our results demonstrate that the genetic programs of the growing myoblastic cells and the intact muscle fibers in mdx mice are not identical.
Effect of Introducing Dystrophin on the Gene Expression in Mdx-sm Cells
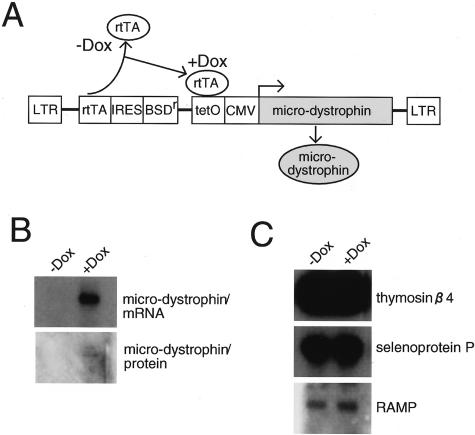
Dystrophin is known to be expressed in myotubes and mature muscle fibers but not in proliferating myoblasts. To exclude the possibility that the altered gene expression in mdx-sm cells is directly linked to the mutation in the dystrophin gene or to the lack of dystrophin mRNA or protein, we established a stable transfectant of mdx-sm cells that expresses the microdystrophin cDNA under the doxycyclin (Dox)-inducible promoter (Figure 2A). As shown in Figure 2B, 48 hours after the addition of Dox to the culture medium, microdystrophin mRNA and protein were produced in the mdx-sm transfectant cells. However, the expression levels of the genes that show disparate expression patterns in mdx mice, including thymosin β4, selenoprotein P, and RAMP, were not altered by the addition of Dox (Figure 2C, data not shown). These results suggest that the differential gene expression between mdx-sm and B10-sm cells is independent of dystrophin levels.
Figure 2.
Effect of enforced expression of microdystrophin protein in mdx-sm cell line. A: Structure of the pLRT vector carrying a mouse microdystrophin gene under the Dox-inducible promoter. B: Dox-inducible expression of microdystrophin mRNA and protein in mdx-sm cells transfected with pLRT-microdystrophin. C: Effect of introducing microdystrophin into mdx-sm cells on the expression of some of the mdx up- and down-regulated genes. The levels of the mRNAs specific for thymosin β4, selenoprotein P, and RAMP before and after adding Dox were verified by Northern blot analyses.
Primary Sequence and Expression of RAMP
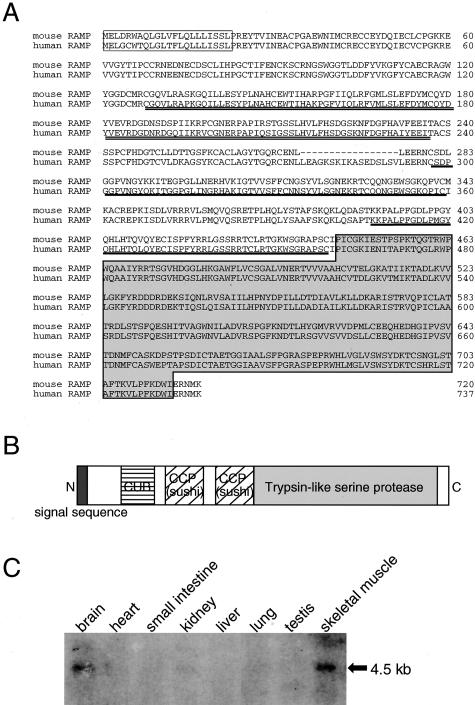
Because myoblasts play a central role in muscle regeneration, we hypothesized that the 13 differentially expressed genes in mdx-sm cells might play a role in the progression of muscle wasting seen in DMD patients and/or the milder myopathy in mdx mice. We especially focused on the RAMP gene because we found its mRNA expression was enhanced in regenerating muscle fibers (see Figure 4). Mouse RAMP cDNA in the database is 3085 bp in length and its predicted open reading frame codes for 720 amino acid residues (Figure 3A). A BLAST search of the database revealed that mouse RAMP protein shows 88% identity at the amino acid level with the uncharacterized human protein DKFZP586H2131 (accession number, MN 015430), which we refer to as human RAMP in this study. As shown schematically in Figure 3B, mouse RAMP protein contains a putative signal peptide at its N-terminal region (1 to 22),33 the CUB domain (122 to 236),34 two complement control protein (CCP) modules (also known as Sushi domains) (280 to 342 and 389 to 442),35 and a trypsin-like serine protease domain (444 to 715).36 CUB and CCP/Sushi domains are often found in developmentally regulated proteins and cell adhesion molecules, respectively. These subdomain structures are conserved in human RAMP as well. The calculated molecular mass of mouse RAMP was 82.1 kd with an isoelectric point of 7.3. In the normal adult mouse, RAMP mRNA is only detectable in the brain and skeletal muscle as a single band at the position of 4.5 kb (Figure 3C). Thus, it appears that RAMP is a novel secreted protease that potentially plays some functions in skeletal muscle.
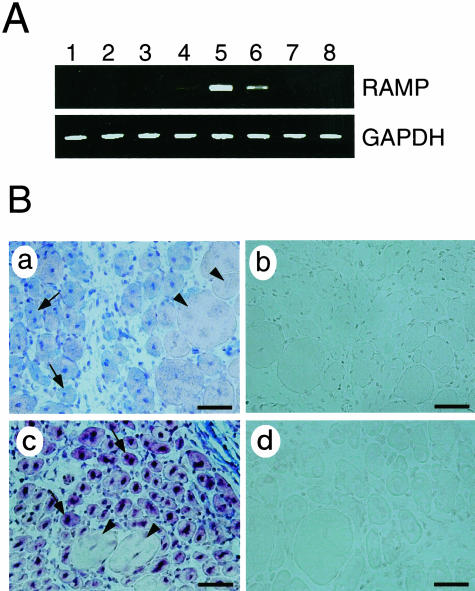
Figure 4.
Up-regulation of RAMP mRNA expression in skeletal muscle after injury. A: Gastrocnemius muscle of mice was dissected 5 hours (lane 1) or 1 (lane 2), 2 (lane 3), 3 (lane 4), 4 (lane 5), 7 (lane 6), 10 (lane 7), or 14 (lane 8) days after injuring the muscle with a needle. Total RNA from each sample was extracted, reverse-transcribed, and amplified by 30 cycles of PCR using RAMP-specific primers. RT-PCR for GAPDH cDNA was performed to verify that equal amounts of template cDNA were used. B: Detection of RAMP mRNA in regenerating muscle fibers by in situ hybridization. Transverse cryosections of the tibialis anterior muscle of C57BL/10 mice harvested 6 days after crush injury were hybridized with digoxigenin-labeled anti-sense (a) or sense (b) cRNA probes for RAMP. Tibialis anterior muscle sections prepared from mdx mice after cardiotoxin injection were also hybridized with digoxigenin-labeled anti-sense (c) or sense (d) cRNA probes for RAMP. Specific signals for RAMP mRNA were detected as blue (a) or brown paints (c) in the centrally nucleated muscle fibers (arrows in a and c), but not in mature fibers (arrowheads in a and c). Scale bars, 50 μm.
Figure 3.
Sequence and tissue distribution of RAMP. A: Sequences of human and mouse RAMP proteins. Predicted signal sequences are boxed. CUB and CCP/Sushi domains are indicated by a double line and a solid line, respectively. The trypsin-like serine protease domain is shown by a light gray box. B: Schematic representation of the structural motifs in RAMP. C: Predominant expression of RAMP mRNA in skeletal muscle and brain. Two μg of poly(A)+ RNA prepared from various organs of adult C57BL/6 mouse were electrophoresed, blotted, and hybridized with 32P-labeled mouse RAMP cDNA. The band size of the detected transcript is shown.
Induction of RAMP mRNA in Regenerating Skeletal Muscle Fiber
We next examined whether the expression of RAMP mRNA is changed in regenerating skeletal muscle fibers. A crush injury of gastrocnemius muscle was induced in normal mice and the expression of RAMP mRNA was measured. RAMP expression gradually increased throughout the days after the injury, reaching the highest levels 4 days after the injury, and then being reduced to the baseline within 1 to 2 weeks (Figure 4A). Consistent with the transient up-regulation of RAMP mRNA in the muscle injury model, in situ hybridization assays revealed that RAMP mRNA was specifically detected in centrally nucleated regenerating muscle fibers (shown by arrows), but not in unaffected fibers (arrowheads) (Figure 4B, a). In the tibialis anterior muscle of mdx mice after cardiotoxin injection, expression of RAMP was again specifically observed in regenerating muscle fibers (arrows) (Figure 4B, c). Thus, the RAMP gene is induced by the injury of skeletal muscle.
Expression of RAMP mRNA in Skeletal Muscle Cell Lines Derived from BMD and DMD Patients
To evaluate the importance of RAMP in DMD pathology, we established skeletal muscle cell lines derived from biopsies of an unaffected donor and BMD/DMD patients. The strategy used to achieve this was the same as that used to establish the mouse muscle cell lines except that an amphotropic retrovirus was used and the cells were exposed to the virus for a longer period. Continuously growing cells at 32.5°C were obtained from one normal, two BMD, and six DMD biopsies within 1 to 2 months after introducing SV40 tsT. In contrast, uninfected parallel control cultures exhibited senescence. Similar to the mdx-sm cell line, the human BMD/DMD-derived muscle cell lines differentiated into myotubes when cultured at 39.5°C. Figure 5 shows a representative BMD cell line, BMD-sm1, during its undifferentiated growth (b) and after it has been placed in differentiation-inducing conditions (a). Immunohistochemical analysis demonstrated that there was a high frequency of desmin-positive myotubes in the differentiated BMD-sm1 cell line (Figure 5A, d). The other eight human patient-derived cell lines proliferated similarly and also possessed myogenic properties (data not shown), although their growth rates were very low compared to the mouse cell line. By designing specific sets of primers for human orthologs to the mdx-up and -down genes, we examined the mRNA expression of these genes in the nine human muscle cell lines by RT-PCR. We found that relative to the unaffected donor-derived cell line, mRNAs for Arg/Gly amidinotransferase and EST-MNCb4008 were always increased in the BMD/DMD patient-derived cell lines (data not shown, Table 1). In contrast, all of the BMD/DMD patient-derived cell lines showed lower RAMP mRNA expression levels relative to the control line (Figure 5B). That both the mdx-sm- and the BMD/DMD-derived cell lines show an attenuation of RAMP expression suggests that RAMP could be involved in the pathogenesis of DMD.
Figure 5.
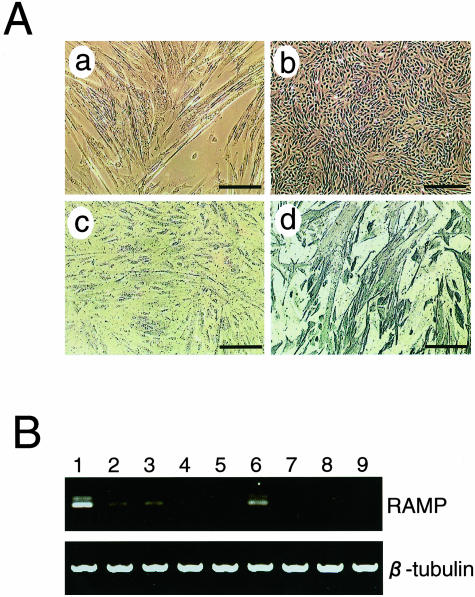
Expression of RAMP mRNA in cell lines derived from human BMD and DMD patients. A: Myotube differentiation capacity of BMD-sm1, a representative cell line derived from a BMD patient’s skeletal muscle. The morphology is shown by phase contrast microscopy of the BMD-sm1 cell line cultured at 39.5°C (a) or 32.5°C (b). BMD-sm1 cells cultured at 39.5°C were immunostained with (d) or without (c) anti-desmin antibody. B: RT-PCR analysis of RAMP mRNA expression in nine human cell lines derived from skeletal muscle biopsies of patients. Total RNAs were extracted from the control (lane 1), two BMD (lanes 2 and 3), and six DMD (lanes 4 to 9) human muscle cell lines, reverse-transcribed, and amplified by 30 cycles of PCR using specific primers for human RAMP cDNA. Ethidium bromide-staining patterns of DNA bands are shown with β-tubulin, which shows that equal amounts of template cDNA were loaded. Scale bars, 150 μm.
Discussion
In this study, we identified 19 genes that are differentially expressed by newly established skeletal muscle cell lines from mdx and control B10 mice. We also showed that the expression of the novel gene RAMP, which is down-regulated in the mdx line, was frequently impaired in the muscle cell lines derived from six DMD and two BMD patients.
Gene expression studies using mdx mice or DMD biopsies have been published before10–13,16,17 but unlike these studies, we used immortalized skeletal muscle cell lines as the initial materials for the cDNA microarray and Northern blot analyses. The immortalization was achieved by conditional transformation of the primary cells by infecting them with the SV40 tsT-bearing retrovirus vector. This method has been used successfully to expand target cell types before.19 It has been reported that myogenic cell lines established by the SV40 large T antigen retain their differentiation capacity in vitro.37,38 Supporting this is that the myoblastic cell lines established in this study possess myogenic differentiation activity. It is also known that rodent cells can be fully immortalized by the SV40 T antigen.39 However, as the life span of human primary cells in vitro is dependent on their telomerase activity, human cells carrying the SV40 large T antigen will eventually undergo senescence and crisis because of telomere shortening. Nevertheless, our approach yielded sufficient numbers of progeny human myoblastic cells from single biopsies from DMD or BMD patients.
The skeletal muscle cell line that was established from mdx mice (mdx-sm) differentiated more frequently into a myotube-like structure in vitro than the line derived from the control C57BL/10 mice (B10-sm) (Figure 1A). That MEF2C expression in mdx-sm cells is enhanced (Table 1, data not shown) confirms that the myogenic differentiation program in these cells is engaged. This property could reflect the status of the myoblastic precursor cells in the mdx mouse muscle at the time of virus infection. In adult mice, skeletal muscle stem cells (satellite cells) that reside beneath the basal lamina of muscle fibers are mitotically quiescent and are only activated in response to mechanical stimuli such as exercise and injury.40 In the process of muscle regeneration, satellite cells are believed to generate myoblasts that proliferate and differentiate to make myotubes before fusing with existing myofibers. In mdx mice, the destruction of muscle fibers caused by the lack of dystrophin and compensatory muscle regeneration are thought to occur continuously.9 As is consistent with a previous report, the injury-response signaling pathway mediated by JNK1 MAP kinase may thus be activated in the majority of myoblasts in mdx mice.40 All of the subclonal cell lines from the original bulk culture of mdx-sm produced desmin+ myotube-like structures at a higher rate (data not shown). Because retrovirus infection occurs only in dividing cells in culture, the mdx-sm line may represent a proliferating subpopulation of myogenic precursor cells in mdx skeletal muscle. This may also be true for the human patient-derived muscle cell lines. Thus, the genetic programs in these cell lines are likely to represent those that are present in most, if not all, myoblasts in intact skeletal muscle.
The comparison of the mdx-sm and B10-sm cell lines revealed that 12 genes are up-regulated and 7 genes are down-regulated in the mdx-sm line (Figure 1B, Table 1). Dystrophin is expressed in muscle fibers and mature myotubes but not in proliferating myoblasts. To verify that the differential gene expression in mdx-sm cells is not because of the absence of dystrophin, we introduced a microdystrophin gene lacking the 4th to 23rd rod domains into mdx-sm cells and induced these cells to express the truncated dystrophin protein. Microdystrophin has been proven to be fully functional in restoring myopathy in mdx mice.23,41 However, the presence of this protein in mdx-sm cells did not affect the expression of the 19 genes whose expression patterns are altered in mdx-sm cells (Figure 2B, data not shown). Thus, the altered gene expression pattern in mdx-sm cells is the result of a dystrophin-independent event in the myoblasts of mdx mice.
Among the 12 up-regulated genes in the mdx-sm cell line, Arg/Gly amidinotransferase is of particular interest because it is a first and rate-limiting enzyme for the creatine biosynthesis that is a major energy source in skeletal muscle.42 A previous study has shown that Arg/Gly amidinotransferase mRNA is rapidly induced in kidney in vivo by adding creatine and growth hormone.29 It is reasonable to suppose that skeletal muscle cells of mdx mice also produce this enzyme more abundantly in response to the creatine and creatine kinase that is released by disrupted muscle fibers. We found that all DMD- and BMD-derived muscle cell lines also produce a higher level of Arg/Gly amidinotransferase (Table 1, data not shown). This suggests that the measurement of mRNA for this enzyme in muscle cells as well as detection of higher creatine kinase levels in serum may be useful in the diagnosis of DMD and other myopathies.
Increased expression of thymosin β4 in the mdx mouse muscle has been reported in previous studies.12 We found that its expression was up-regulated in mdx-sm cells and mdx muscle tissue, but not in the DMD patient-derived cell lines. Thymosin β4 is a bifunctional protein that sequesters G-actin in the cytoplasm and stimulates the migration of endothelial cells and monocytes once secreted outside of cells.43,44 The acetylated tetrapeptide acSDKP that is proteolytically released from the N-terminus of thymosin β4 inhibits proliferation of hematopoietic progenitor cells and enhances angiogenesis.45 Presumably, thymosin β4 may play some roles in the regenerating muscle area by acting on inflammation-associated cells. A study is underway to determine whether higher amounts of thymosin β4 could ameliorate DMD pathogenesis.
The gene products of SCHIP-1 and mc7 are structural proteins in the muscle and brain. It has been suggested that SCHIP-1 links membrane proteins to the cytoskeleton26 and that mc7 directly binds to the C-terminal region of dystrophin (GenBank data base), although clear evidence for these functions is not yet available. The relevance of the up-regulation of these two genes in the mdx-sm cell line remains uncertain. However, it is tempting to speculate that the expression of sarcolemmal membrane-associated proteins is induced in the absence of dystrophin by a compensatory mechanism. The expression of utrophin in mdx mice is the most famous example of this. Recent gene profiling studies of mdx mice and DMD biopsies also revealed such tendencies.10,17
The selenoprotein P gene is also down-regulated in the mdx-sm cell line. This down-regulation could affect the anti-oxidant defense of cells. Selenoproteins possess thioredoxin reductase activity that neutralizes the cytotoxic response of cells to the oxidant stress mediated by thioredoxin.46 Recently, 10 families with congenital muscular dystrophy were shown to carry mutations in the selenoprotein N gene.46 Moreover, there are reports showing that selenium deficiency is associated with muscular dystrophy in animals and cardiomyopathy in humans.47,48 However, we found that the overall mRNA level of selenoprotein P in mdx mice was higher than that in control mice, as has also been shown in a previous report.10 The presence of increased amounts of selenoprotein P as well as RAMP in adult mdx mice may be related to the milder skeletal myopathic phenotype observed in this model.
Of the seven down-regulated genes in mdx-sm cells, RAMP was the only one whose expression was always lower in muscle cell lines from six DMD and two BMD patients compared to the expression in a line from an unaffected control. Although the normal human myoblastic cell line used in this study was derived from a relatively aged donor, this difference appears to be significant because most of the other genes that showed differential expression in the mdx-sm line had a similar pattern of expression between the BMD/DMD patient-derived cell lines and the unaffected control cell line (Table 1). RAMP is a novel secreted protease that carries three major molecular signatures (Figure 3B). One of these is the CUB domain, which is often found in the extracellular domain of developmentally regulated proteins. Examples of these are bone morphogenic protein 1 (a metalloendopeptidase that induces cartilage and bone formation) and neuropillin (a calcium-independent cell adhesion molecule that functions during the formation of neuronal and vascular circuits). Another molecular signature is the CCP modules/Sushi domain. These exist in a wide variety of complement and adhesion proteins. The third significant molecular motif of RAMP is its serine protease domain belonging to the trypsin family at its C-terminus. In normal unchallenged mice, RAMP mRNA is only detectable in the skeletal muscle and brain. Previous studies showed that genes encoding cathepsin S and cathepsin H proteases are up-regulated in mdx muscle,11,12 but RAMP-related proteases have not been described previously. The reason why the RAMP gene has not been identified in the microarray studies in the past by using mdx mice is probably because its expression level is low and the amount of mRNA in the intact skeletal muscle of mdx and normal mice do not differ significantly. A unique finding of this study is that RAMP mRNA is specifically induced in regenerating muscle fibers found after skeletal muscle injury. As described above, during muscle regeneration, myoblasts differentiate into myotubes and the cells then fuse with existing muscle fiber. Accumulating evidence from previous publications suggests that an elevated calcium concentration in regenerating muscle activates calcineurin, which dephosphorylates nuclear factors of activated T cells (NF-ATs).49,50 Specific NF-AT isoforms in skeletal muscle play crucial roles in the myogenic differentiation and myoblast fusion events. As with the interleukin-4 induction that triggers myotube fusion in regenerating muscle areas,51 RAMP mRNA is induced in small centrally nucleated myofibers (Figure 4B). Thus, RAMP could be an important regulator of the muscle regeneration process. Further analysis of its enzymatic targets and biological activities in skeletal muscle would shed light on this novel protease and enhance our understanding of the processes involved in muscle regeneration and the malignant progression of DMD.
Acknowledgments
We thank Drs. Yuko Miyagoe-Suzuki and Shin-ichi Takeda (National Institute of Neuroscience) for valuable advice; and Drs. Norman Drinkwater (University of Wisconsin Medical School) and Masatoshi Hagiwara (Tokyo Medical and Dental University) for providing us with the pMESVts and pLRT-X retrovirus vectors, respectively.
Footnotes
Address reprint requests to Takahiko Hara, Ph.D., Department of Tumor Biochemistry, The Tokyo Metropolitan Institute of Medical Science, Tokyo Metropolitan Organization for Medical Research, 3-18-22 Honkomagome, Bunkyo-ku, Tokyo 113-8613, Japan. E-mail: thara@rinshoken.or.jp.
Supported by grants-in-aid for Research on Nervous and Mental Disorders (13B-1) and for Research in Brain Science (H12-brain-028) from the Ministry of Health, Labor, and Welfare of Japan; a grant-in aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (14770106); and a grant-in-aid from the Nakatomi Foundation.
References
- Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. Dystrophin and the membrane skeleton. Curr Opin Cell Biol. 1993;5:82–87. doi: 10.1016/s0955-0674(05)80012-2. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Xia H, Aldape K, Bredt DS. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong PY, Turner PR, Denetclaw WF, Steinhardt RA. Increased activity of calcium leak channels in myotubes of Duchenne human and mdx mouse origin. Science. 1990;250:673–676. doi: 10.1126/science.2173137. [DOI] [PubMed] [Google Scholar]
- Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- Dangain J, Vrbova G. Muscle development in mdx mutant mice. Muscle Nerve. 1984;7:700–704. doi: 10.1002/mus.880070903. [DOI] [PubMed] [Google Scholar]
- Tkatchenko AV, Le Cam G, Leger JJ, Dechesne CA. Large-scale analysis of differential gene expression in the hindlimb muscles and diaphragm of mdx mouse. Biochim Biophys Acta. 2000;1500:17–30. doi: 10.1016/s0925-4439(99)00084-8. [DOI] [PubMed] [Google Scholar]
- Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Guo W, Andrade FH. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum Mol Genet. 2002;11:263–272. doi: 10.1093/hmg/11.3.263. [DOI] [PubMed] [Google Scholar]
- Tseng BS, Zhao P, Pattison JS, Gordon SE, Granchelli JA, Madsen RW, Folk LC, Hoffman EP, Booth FW. Regenerated mdx mouse skeletal muscle shows differential mRNA expression. J Appl Physiol. 2002;93:537–545. doi: 10.1152/japplphysiol.00202.2002. [DOI] [PubMed] [Google Scholar]
- Rouger K, Le Cunff M, Steenman M, Potier MC, Gibelin N, Dechesne CA, Leger JJ. Global/temporal gene expression in diaphragm and hindlimb muscles of dystrophin-deficient (mdx) mice. Am J Physiol. 2002;283:C773–C784. doi: 10.1152/ajpcell.00112.2002. [DOI] [PubMed] [Google Scholar]
- Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 2002;157:137–148. doi: 10.1083/jcb.200108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, Khurana TS. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420:418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslett JN, Sanoudou D, Kho AT, Bennett RR, Greenberg SA, Kohane IS, Beggs AH, Kunkel LM. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc Natl Acad Sci USA. 2002;99:15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinset C, Montarras D. San Diego: Academic Press, Inc.; Cell Systems for ex Vivo Studies of MyogenesisA Protocol for the Isolation of Stable Muscle Cell Populations from Newborn to Adult Mice. 1994 [Google Scholar]
- Lee GH, Ogawa K, Drinkwater NR. Conditional transformation of mouse liver epithelial cells. An in vitro model for analysis of genetic events in hepatocarcinogenesis. Am J Pathol. 1995;147:1811–1822. [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Nagasugi Y, Azuma T, Kato M, Sugano S, Hashimoto K, Masuho Y, Muramatsu M, Seki N. Isolation of novel mouse genes differentially expressed in brain using cDNA microarray. Biochem Biophys Res Commun. 2000;275:532–537. doi: 10.1006/bbrc.2000.3330. [DOI] [PubMed] [Google Scholar]
- Maeda S, Otsuka M, Hirata Y, Mitsuno Y, Yoshida H, Shiratori Y, Masuho Y, Muramatsu M, Seki N, Omata M. cDNA microarray analysis of Helicobacter pylori-mediated alteration of gene expression in gastric cancer cells. Biochem Biophys Res Commun. 2001;284:443–449. doi: 10.1006/bbrc.2001.5006. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Yuasa K, Yoshimura M, Yokota T, Ikemoto T, Suzuki M, Dickson G, Miyagoe-Suzuki Y, Takeda S. Micro-dystrophin cDNA ameliorates dystrophic phenotypes when introduced into mdx mice as a transgene. Biochem Biophys Res Commun. 2002;293:1265–1272. doi: 10.1016/S0006-291X(02)00362-5. [DOI] [PubMed] [Google Scholar]
- Watsuji T, Okamoto Y, Emi N, Katsuoka Y, Hagiwara M. Controlled gene expression with a reverse tetracycline-regulated retroviral vector (RTRV) system. Biochem Biophys Res Commun. 1997;234:769–773. doi: 10.1006/bbrc.1997.6705. [DOI] [PubMed] [Google Scholar]
- Hara T, Tamura K, de Miguel MP, Mukouyama Y, Kim H, Kogo H, Donovan PJ, Miyajima A. Distinct roles of oncostatin M and leukemia inhibitory factor in the development of primordial germ cells and Sertoli cells in mice. Dev Biol. 1998;201:144–153. doi: 10.1006/dbio.1998.8990. [DOI] [PubMed] [Google Scholar]
- Goutebroze L, Brault E, Muchardt C, Camonis J, Thomas G. Cloning and characterization of SCHIP-1, a novel protein interacting specifically with spliced isoforms and naturally occurring mutant NF2 proteins. Mol Cell Biol. 2000;20:1699–1712. doi: 10.1128/mcb.20.5.1699-1712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutou E, Matsas R, Mamalaki A. Isolation of a mouse brain cDNA expressed in developing neuroblasts and mature neurons. Brain Res Mol Brain Res. 2001;86:153–167. doi: 10.1016/s0169-328x(00)00281-3. [DOI] [PubMed] [Google Scholar]
- Huff T, Muller CS, Otto AM, Netzker R, Hannappel E. Beta-thymosins, small acidic peptides with multiple functions. Int J Biochem Cell Biol. 2001;33:205–220. doi: 10.1016/s1357-2725(00)00087-x. [DOI] [PubMed] [Google Scholar]
- Guthmiller P, Van Pilsum JF, Boen JR, McGuire DM. Cloning and sequencing of rat kidney L-arginine:glycine amidinotransferase. Studies on the mechanism of regulation by growth hormone and creatine. J Biol Chem. 1994;269:17556–17560. [PubMed] [Google Scholar]
- Smith JB, Herschman HR. The glucocorticoid attenuated response genes GARG-16, GARG-39, and GARG-49/IRG2 encode inducible proteins containing multiple tetratricopeptide repeat domains. Arch Biochem Biophys. 1996;330:290–300. doi: 10.1006/abbi.1996.0256. [DOI] [PubMed] [Google Scholar]
- Mostert V. Selenoprotein P: properties, functions, and regulation. Arch Biochem Biophys. 2000;376:433–438. doi: 10.1006/abbi.2000.1735. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- Norman DG, Barlow PN, Baron M, Day AJ, Sim RB, Campbell ID. Three-dimensional structure of a complement control protein module in solution. J Mol Biol. 1991;219:717–725. doi: 10.1016/0022-2836(91)90666-t. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouly V, Edom F, Decary S, Vicart P, Barbert JP, Butler-Browne GS. SV40 large T antigen interferes with adult myosin heavy chain expression, but not with differentiation of human satellite cells. Exp Cell Res. 1996;225:268–276. doi: 10.1006/excr.1996.0176. [DOI] [PubMed] [Google Scholar]
- Berghella L, De Angelis L, Coletta M, Berarducci B, Sonnino C, Salvatori G, Anthonissen C, Cooper R, Butler-Browne GS, Mouly V, Ferrari G, Mavilio F, Cossu G. Reversible immortalization of human myogenic cells by site-specific excision of a retrovirally transferred oncogene. Hum Gene Ther. 1999;10:1607–1617. doi: 10.1089/10430349950017617. [DOI] [PubMed] [Google Scholar]
- Manfredi JJ, Prives C. The transforming activity of simian virus 40 large tumor antigen. Biochim Biophys Acta. 1994;1198:65–83. doi: 10.1016/0304-419x(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk SM, Walsh GS, Balazsi K, Seale P, Sandoz J, Hierlihy AM, Rudnicki MA, Chamberlain JS, Miller FD, Megeney LA. Activation of JNK1 contributes to dystrophic muscle pathogenesis. Curr Biol. 2001;11:1278–1282. doi: 10.1016/s0960-9822(01)00397-9. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Hauser MA, DelloRusso C, Duan D, Crawford RW, Phelps SF, Harper HA, Robinson AS, Engelhardt JF, Brooks SV, Chamberlain JS. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- Carlson M, Van Pilsum JF. S-adenosylmethionine: guanidinoacetate N-methyltransferase activities in livers from rats with hormonal deficiencies or excesses. Proc Soc Exp Biol Med. 1973;143:1256–1259. doi: 10.3181/00379727-143-37512. [DOI] [PubMed] [Google Scholar]
- Malinda KM, Goldstein AL, Kleinman HK. Thymosin beta 4 stimulates directional migration of human umbilical vein endothelial cells. EMBO J. 1997;11:474–481. doi: 10.1096/fasebj.11.6.9194528. [DOI] [PubMed] [Google Scholar]
- Young JD, Lawrence AJ, MacLean AG, Leung BP, McInnes IB, Canas B, Pappin DJ, Stevenson RD. Thymosin beta 4 sulfoxide is an anti-inflammatory agent generated by monocytes in the presence of glucocorticoids. Nat Med. 1999;5:1424–1427. doi: 10.1038/71002. [DOI] [PubMed] [Google Scholar]
- Liu JM, Lawrence F, Kovacevic M, Bignon J, Papadimitriou E, Lallemand JY, Katsoris P, Potier P, Fromes Y, Wdzieczak-Bakala J. The tetrapeptide AcSDKP, an inhibitor of primitive hematopoietic cell proliferation, induces angiogenesis in vitro and in vivo. Blood. 2003;101:3014–3020. doi: 10.1182/blood-2002-07-2315. [DOI] [PubMed] [Google Scholar]
- Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, Muntoni F, Topaloglu H, Guicheney P. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet. 2001;29:17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- Oldfield JE. Selenium in animal nutrition: the Oregon and San Joaquin Valley (California) experiences—examples of correctable deficiencies in livestock. Biol Trace Elem Res. 1989;20:23–29. doi: 10.1007/BF02919095. [DOI] [PubMed] [Google Scholar]
- Ge K, Xue A, Bai J, Wang S. Keshan disease—an endemic cardiomyopathy in China. Virchows Arch A Pathol Anat Histopathol. 1983;401:1–15. doi: 10.1007/BF00644785. [DOI] [PubMed] [Google Scholar]
- Abbott KL, Friday BB, Thaloor D, Murphy TJ, Pavlath GK. Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol Biol Cell. 1998;9:2905–2916. doi: 10.1091/mbc.9.10.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling U, Tureckova J, Lim HW, De Windt LJ, Rotwein P, Molkentin JD. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol Cell Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]