Abstract
We observed that in urothelium, both cornifying and noncornifying forms of squamous metaplasia are accompanied by changes in the localization of the nuclear hormone receptors, peroxisome proliferator activated receptor γ (PPAR-γ) and retinoid X receptor (RXR-α). To obtain objective evidence for a role for PPAR-γ-mediated signaling in urothelial differentiation, we examined expression of the cytokeratin isotypes CK13, CK20, and CK14 as indicators of transitional, terminal transitional, and squamous differentiation, respectively, in cultures of normal human urothelial cells. In control culture conditions, normal human urothelial cells showed evidence of squamous differentiation (CK14+, CK13−, CK20−). Treatment with the high-affinity PPAR-γ agonist, troglitazone (TZ), resulted in gain of CK13 and loss of CK14 protein expression. The effect of TZ was significantly augmented when the autocrine-stimulated epidermal growth factor receptor pathway was inhibited and this resulted in induction of CK20 expression. The RXR-specific inhibitors PA452, HX531, and HX603 inhibited the TZ-induced CK13 expression, supporting a role for RXR in the induction of CK13 expression. Thus, signaling through PPAR-γ can mediate transitional differentiation of urothelial cells and this is modulated by growth regulatory programs.
Urothelium, the epithelium lining the bladder and associated urinary tract is normally a transitional epithelium, but has the capacity to undergo either squamous, or more rarely, glandular metaplasia. This plasticity of differentiation is thought to reflect the embryonic derivation of urothelium from the urogenital sinus epithelium, which also gives rise to vaginal-type stratified squamous and prostatic-type glandular epithelia.1 The precise molecular mechanisms and factors that regulate the induction and type of differentiation in urothelium are poorly understood.
The cytokeratins (CKs) are widely regarded as markers par excellence of epithelial cytodifferentiation. Not only is their expression restricted primarily to cells committed to an epithelial lineage, but distinct CK isotype expression profiles are associated with particular epithelial differentiation programs. Furthermore, expression of particular CK isotypes can be associated with specific maturation or pathogenic stage.2
Transitional epithelium shows a well-characterized progression of changes in CK isotype expression from basal through to the terminally differentiated superficial cells.3 Whereas CK5 and CK17 are basally expressed and CK13 is present in all but the superficial cells, the expression of CK20 is restricted to the superficial cell layer, where it is recognized as a highly sensitive marker of normal urothelial cytodifferentiation.4–7 Urothelial expression of CK14, which is expressed in the basal compartment of all stratified squamous epithelia, has been correlated with squamous differentiation of urothelium in vivo.8
Experimental and clinical investigations have demonstrated that under conditions of dietary vitamin A deficiency, urothelium undergoes squamous metaplasia in vivo.9–11 Retinoic acid (RA) effects are transduced by two distinct classes of nuclear hormone receptors, retinoid A receptors (RAR) and retinoid X receptors (RXR). RXR binds 9-cis-RA with high affinity, whereas RARs interact with 9-cis-, 13-cis-, and all-trans RA.12 We have shown previously that normal human urothelial (NHU) cells grown in vitro in the absence of exogenous retinoids express a squamous-associated CK13−, CK14+ phenotype, but revert to a transitional CK13+, CK14− phenotype on treatment with the RAR ligand, 13-cis-RA.13 Our study found no expression of CK20, suggesting that although differentiation had been redirected from a squamous to a transitional pathway, there was no induction of terminal differentiation.13 Another nuclear hormone receptor, peroxisome proliferator activated receptor (PPAR)-γ, has been implicated in the differentiation of adipocytes.14 When PPAR-γ is activated it forms a heterodimer with the nuclear hormone receptor, RXR. This nuclear hormone complex can bind to the peroxisome proliferator response element (PPRE) and induce gene transactivation.15
A role has been proposed for cell surface [interferon-γ, transforming growth factor-β, epidermal growth factor (EGF)] and nuclear hormone (RAR, TH3) receptor signaling in modulating patterns of CK expression in other epithelial tissues.16 In this study, we have investigated how signaling through PPAR-γ affects urothelial cytodifferentiation, by assessing the effects of receptor activation on CK expression. Because proliferation through the epidermal growth factor receptor (EGFR) pathway may inhibit differentiation,17 the effects on CK expression by activation of PPAR-γ were performed in conjunction with inhibitors of EGFR.
Materials and Methods
Materials
PPAR-γ was activated by the high-affinity agonist troglitazone (TZ), a member of the thiazolidinedione class of anti-diabetic drugs.18 TZ was provided as a gift by Parke-Davis Pharmaceutical Research (Ann Arbor, MI). Antagonists for PPAR-γ, bisphenol A diglycidyl ether (BADGE19), was obtained from Tocris Cookson Ltd. (Bristol, UK) and GW966220 was provided as a gift by GlaxoSmithKline (Worthing, UK). The RXR antagonists, HX531, HX603, and PA45221,22 were a kind gift from Dr. Hiroyuki Kagechika (University of Tokyo, Tokyo, Japan). The EGFR inhibitors, PD153035 and AG1478, were purchased from Calbiochem-Novabiochem Biosciences Ltd. (Nottingham, UK). [32P]CTP was from Amersham Pharmacia Biotech Ltd. (Little Chalfont, UK). mRNA extracted from whole tissue colon, ureter, fetal bladder, and breast was obtained from Stratagene, Amsterdam, The Netherlands. All agonists and antagonists were dissolved in dimethyl sulfoxide and solvent controls were included in all experiments, in which the maximum concentration of dimethyl sulfoxide was 0.001%.
Antibodies
Mouse monoclonal antibodies were used to the following specificities: CK20 (clone IT-Ks 20.3; Chemicon International Ltd., Harrow, UK); CK13 (clone IC7; ICN Biomedicals Inc., Basingstoke, UK); CK14 (clone LL002; Serotec Ltd. Oxford, UK), CK19 (clone LP2K, Central Resources, Cancer Research, London, UK), β-actin (Sigma-Aldrich Ltd., Poole, UK), PPAR-γ (clone E-8; Santa Cruz Biotechnology Inc., supplied by Autogen Bioclear Ltd., Calne, UK). Rabbit anti-RXR-α (RXR-α) (D20) was obtained from Santa Cruz.
Tissues
Surgical specimens of ureter and renal pelvis were obtained from patients with no histological evidence of urothelial dysplasia or malignancy. Three specimens of urothelium were included that showed histological evidence of squamous metaplasia, including one case of cornifying and two cases of noncornifying squamous differentiation. The collection of surgical specimens was approved by the relevant Local Research Ethics Committees and had full patient consent, as required. Tissues were collected in Hanks’ balanced salt solution containing 10 mmol/L HEPES, pH 7.6, and 20 kIU aprotinin (Trasylol; Bayer plc, Newbury, UK), as described.23,24 Representative pieces of each tissue sample were processed into paraffin wax for histology and the remaining sample (normal urothelium) was used to establish NHU cell lines using methods that have been described in detail elsewhere.13,23
Cell Culture
NHU cell lines were maintained in keratinocyte serum-free medium, containing bovine pituitary extract and EGF at the manufacturer’s recommended concentrations (Invitrogen Ltd., Paisley, UK) and supplemented with 30 ng/ml cholera toxin (Sigma).12,17 NHU cell lines from three independent donors were used for these studies between passages 3 and 5. For experiments, cells were seeded at 9 × 105 cells/ml and allowed to grow to 70% confluence before treatment. Preliminary studies (not reported here) performed to optimize ligand concentrations showed that the minimum time of exposure to TZ to induce maximal effect was 24 hours. As appropriate to the experiment (see legends to Figures), cells were pretreated with PPAR-γ inhibitors for 3 hours before treatment with PPAR-γ agonist, which was removed after 24 hours. Any EGFR inhibitors were added at this stage. The medium was replaced every 2 to 3 days and cells were maintained in the presence of appropriate inhibitors until analysis at day 6. All cultures were maintained at 37°C in a humidified atmosphere of 5% CO2 in air.
Immunohistochemistry
The immunoreactivity of antigens masked by tissue processing was restored by boiling sections for 10 minutes in 10 mmol/L citric acid buffer, pH 6.0, in a microwave oven which, in the case of all antibodies except anti-RXR-α, was preceded by digestion for 1 minute in 0.1% (w/v) trypsin in 0.1% (w/v) CaCl2, pH 7.6. Endogenous peroxidase activity in tissue sections was blocked by incubation with 3% (v/v) hydrogen peroxide solution for 10 minutes and endogenous avidin-binding sites were blocked using an avidin/biotin kit (Vector Laboratories, Peterborough, UK) according to the manufacturer’s protocol.
Immunohistochemistry for CKs was performed using an indirect streptavidin ABC immunoperoxidase method (Dako Cytomation Ltd., Ely, UK). For the detection of low-density PPAR-γ and RXR-α antigens, a more sensitive tyramide-based catalyzed signal amplification method (catalyzed signal amplification peroxidase system, Dako Cytomation Ltd.) was used. The manufacturer’s recommendations were followed for catalyzed signal amplification except that to optimize the signal-to-noise ratio, the primary antibody incubation time was extended to 30 minutes and the secondary antibody incubation and amplification stages were reduced to 5 minutes each. All labeling series were run with relevant negative and positive controls. Slides were counterstained with hematoxylin, dehydrated, and mounted in DPX (from Fisher Scientific UK, Loughborough, UK).
Immunofluorescence
Cells grown on slides were fixed in a 1:1 mixture of methanol and acetone, air-dried, and incubated with titrated primary antibody for 16 hours at 4°C, before washing and incubation in secondary antibody conjugated to Alexa 488 (Molecular Probes, supplied by Cambridge Bioscience, Cambridge, UK). Hoechst 33258 (0.1 μg/ml; Sigma-Aldrich Ltd.) was included in the penultimate wash to visualize nuclei. Slides were observed on an Olympus BX60 microscope under epifluorescence illumination.
Ribonuclease Protection Assays
To extract RNA from cell monolayers, Trizol (Invitrogen Ltd.) was added to the cell monolayer (1 ml/10 cm2) and the cell lysate was scraped into a microcentrifuge tube. RNA was extracted as recommended by the manufacturer. Part-length cDNA fragments of the coding region for human CK13 (KRT13), CK14 (KRT14), and CK20 (KRT20) were amplifed by polymerase chain reaction (PCR) using the following primer sets: KRT13: forward primer, TCCACACCAAGAGTGCAGAG and reverse primer, TCTGGCACTCCATCTCACTG; KRT14: forward primer, TTCTGAACGAGATGCGTGAC and reverse primer, GCAGCTCAATCT-CCAGGTTC; KRT20: forward primer, AAGCCTCCAAGAAATGCCTC and reverse primer, CTGTGAGATCGCTCCCATAG. The PCR products for the CKs were cloned into pGEM-T Easy (Promega, Southampton, UK). GAPDH25 was used as an internal riboprobe control. 32P-labeled anti-sense transcripts of the cDNAs of interest were generated from linearized plasmids using an In Vitro transcription kit (Promega), according to the manufacturer’s protocol. After DNase treatment, riboprobes were purified by passage through Chromaspin 30-DEPC columns (BD Biosciences Clontech UK, Oxford, UK).
Approximately 2 fmol of each riboprobe were mixed and hybridized to 5 μg of total RNA using a RPAIII kit (Ambion Ltd., Huntingdon, UK), according to the manufacturer’s protocols. Products were separated on 5% denaturing polyacrylamide gels (Sequagel; Flowgen, Lichfield, UK), visualized by autoradiography and quantified by means of a phosphorimager (GS-525 Molecular Imager System; Bio-Rad, Hemel Hempstead, UK). Signal intensity was corrected for loading inaccuracies by normalizing against the GAPDH signal and changes in intensity between experimental conditions were presented as fold changes; where no signal was detected the fold-change was left blank.
Immunoblotting
Cell cultures were lysed in 25 mmol/L Hepes (pH 7.4), 125 mmol/L NaCl, 10 mmol/L NaF, 10 mmol/L sodium orthovanadate, 10 mmol/L sodium pyrophosphate, 0.2% (w/v) sodium dodecyl sulfate, 0.5% (w/v) sodium deoxycholate, 1% (w/v) Triton X-100, 1 μg/ml aprotinin, 10 μg/ml leupeptin, and 100 μg/ml phenylmethyl sulfonyl fluoride. Lysates were sheared by passing 3 times through a 21-gauge needle and left on ice for 30 minutes, before microcentrifugation at 10,000 × g for 30 minutes at 4°C. The protein concentrations of supernatants were measured by the Bradford assay (Pierce, supplied by Perbio Science UK Ltd., Cheshire, UK). Cell extracts were resolved electrophoretically on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were incubated with primary antibodies for 16 hours at 4°C. Bound antibody was detected with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence using the ECL detection kit (Amersham Pharmacia).
Rapid-Amplification of cDNA Ends (RACE) and Cloning
RACE was performed using the 5′-Full RACE Core Set from Takara Biomedicals (Takara Shuzo Co. Ltd., Shiga, Japan), using the recommended protocol, which is briefly outlined. First strand cDNA was synthesized from colon RNA (5 μg) (Stratagene), using 5 U/μl AMV reverse transcriptase, 200 μmol/L 5′ P-GTTCTGCATGGCC primer, 40 U/μλ RNase inhibitor in room temperature buffer and incubated for 10 minutes at 30°C, 60 minutes at 50°C, 2 minutes at 80°C, and cooled to 4°C. The RNA template was digested using 60 U/μl RNase H at 30°C for 1 hour and the cDNA was purified by ethanol precipitation. The cDNA was circularized using 40 U/μl T4 RNA ligase with 40% (w/v) PEG 6000 in RNA (ssDNA) ligation buffer and incubated for 18 hours at 15°C. PCR was performed using the ligated cDNA, 25 mmol/L MgCl2, 10 mmol/L dNTP, 50 μmol/L primers (3′3 GGACCTGTTTGTTGGCAATG and 5′3 CTGTGAGATCGCTCCCATAG) and Q-solution and Taq polymerase obtained from Qiagen Ltd. (Crawley, West Sussex, UK) using 94°C for 3 minutes followed by 94°C for 30 seconds; 50 to 65°C for 30 seconds, and 72°C for 2 minutes for 25 cycles. A second round of PCR was performed using the first round PCR products (diluted 1 in 10) as template, under the same conditions, using nested primers 3′4 CAATGAGAAAATGGCCATGC and 5′4 CGTGTGTCTGGAGTTGGAGA. The PCR products were purified using QIAquick Nucleotide Removal (Qiagen Ltd.) and visualized on a 1.5% agarose gel. The PCR product was ligated into pGEM-T Easy vector overnight at 4°C, using rapid ligation buffer and T4 DNA ligase obtained from Promega and transformed into JM109 competent cells as outlined in the manufacturer’s protocol. The transformed cells were plated on to LB broth plates for 18 hours at 37°C with 50 μg/ml ampicillin selection. The plasmids were purified using the QIAprepSpin MiniPrep Kit (Qiagen Ltd.) and the insert was sequenced after excision with EcoRI.
Statistical Analysis
Comparisons between groups were analyzed using two-tailed Student’s t-test. Differences were considered significant when P < 0.05.
Results
Immunohistology
The expression of PPAR-γ and RXR-α was examined in specimens of normal urothelium of both bladder and ureteric origins. PPAR-γ expression was nuclear and occasionally focally cytoplasmic (Figure 1a). The nuclear localization was differentiation-associated, with the most intense labeling confined to the nuclei of the superficial cell layer. When present, cytoplasmic labeling was generally identified in all cell layers, but most conspicuous in the superficial cells. RXR-α labeling patterns were similar to PPAR-γ, with nuclear expression that varied in intensity (Figure 1b). There was focal negativity of suprabasal and basal cell nuclei and a more pronounced cytoplasmic labeling than seen with PPAR-γ.
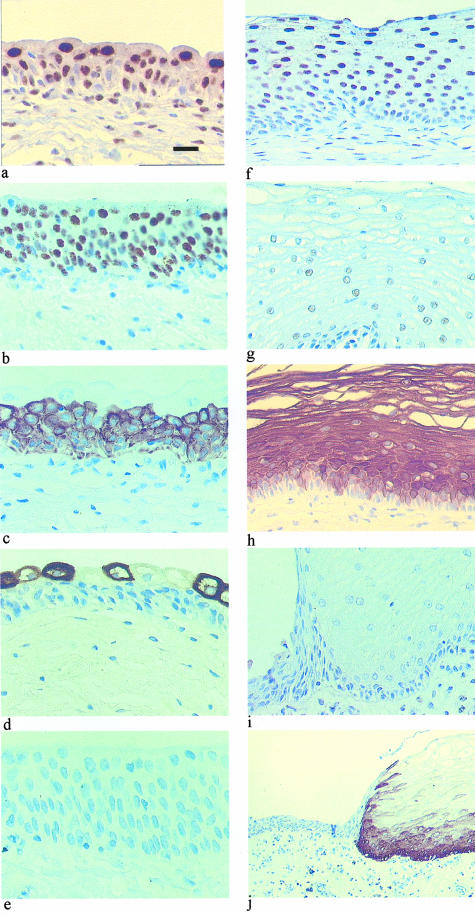
Figure 1.
Immunohistochemical labeling for PPAR-γ (a and f), RXR-α (b and g), CK13 (c and h), CK20 (d and i), and CK14 (e and j) in normal human urothelium (a to e) and cornifying squamous metaplasia (f to j). Scale bar: 50 μm (a to i); 125 μm (i); and 440 μm (j).
Urothelia from the urinary bladder and the ureter were reactive with antibodies to CK13 and CK20. Expression of CK13 was confined to the cytoplasm of basal and intermediate cells, with superficial cells spared (Figure 1c). CK20 expression was almost exclusively confined to the superficial cell layer, with a variable number of superficial cells displaying no reaction (Figure 1d). There was no labeling of CK14 in any of the control cases (Figure 1e).
In all three cases of squamous metaplasia, PPAR-γ showed diffuse nuclear expression involving all suprabasal cell layers (Figure 1f), whereas expression of RXR-α was very weak to absent in cornifying squamous metaplasia (Figure 1g). In areas of metaplasia, associated with intercellular prickles, CK14 was expressed throughout the epithelium and CK13 was basally excluded (Figure 1, h and j); CK20 was not expressed (Figure 1i). The switch in CK expression was especially apparent at the junction between normal and squamous epithelia, where CK14 positivity was always accompanied by CK13 negativity of the basal compartment, whereas CK14-negative regions showed basal CK13 expression (Figure 2). In addition, none of the three cases of squamous metaplasia showed expression of uroplakins UPIa, UPIb, UPIII, or of the simple epithelial components CK8, CK18, and CK7 usually expressed by urothelium (not shown).
Figure 2.
Immunohistochemical labeling for CK13 (a and b) and CK14 (c and d) in normal human urothelium (a and c) and a region of noncornifying squamous metaplasia within the same specimen (b and d). Scale bar, 20 μm.
Effects of TZ on CK Isotype Expression
We have previously shown that NHU cells grown in culture show changes in CK isotype expression compared to urothelium in situ, with maintenance of CK7, CK8, CK17, CK18, and CK19, loss of CK13 and CK20, and de novo CK14 expression.13 To investigate the effects of the PPAR-γ agonist, TZ, on the expression of CK isotypes, NHU cells were treated with 1 μmol/L TZ and analyzed for up to 6 days for expression of PPAR-γ, RXR-α, CK13, CK14, CK19, and CK20 (Table 1).
Table 1.
Summary of the Effects of TZ (1 μmol/L) and PD153035 (1 μmol/L) on CK Expression in NHU Cells
| Expression | Treatment | CK13 | CK14 | CK20 |
|---|---|---|---|---|
| Antigen | TZ | ↑ | ↓↓ | ↑ |
| PD153035 | ↑ | → | ↑ | |
| TZ and PD153035 | ↑↑↑ | ↓↓ | ↑↑↑ | |
| Protein | TZ | — | ↓↓ | n/a |
| PD153035 | — | → | n/a | |
| TZ and PD153035 | ↑↑↑ | ↓↓ | n/a | |
| mRNA | TZ | ↑ | → | → |
| PD153035 | ↑ | → | → | |
| TZ and PD153035 | ↑↑↑ | → | ↑↑↑ |
The change in expression was assessed subjectively as either increasing (↑) or decreasing (↓) by a small (↑) to large (↑↑↑) extent. The following were represented as, no change (→), no expression (–), and not assessed (n/a).
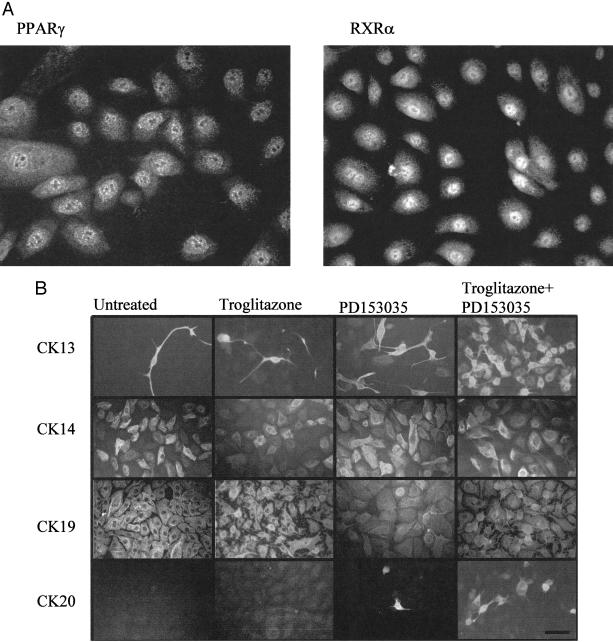
In NHU cells, PPAR-γ and RXR-α were shown to be expressed in the nucleus, with weaker cytoplasmic expression (Figure 3A). By immunofluorescence in untreated control cells, CK19 was expressed homogeneously by all cells, CK14 was expressed by at least 60% of cells ranging in intensity from weak to strong, CK13 was expressed by rare dispersed cells (<2.0%), but CK20 was never expressed (Figure 3B). Treatment with either TZ or PD153035 alone resulted in a small increase in the percentage of CK13+ cells (TZ, 6.3% ± 4.9 SD; PD153035, 15.9% ± 0.4 SD; P < 0.01) and the appearance of rare, dispersed CK20+ cells. A synergistic effect was observed when cells were pretreated with TZ and maintained with the EGFR inhibitor, PD153035, with 68.0% ± 3.5 SD (P < 0.01) expressing CK13 and 3.7% ± 1.2 SD cells showing intense and unequivocal expression of CK20 expression (Figure 3B). CK14 expression was inhibited by TZ and PD153035 (21.0% ± 3.2 SD), but there was no apparent effect on the number of CK14+ cells when cells were treated with either TZ or PD153035 alone (43.7% ± 2.8 SD and 45.0% ± 4.0 SD, respectively). However, a reduction in the intensity of CK14 fluorescence was noted when cells were treated with TZ alone. Therefore, it was important to confirm and quantify these results by immunoblotting.
Figure 3.
Localization of antigens by immunofluorescence microscopy. A: PPAR-γ and RXR-α localization in NHU cells. NHU cells were seeded at 2 × 105 cells/ml on to glass slides and fixed after 24 hours. B: Treatment of NHU cells with TZ and an EGFR inhibitor resulted in the synergistic induction of CK13 and CK20 protein expression, above that induced by either mediator alone. NHU cells were seeded at 2 × 105 cells/ml onto glass slides, allowed to attach and treated for 24 hours with or without TZ (1 μmol/L), before replacement with medium with or without PD153035 (1 μmol/L), as detailed in the figure. Medium was changed every 3 days. The slides were fixed after 6 days and immunofluorescence performed for CK13, CK14, CK19, and CK20. The percentage of cells positive for CK13, CK14, and CK20 (mean ± SD) were determined from three fields of view (minimum of 50 cells per field), from three independent cell lines, using Hoechst 33258 staining of nuclei to determine total number of cells. Exposure times were kept constant for comparison purposes. Scale bar, 50 μm.
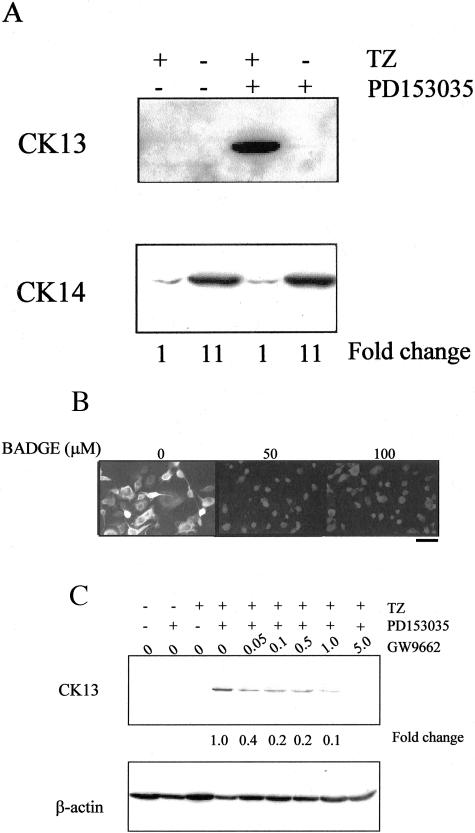
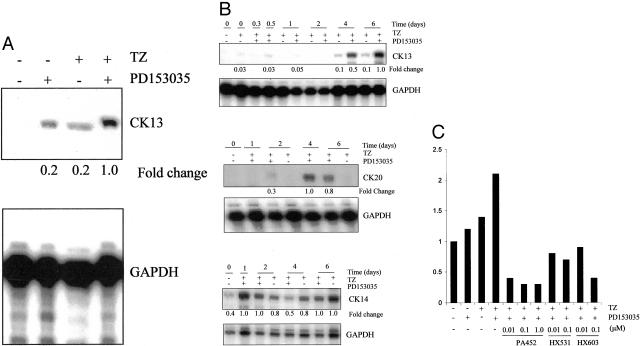
CK13 expression was induced by combined treatment with TZ and PD153035 at 6 days (Figure 4A). No CK13 protein expression was detected by immunoblotting in cells treated with either TZ or PD153035 alone, reflecting the small proportion of cells that were positive by immunofluorescence (Figure 3B). CK14 expression was reduced, relative to control cultures, by TZ in the presence (11-fold decrease at 6 days) or absence (11-fold decrease at 6 days) of PD153035, but PD153035 alone had no effect on CK14 protein expression at either time point (summarized in Table 1).
Figure 4.
Synergistic effect of TZ and PD153035 on CK13 and CK14 protein expression. A: NHU cells were treated with or without TZ (1 μmol/L) for 24 hours, followed by PD153035 (1 μmol/L) for 6 days. Equal amounts of protein (40 μg/lane) were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel, electrophoresed, and transferred to nitrocellulose membrane. Membranes were incubated with antibodies to CK13 and CK14. Representative of one of three similar immunoblotting experiments. For analysis of relative expression of CK13, cells treated with both TZ and PD153035 were taken to be 1.0, whereas for CK14 expression, cells treated with TZ alone were taken to be 1.0. B: Inhibition of TZ-mediated effect by PPAR-γ antagonist. NHU cells were seeded at 2 × 105 cells/ml onto glass slides, allowed to attach, and pretreated for 1 hour at 0, 50, and 100 μmol/L concentrations of BADGE, followed by 24 hours with TZ (1 μmol/L) and BADGE at the same concentrations. The medium was replaced to contain PD153035 (1 μmol/L) and BADGE at the same concentrations and subsequently every 3 days. After 6 days the slides were fixed and immunofluorescence for CK13 was performed as outlined in the Materials and Methods. Exposure times were kept constant for comparison purposes. C: NHU cells were treated with or without GW9662 for 3 hours and then in the presence of GW9662 and TZ (1 μmol/L) for 24 hours, followed by GW9662 and PD153035 (1 μmol/L) for 6 days. Cell lysates were analyzed by immunoblotting for CK13. For comparison of relative expression, treatment with TZ and PD153035 was taken to be 1.0. The membrane was reprobed with β-actin to demonstrate equal loading of protein. Scale bar, 30 μm (B).
To demonstrate that the TZ-mediated increase on CK13 expression was because of specific activation of PPAR-γ, cells were treated with TZ and PD153035 as above, but in the presence or absence of the PPAR-γ-specific inhibitor, BADGE.19 CK13 protein expression induced by TZ and PD153035 was totally blocked by BADGE (Figure 4B), as were changes in CK14 and CK20 expression (data not shown). Similar results were obtained using another specific antagonist of PPAR-γ, GW9662,20 at 1 to 5 μmol/L concentration (Figure 4C). This suggests that the observed changes in CK expression were because of the specific activation of PPAR-γ.
Expression of CK13, CK14, and CK20 Transcripts
When cells were treated with TZ or PD153035 alone, there was a slight increase in CK13 mRNA expression; this was greatly increased (fivefold) when cells were treated with both TZ and PD153035 (Figure 5A). In agreement with the immunofluorescence and immunoblotting data, CK13 and CK20 mRNA expression levels were maximally induced when NHU cells were treated with both TZ and PD153035 (Figure 5B). The greatest induction of CK13 and CK20 transcripts occurred between 4 and 6 days. CK14 mRNA expression showed no clear change between cells treated with TZ and maintained in the presence or absence of PD153035.
Figure 5.
Ribonuclease protection assay to show synergy between TZ and inhibition of EGFR in the induction of CK13 and CK20 mRNA. A: NHU cells were treated for 24 hours in the presence or absence of TZ (1 μmol/L) and then for 4 days in the presence or absence of medium containing PD153035 (1 μmol/L). Total RNA was extracted and 5 μg was hybridized with 32P-labeled human CK13 and GAPDH riboprobes, electrophoresed, and quantified as described in the Materials and Methods. Band intensity was quantified by means of a phosphorimager and normalized against the GAPDH signal, which was included as an internal standard for loading efficiency. For comparison of relative expression, the cells treated with PD153035 and TZ cells were taken to be 1.0. Representative of three similar experiments. B: NHU cells were treated for 24 hours in the presence of TZ (1 μmol/L). Next, the cells were treated with PD153035 (1 μmol/L) and samples were taken at the times indicated. Controls that omitted PD153035 were included at days 2 and 6 to show the effect of TZ alone. Total RNA was extracted and 5 μg was hybridized with 32P-labeled human CK13, CK14, CK20, and GAPDH riboprobes. The samples were electrophoresed on a 5% polyacrylamide gel. Band intensity was quantified by means of a phosphorimager and normalized as described in A. The maximum level of expression was taken to be 1.0 and all of the other samples were expressed relative to the maximum. Representative of three similar experiments. C: NHU cells were pretreated for 1 hour with or without PA452, HX531, or HX603 at concentrations indicated, before incubation in the presence or absence of TZ and PA452, HX531, or HX603 for 24 hours, as indicated. Next, cells were either treated with or without PD153035 (1 μmol/L) and PA452, HX531, or HX603 for 4 days changing the medium and inhibitors every 2 days. Total RNA was extracted and 5 μg was hybridized to the CK13 riboprobe, electrophoresed, and band intensity was quantified by means of a phosphorimager and normalized as described in A. Expression levels of untreated NHU cells was taken to be 1.0 and all other samples were compared to the untreated cells.
To address the role of RXR, which is an obligate partner in the transcription factor formed on activation of PPAR-γ, NHU cells were treated with the RXR inhibitors, PA452, HX531, and HX603, at concentrations specific for inhibition of RXR (Figure 5C).21,22,26 There was an approximate threefold inhibition in the induction of CK13 mRNA by TZ and PD153035 brought about by the presence of PA452 (0.01 to 1.0 μmol/L) or HX603 (1 μmol/L), and a lesser inhibition was seen with HX531 (Figure 5C). Of the three inhibitors PA452 has been reported to be the most RXR-specific inhibitor.26
When used in combination on NHU cells, both TZ and PD153035 showed dose-dependent effects on CK13 mRNA expression, with maximum effect observed at 0.5 to 1.0 μmol/L PD153035 and 5 μmol/L TZ. An alternative EGF receptor inhibitor, AG1478, also enhanced CK13 mRNA expression in TZ-treated cells in a dose-dependent manner, with the maximal effect between 1 to 5 μmol/L. Treatment of NHU cells with the weak natural PPAR-γ ligand 15-dPGJ2, in the presence of PD153035, had no effect on the expression of CK13 mRNA and was inhibitory at higher concentrations. Furthermore, when cells were treated with the PPAR-α ligand, clofibrate (CF; EC50 = 55 μmol/L27), CF had no effect on CK13 mRNA expression, even when cells were treated with PD153035, suggesting that the effect was PPAR-γ-specific (data seen by referees).
Transcriptional Start Site of CK20
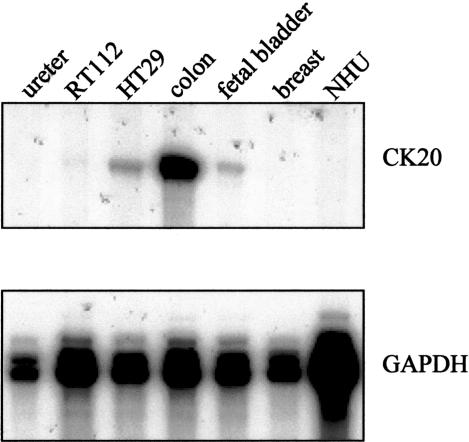
To determine whether PPAR-γ regulates CK gene expression directly or indirectly, we needed to know if there are any PPRE binding sites within the promoter regions of CK13 or CK20. The transcriptional start site for CK13 has been reported,28 but that of CK20 is unknown. A riboprobe designed to span the translational ATG start site of CK20 was used in a ribonuclease protection assay and demonstrated that CK20 was highly expressed by colon, moderately by the colonic HT29 cell line and whole fetal bladder, and very weakly by the urothelial cell carcinoma-derived RT112 cell line. There was no CK20 detected in ureteric urothelium, breast, or cultured NHU cells (Figure 6). The CK20 band was consistently 235 bp in size, suggesting that the transcriptional start site was 235 bp upstream of the 3′ oligonucleotide end point and 54 bp upstream of the start ATG. The start site was confirmed by RACE on colon-derived RNA, using nested primers in the region of interest and the same site was also identified using promoter prediction software (www.fruitfly.org).
Figure 6.
Identification of CK20 transcriptional start site. Total RNA was extracted from ureter, human transitional cell carcinoma cell line RT112, and human colon adenocarcinoma cell line HT29. Colon, fetal bladder, and breast RNA were purchased. Five μg of total RNA was hybridized with a riboprobe that spanned the ATG start site of CK20 and analyzed by ribonuclease protection assays. Protected fragments of 235 bp were electrophoresed on a 5% polyacrylamide gel and visualized by autoradiography.
The 5′ upstream 1000-bp sequences of CK20 and CK1328 were analyzed to identify predicted transcription factor binding sites using the AliBaba2 software (www.gene-regulation.com). No PPRE sites were found in either of the sequences, but there were a number of transcription factor binding sites common to both genes, including AP-1 (one in CK13, two in CK20), nuclear factor-κB (two in CK13, two in CK20), and C/EBPα (four in CK13, two in CK20) in the 1000-bp sequence upstream of the transcriptional start site.
Discussion
This is the first evidence to indicate that activation of PPAR-γ can modulate the pattern of CK isotype expression in epithelial cells and by implication, affect the differentiation state. Although PPAR-γ-activated signaling has been implicated in the regulation of differentiation of adipocytes29 and carcinomas of prostate, colon, pancreas, and breast,30–33 to date there has been no direct evidence that it may play a role in regulating the differentiation of normal epithelial cells. Expression of PPAR-γ has been described in the developing urothelium of the mouse urogenital sinus and in the mature urothelium of mice, rabbits and man.34,35 Although activation of PPAR-γ has been shown to suppress growth of normal and malignant urothelial cells in vitro,36 it has not previously been associated with urothelial cytodifferentiation. In view of the requirement for both PPAR-γ and RXR-α to activate the PPAR-γ pathway, it was noteworthy that in urothelium, both cornifying and noncornifying forms of squamous metaplasia were accompanied by a pronounced reduction in RXR-α expression and a loss of accentuation of PPAR-γ expression by superficial cells.
The growth of NHU cells in vitro represents a model for the regenerative phenotype acquired by urothelial cells in vivo in response to injury.37,38 This highly proliferative and migratory phenotype is driven through an EGFR autocrine loop (manuscript in preparation). Our data shows that when the EGFR pathway is inhibited, activation of the PPAR-γ signaling pathway acts synergistically to switch human urothelial cells from a squamous metaplastic phenotype and promote transitional differentiation, as indicated by the induction of CK13 and CK20 expression, at the expense of the squamous differentiation marker, CK14. This CK13-CK14 switch is characteristic of squamous metaplasia of the bladder of both cornifying and noncornifying types, in which it appears that CK14 expression is mutually exclusive of basal CK13 expression.
The requirement for the EGFR pathway to be blocked for TZ to effect a change in CK expression is intriguing as it implies dominance of proliferation over differentiation and a requirement to inhibit proliferation to permit differentiation. The most likely mechanism is the mitogen-activated protein kinase (MAPK) pathway, which is downstream of EGFR activation and has previously been shown to phosphorylate and inhibit PPAR-γ activation.39,40
The effects mediated by TZ were demonstrated to be PPAR-γ-specific because they were blocked by two independent PPAR-γ-specific inhibitors. By contrast, 15-dPGJ2, which is purported to be a weak natural ligand for PPAR-γ, had no effect on CK13 mRNA expression, even in the presence of the EGF inhibitor. Although the different effects could be because of the recruitment of different co-activators by different PPAR-γ ligands, this seems unlikely and we are inclined to accept recent suggestions that the PGJ2 derivatives are not effective PPAR-γ ligands because of the low potencies in comparison to the synthetic ligands.41
Activation of the PPAR-γ pathway requires heterodimerization of ligand-bound PPAR-γ with the RXR to form a transcription factor that binds specific PPRE in the promoters of target genes.42–44 The response to PPAR-γ activation has been reported to be synergized by co-activation of the RXR partner.45,46 Our data suggest that although the NHU cells were grown in a retinoid-free culture medium, the RXR-α is ligand-bound, as RXR inhibitors whose mechanism of action is to inhibit the ligand-bound RXR complex26 suppressed the induction of CK13 mRNA by TZ and PD153035. The nature of the predicted RXR-α ligand is unknown, although recent reports suggest that n-3 polyunsaturated fatty acids may act as endogenous RXR-α ligands.47 Therefore, it appears that our data supports a model in which RXR-α is ligand-bound in NHU cells and contributes to the TZ induction of CK13 mRNA via the formation of a fully activated RXR-α-PPAR-γ heterodimer complex.
It has been reported that CK14 gene expression is repressed by keratin response elements that act as a docking platform for nuclear receptors such as RAR, T3R, and GR.16,48,49 Although our results indicate that CK13 and CK20 expression was transcriptionally regulated, it seems unlikely that this was the underlying mechanism involved in the PPAR-γ-mediated down-regulation of the CK14 polypeptide because there was no accompanying change in KRT14 gene transcription, suggesting that the decrease in CK14 expression was regulated at the levels of protein translation and degradation.
The long time delay between PPAR-γ activation and the induction of CK13 and CK20 expression, and the lack of PPRE sites in the promoter regions of CK13 and CK20, has led us to postulate that activated PPAR-γ-RXR-α heterodimers directly induce the expression of a transcription factor, which in turn binds to the promoter region of CK13 and CK20 to induce expression. PPAR-γ clearly plays a central role in the induction of CK13 and CK20 expression because the induction is blocked when PPAR-γ-specific inhibitors are used. This mechanism is plausible because PPAR-γ has been demonstrated to regulate the expression of a number of transcription factors, including AP-1, LXR, and STAT proteins.50–52 We have identified a number of transcription factor binding sites that are present in the upstream promoter regions of both CK13 and CK20 genes and could be potential candidates regulated by PPAR-γ. As well as the transcription factor binding sites for AP-1 and nuclear factor-κB, components of which have been reported to be down-regulated by PPAR-γ, a number of other common sites were identified. These included binding sites for the transcription factor C/EBP, which has been reported to be involved in differentiation.53,54
In conclusion, specific activation of PPAR-γ can induce expression of CK13 and CK20 genes in NHU cells, but requires that the EGFR pathway is inactive. Transitional cytodifferentiation in urothelium is accompanied by expression of the urothelium-specific uroplakin genes and our studies indicate that these genes are also induced by activation of the PPAR-γ pathway.17 Determining the role of PPAR-γ in urothelial differentiation and the balance with proliferation will provide a greater understanding of the molecular processes involved in regulating the differentiation of urothelium and other epithelia.
Acknowledgments
We thank Dr. John Johnson of Parke-Davis for supplying the troglitazone, Dr. Hiroyuki Kagechika of University of Tokyo for providing the RXR inhibitor compounds, Dr. Timothy Wilson for GW9662, Miss Kristy Driver for her assistance with immunofluorescence experiments, and our clinical colleagues who aided in the supply of tissues for research.
Footnotes
Address reprint requests to Professor J. Southgate, Jack Birch Unit of Molecular Carcinogenesis, Department of Biology, University of York, York YO10 5YW, United Kingdom. E-mail: js35@york.ac.uk.
Supported by Wellcome Trust (grant GR061262MA) and York Against Cancer (grant to J.S.).
References
- Wu RL, Osman I, Wu XR, Lu ML, Zhang ZF, Liang FX, Hamza R, Scher H, Cordon-Cardo C, Sun TT. Uroplakin II gene is expressed in transitional cell carcinoma but not in bilharzial bladder squamous cell carcinoma: alternative pathways of bladder epithelial differentiation and tumor formation [published erratum appears in Cancer Res 1998 Jul 1;58(13):2904]. Cancer Res. 1998;58:1291–1297. [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Southgate J, Harnden P, Trejdosiewicz LK. Cytokeratin expression patterns in normal and malignant urothelium: a review of the biological and diagnostic implications. Histol Histopathol. 1999;14:657–664. doi: 10.14670/HH-14.657. [DOI] [PubMed] [Google Scholar]
- Moll R, Lowe A, Laufer J, Franke WW. Cytokeratin 20 in human carcinomas. A new histodiagnostic marker detected by monoclonal antibodies. Am J Pathol. 1992;140:427–447. [PMC free article] [PubMed] [Google Scholar]
- Moll R, Schiller DL, Franke WW. Identification of protein IT of the intestinal cytoskeleton as a novel type I cytokeratin with unusual properties and expression patterns. J Cell Biol. 1990;111:567–580. doi: 10.1083/jcb.111.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnden P, Eardley I, Joyce AD, Southgate J. Cytokeratin 20 as an objective marker of urothelial dysplasia. Br J Urol. 1996;78:870–875. doi: 10.1046/j.1464-410x.1996.23511.x. [DOI] [PubMed] [Google Scholar]
- Harnden P, Mahmood N, Southgate J. Expression of cytokeratin 20 redefines urothelial papillomas of the bladder. Lancet. 1999;353:974–977. doi: 10.1016/S0140-6736(98)05383-5. [DOI] [PubMed] [Google Scholar]
- Harnden P, Southgate J. Cytokeratin 14 as a marker of squamous differentiation in transitional cell carcinomas. J Clin Pathol. 1997;50:1032–1033. doi: 10.1136/jcp.50.12.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbach SB, Howe PR. Tissue changes following deprivation of fat-soluble A vitamin. J Exp Med. 1925;42:753–777. doi: 10.1084/jem.42.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks RM. Hyperplasia and cornification of the transitional epithelium in the vitamin A-deficient rat. Changes in fine structure of the cells. J Ultrastruct Res. 1968;22:206–230. doi: 10.1016/s0022-5320(68)90016-6. [DOI] [PubMed] [Google Scholar]
- Molloy CJ, Laskin JD. Effect of retinoid deficiency on keratin expression in mouse bladder. Exp Mol Pathol. 1988;49:128–140. doi: 10.1016/0014-4800(88)90027-5. [DOI] [PubMed] [Google Scholar]
- Altucci L, Gronemeyer H. The promise of retinoids to fight against cancer. Nat Rev Cancer. 2001;1:181–193. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- Southgate J, Hutton KA, Thomas DF, Trejdosiewicz LK. Normal human urothelial cells in vitro: proliferation and induction of stratification. Lab Invest. 1994;71:583–594. [PubMed] [Google Scholar]
- Morrison RF, Farmer SR. Hormonal signaling and transcriptional control of adipocyte differentiation. J Nutr. 2000;130:3116S–3121S. doi: 10.1093/jn/130.12.3116S. [DOI] [PubMed] [Google Scholar]
- Blanquart C, Barbier O, Fruchart JC, Staels B, Glineur C. Peroxisome proliferator-activated receptors: regulation of transcriptional activities and roles in inflammation. J Steroid Biochem Mol Biol. 2003;85:267–273. doi: 10.1016/s0960-0760(03)00214-0. [DOI] [PubMed] [Google Scholar]
- Tomic-Canic M, Day D, Samuels HH, Freedberg IM, Blumenberg M. Novel regulation of keratin gene expression by thyroid hormone and retinoid receptors. J Biol Chem. 1996;271:1416–1423. doi: 10.1074/jbc.271.3.1416. [DOI] [PubMed] [Google Scholar]
- Varley CL SJ, Lee W-C, Holder J, Diggle C, Selby PJ, Trejdosiewicz LK, Southgate J: Role of PPARγ and EGFR signalling in the urothelial terminal differentiation programme. J Cell Sci (in press) [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Wright HM, Clish CB, Mikami T, Hauser S, Yanagi K, Hiramatsu R, Serhan CN, Spiegelman BM. A synthetic antagonist for the peroxisome proliferator-activated receptor gamma inhibits adipocyte differentiation. J Biol Chem. 2000;275:1873–1877. doi: 10.1074/jbc.275.3.1873. [DOI] [PubMed] [Google Scholar]
- Leesnitzer LM, Parks DJ, Bledsoe RK, Cobb JE, Collins JL, Consler TG, Davis RG, Hull-Ryde EA, Lenhard JM, Patel L, Plunket KD, Shenk JL, Stimmel JB, Therapontos C, Willson TM, Blanchard SG. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry. 2002;41:6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- Ebisawa M, Umemiya H, Ohta K, Fukasawa H, Kawachi E, Christoffel G, Gronemeyer H, Tsuji M, Hashimoto Y, Shudo K, Kagechika H. Retinoid X receptor-antagonistic diazepinylbenzoic acids. Chem Pharm Bull (Tokyo) 1999;47:1778–1786. doi: 10.1248/cpb.47.1778. [DOI] [PubMed] [Google Scholar]
- Ebisawa M, Ohta K, Kawachi E, Fukasawa H, Hashimoto Y, Kagechika H. Novel retinoidal tropolone derivatives. Bioisosteric relationship of tropolone ring with benzoic acid moiety in retinoid structure. Chem Pharm Bull (Tokyo) 2001;49:501–503. doi: 10.1248/cpb.49.501. [DOI] [PubMed] [Google Scholar]
- Southgate J, Masters JR, Trejdosiewicz LK. Culture of human urothelium. Freshney RI, Freshney MG, editors. New York: J Wiley and Sons, Inc.; Culture of Epithelial Cells. 2002:pp 381–400. [Google Scholar]
- Southgate J, Hutton KA, Thomas DF, Trejdosiewicz LK. Normal human urothelial cells in vitro: proliferation and induction of stratification. Lab Invest. 1994;71:583–594. [PubMed] [Google Scholar]
- Lobban ED, Smith BA, Hall GD, Harnden P, Roberts P, Selby PJ, Trejdosiewicz LK, Southgate J. Uroplakin gene expression by normal and neoplastic human urothelium. Am J Pathol. 1998;153:1957–1967. doi: 10.1016/S0002-9440(10)65709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kawada T, Yamamoto T, Goto T, Taimatsu A, Aoki N, Kawasaki H, Taira K, Yokoyama KK, Kamei Y, Fushiki T. Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma. J Biol Chem. 2002;277:16906–16912. doi: 10.1074/jbc.M200585200. [DOI] [PubMed] [Google Scholar]
- Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- Waseem A, Alam Y, Dogan B, White KN, Leigh IM, Waseem NH. Isolation, sequence and expression of the gene encoding human keratin 13. Gene. 1998;215:269–279. doi: 10.1016/s0378-1119(98)00297-2. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Kubota T, Koshizuka K, Williamson EA, Asou H, Said JW, Holden S, Miyoshi I, Koeffler HP. Ligand for peroxisome proliferator-activated receptor gamma (troglitazone) has potent antitumor effect against human prostate cancer both in vitro and in vivo. Cancer Res. 1998;58:3344–3352. [PubMed] [Google Scholar]
- Sarraf P, Mueller E, Jones D, King FJ, DeAngelo DJ, Partridge JB, Holden SA, Chen LB, Singer S, Fletcher C, Spiegelman BM. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat Med. 1998;4:1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- Mueller E, Sarraf P, Tontonoz P, Evans RM, Martin KJ, Zhang M, Fletcher C, Singer S, Spiegelman BM. Terminal differentiation of human breast cancer through PPAR gamma. Mol Cell. 1998;1:465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- Toyota M, Miyazaki Y, Kitamura S, Nagasawa Y, Kiyohara T, Shinomura Y, Matsuzawa Y. Peroxisome proliferator-activated receptor gamma reduces the growth rate of pancreatic cancer cells through the reduction of cyclin D1. Life Sci. 2002;70:1565–1575. doi: 10.1016/s0024-3205(01)01524-7. [DOI] [PubMed] [Google Scholar]
- Guan Y, Zhang Y, Davis L, Breyer MD. Expression of peroxisome proliferator-activated receptors in urinary tract of rabbits and humans. Am J Physiol. 1997;273:F1013–F1022. doi: 10.1152/ajprenal.1997.273.6.F1013. [DOI] [PubMed] [Google Scholar]
- Jain S, Pulikuri S, Zhu Y, Qi C, Kanwar YS, Yeldandi AV, Rao MS, Reddy JK. Differential expression of the peroxisome proliferator-activated receptor gamma (PPARgamma) and its coactivators steroid receptor coactivator-1 and PPAR-binding protein PBP in the brown fat, urinary bladder, colon, and breast of the mouse. Am J Pathol. 1998;153:349–354. doi: 10.1016/s0002-9440(10)65577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiro KI, Hayashi Y, Kita A, Tamatani T, Chlenski A, Usuda N, Hattori K, Reddy JK, Oyasu R. Role of peroxisome proliferator-activated receptor gamma and its ligands in non-neoplastic and neoplastic human urothelial cells. Am J Pathol. 2001;159:591–597. doi: 10.1016/s0002-9440(10)61730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southgate J, Harnden P, Selby PJ, Thomas DFM, Trejdosiewicz LK. Urothelial tissue regulation: unraveling the role of the stroma. Adv Exp Med Biol. 1999;462:19–30. [PubMed] [Google Scholar]
- Smith BA, Kennedy W, Harnden P, Selby PJ, Trejdosiewicz LK, Southgate J. Identification of genes involved in human urothelial cell: matrix interactions: implications for the progression pathways of malignant urothelium. Cancer Res. 2001;61:1678–1685. [PubMed] [Google Scholar]
- Adams M, Reginato MJ, Shao D, Lazar MA, Chatterjee VK. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J Biol Chem. 1997;272:5128–5132. doi: 10.1074/jbc.272.8.5128. [DOI] [PubMed] [Google Scholar]
- Camp HS, Tafuri SR. Regulation of peroxisome proliferator-activated receptor gamma activity by mitogen-activated protein kinase. J Biol Chem. 1997;272:10811–10816. doi: 10.1074/jbc.272.16.10811. [DOI] [PubMed] [Google Scholar]
- Nosjean O, Boutin JA. Natural ligands of PPARgamma: are prostaglandin J(2) derivatives really playing the part? Cell Signal. 2002;14:573–583. doi: 10.1016/s0898-6568(01)00281-9. [DOI] [PubMed] [Google Scholar]
- Gearing KL, Gottlicher M, Teboul M, Widmark E, Gustafsson JA. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc Natl Acad Sci USA. 1993;90:1440–1444. doi: 10.1073/pnas.90.4.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issemann I, Prince RA, Tugwood JD, Green S. The peroxisome proliferator-activated receptor: retinoid X receptor heterodimer is activated by fatty acids and fibrate hypolipidaemic drugs. J Mol Endocrinol. 1993;11:37–47. doi: 10.1677/jme.0.0110037. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Yajima Y, Kawashima S, Tanaka K, Kagechika H. Synergistic potentiation of thiazolidinedione-induced ST 13 preadipocyte differentiation by RAR synergists. Biochem Biophys Res Commun. 2001;280:646–651. doi: 10.1006/bbrc.2000.4172. [DOI] [PubMed] [Google Scholar]
- Schulman IG, Shao G, Heyman RA. Transactivation by retinoid X receptor-peroxisome proliferator-activated receptor gamma (PPARgamma) heterodimers: intermolecular synergy requires only the PPARgamma hormone-dependent activation function. Mol Cell Biol. 1998;18:3483–3494. doi: 10.1128/mcb.18.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Mitschler A, Moras D. Molecular recognition of agonist ligands by RXRs. Mol Endocrinol. 2002;16:987–997. doi: 10.1210/mend.16.5.0823. [DOI] [PubMed] [Google Scholar]
- Radoja N, Komine M, Jho SH, Blumenberg M, Tomic-Canic M. Novel mechanism of steroid action in skin through glucocorticoid receptor monomers. Mol Cell Biol. 2000;20:4328–4339. doi: 10.1128/mcb.20.12.4328-4339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho SH, Radoja N, Im MJ, Tomic-Canic M. Negative response elements in keratin genes mediate transcriptional repression and the cross-talk among nuclear receptors. J Biol Chem. 2001;276:45914–45920. doi: 10.1074/jbc.M103144200. [DOI] [PubMed] [Google Scholar]
- Subbaramaiah K, Lin DT, Hart JC, Dannenberg AJ. Peroxisome proliferator-activated receptor gamma ligands suppress the transcriptional activation of cyclooxygenase-2. Evidence for involvement of activator protein-1 and CREB-binding protein/p300. J Biol Chem. 2001;276:12440–12448. doi: 10.1074/jbc.M007237200. [DOI] [PubMed] [Google Scholar]
- Stephens JM, Morrison RF, Wu Z, Farmer SR. PPARgamma ligand-dependent induction of STAT1, STAT5A, and STAT5B during adipogenesis. Biochem Biophys Res Commun. 1999;262:216–222. doi: 10.1006/bbrc.1999.0889. [DOI] [PubMed] [Google Scholar]
- Patel L, Charlton SJ, Marshall IC, Moore GB, Coxon P, Moores K, Clapham JC, Newman SJ, Smith SA, Macphee CH. PPARgamma is not a critical mediator of primary monocyte differentiation or foam cell formation. Biochem Biophys Res Commun. 2002;290:707–712. doi: 10.1006/bbrc.2001.6263. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A, Budhiraja S, Qian X, Clevidence D, Costa RH, Reichel RR. Retinoic acid-mediated activation of HNF-3 alpha during EC stem cell differentiation. Nucleic Acids Res. 1994;22:2126–2133. doi: 10.1093/nar/22.11.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]