Abstract
The cus determinant of Escherichia coli encodes the CusCFBA proteins that mediate resistance to copper and silver by cation efflux. CusA and CusB were essential for copper resistance, and CusC and CusF were required for full resistance. Replacements of methionine residues 573, 623, and 672 with isoleucine in CusA resulted in loss of copper resistance, demonstrating their functional importance. Substitutions for several other methionine residues of this protein did not have any effect. The small 10-kDa protein CusF (previously YlcC) was shown to be a periplasmic protein. CusF bound one copper per polypeptide. The pink CusF copper protein complex exhibited an absorption maximum at around 510 nm. Methionine residues of CusF were involved in copper binding as shown by site-directed mutagenesis. CusF interacted with CusB and CusC polypeptides in a yeast two-hybrid assay. In contrast to other well-studied CBA-type heavy metal efflux systems, Cus was shown to be a tetrapartite resistance system that involves the novel periplasmic copper-binding protein CusF. These data provide additional evidence for the hypothesis that Cu(I) is directly transported from the periplasm across the outer membrane by the Cus complex.
Copper is required for the function of several important enzymes in Escherichia coli including the cytochrome bo ubiquinole oxidase (3), copper-zinc superoxide dismutase (19), and amine oxidase (11). However, copper can also be extremely toxic, requiring homeostatic mechanisms. In E. coli at least three systems are involved in copper tolerance. First, the P-type ATPase CopA pumps excess copper out of the cytoplasm (16, 53). Second, the multicopper oxidase CueO may protect periplasmic enzymes from copper-mediated damage (21). Third, the cus determinant confers copper and silver resistance (17, 22, 24, 41).
The cus determinant consists of two operons, cusRS (previously ylcA-ybcZ) and cusCFBA (previously ylcBCD-ybdE), transcribed in opposite directions. The cusRS operon encodes a histidine kinase, CusS, and a response regulator, CusR. Transcription initiation of cusCFBA is dependent on the concentration of copper and silver (17, 41). The promoter region of cusCFBA harbors a motif with sequence homology to the DNA binding motif of CopR from Pseudomonas syringae, previously described as a cop box (40). Additionally, if the plasmid-encoded copper resistance determinant pco is present, expression of pcoE is also regulated by cusRS (10, 41).
The cusCFBA operon encodes proteins required for efflux of copper. CusA is a member of the resistance-nodulation-division (RND) protein superfamily (TC 2.A.6.1.1 in Milton Saier's functional-phylogenetic classification system for transmembrane solute transporters), which is a protein family of membrane-bound, proton-driven transporters (56, 57, 64). RND proteins, more than 1,000 amino acid residues in size, are composed of four large domains, two of them hydrophobic and two hydrophilic (18). The two hydrophilic domains are located in the periplasm and may contact the other subunits of the efflux protein complex.
CusB belongs to the family of membrane fusion proteins (MFP) (56), and CusC is an outer membrane factor (OMF) (14). Additionally, there is a small open reading frame, designated cusF, located between cusC and cusB. A similar open reading frame is also present in the sil determinant of Salmonella enterica serovar Typhimurium on plasmid pMG101 (23).
Deletion of cusA or cusCFBA led to the sensitivity of E. coli to silver (17, 24). Under anaerobic conditions, deletion of the cus system also resulted in copper sensitivity. However, under aerobic conditions only the additional deletion of cueO, encoding a multicopper oxidase, led to a copper-sensitive phenotype (22, 47). In this report we characterize the function of the different components of the CusCBA protein complex. Additionally, we identified several essential residues in the CusA protein and purified and characterized CusF, a novel copper-binding protein that interacts with the CusCBA efflux complex.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains GR1 (ΔcueO) and GR15 (ΔcueO ΔcusA::Cm) (22) are derivatives of wild-type strain W3110 (22). All other strains are listed in Table 1. E. coli strains were grown in Luria-Bertani (LB) broth (58). To study copper inhibition of growth, LB medium containing different concentrations of CuCl2 was routinely used. Additionally and to increase the differences between copper-sensitive and copper-resistant bacterial strains, LB medium was adjusted to pH 7.5 with NaOH, and CuSO4 was used instead of CuCl2. To induce cus gene expression from a plasmid tetAp promoter in both copper-containing media, anhydrotetracycline (200 μg/liter) was added. Antibiotics (ampicillin [125 mg/liter], chloramphenicol [15 to 25 μg/liter], and kanamycin [25 mg/liter]) were added where appropriate.
TABLE 1.
MICs of copper for E. coli strain GR1 mutant derivatives in LB medium containing copper chloride
| Bacterial strain | Relevant genotype | Complementing gene in trans | MIC (mM) of CuCl2a |
|---|---|---|---|
| GR1 | ΔcueO | None | 3.25 |
| GR15 | ΔcueO ΔcusA::Cm | None | 1.5b |
| EC950 | ΔcueO ΔcusB | None | 1.5b |
| EC951 | ΔcueO ΔcusC | None | 1.75b |
| ECA067 | ΔcueO ΔcusC ΔtolC::Kan | None | 1.75b |
| EC933 | ΔcueO ΔcusF | None | 2.25 |
| cusA mutant strains | |||
| GR15(pECD738) | ΔcueO ΔcusA::Cm | cusA | 2.75bc |
| GR15(pECD739) | ΔcueO ΔcusA::Cm | cusA (M573I) | 1.5bc |
| GR15(pECD740) | ΔcueO ΔcusA::Cm | cusA (M623I) | 1.25bc |
| GR15(pECD741) | ΔcueO ΔcusA::Cm | cusA (M640I) | 2.75bc |
| GR15(pECD742) | ΔcueO ΔcusA::Cm | cusA (M672I) | 1.25bc |
| GR15(pECD743) | ΔcueO ΔcusA::Cm | cusA (M812I) | 2.5bc |
| GR15(pECD744) | ΔcueO ΔcusA::Cm | cusA (M738I) | 2.75bc |
| GR15(pECD745) | ΔcueO ΔcusA::Cm | cusA (M755I) | 2.5bc |
| GR15(pECD746) | ΔcueO ΔcusA::Cm | cusA (M792I) | 2.75bc |
| GR15(pECD747) | ΔcueO ΔcusA::Cm | cusA (M833I) | 2.5bc |
| GR15(pECD748) | ΔcueO ΔcusA::Cm | cusA (M875I-M878I-M881I) | 2.5bc |
| GR15(pECD768) | ΔcueO ΔcusA::Cm | cusA (D405N) | 1.5c |
| GR15(pECD769) | ΔcueO ΔcusA::Cm | cusA (E412Q) | 1.5c |
| GR15(pECD770) | ΔcueO ΔcusA::Cm | cusA (E412D) | 2.5c |
| GR15(pECD767) | ΔcueO ΔcusA::Cm | cusA (A399D) | 1.75c |
| cusF mutant strains | |||
| EC933(pECD735) | ΔcueO ΔcusF | cusF | 2.75c |
| EC933(pECD736) | ΔcueO ΔcusF | cusA (M69I-M71I) | 2.25c |
The MIC is defined as the lowest concentration at which no growth was observed. Strains were grown overnight and diluted 1:500 into fresh LB medium. After 2 h of growth at 37°C they were plated onto LB agar containing different concentrations (0.5 to 4.0 mM in steps of 0.25 mM) of CuCl2. The cusCFBA operon is fully expressed at copper concentrations above 100 μM (17). Growth was monitored after 16 h at 37°C. All experiments were done at least three times with identical results.
Colonies were small and mucoid at concentrations well below the MIC.
LB agar plates for complementation experiments contained anhydrotetracyline (200 μg/liter) to induce expression of complementing genes.
Genetic techniques.
Standard molecular genetic techniques were used (58). PCRs were performed with the polymerases Taq and Pwo (Roche, Mannheim, Germany). Deletions were generated as described by Datsenko and Wanner (13). The gene for chloramphenicol acetyltransferase was amplified by PCR from plasmid pKD3 (13) and transferred into E. coli strain BW25113 (13) as described by these authors. The deletion was transferred into W3110 wild type and GR1 (ΔcueO) (22) by general P1 transduction to create E. coli strains EC952 (ΔcusF), EC933 (ΔcueO ΔcusF), EC950 (ΔcueO ΔcusB), and EC951 (ΔcueO ΔcusC), respectively. The codons for L20 to Y441 were deleted from the cusC gene, those for A19 to S379 were deleted from cusB, and those for K2 to S101 were deleted from cusF. By use of the same approach but with plasmid pKD4 (13), the tolC gene (deletion from M3 to T484) was replaced by a kanamycin resistance cassette, leading to strain ECA067 (ΔcueO ΔcusC ΔtolC::Kan). Topology analysis was performed as described previously (4) with the cusF gene cloned as a C-terminal ′phoA fusion.
Construction of vectors for expression of CusA and CusF.
To construct the CusA expression vector pECD738, the cusA gene was amplified by PCR from chromosomal DNA and cloned into pASK-IBA3 (IBA GmbH, Göttingen, Germany) by using the restriction sites EcoRI and PstI, introduced by PCR. Plasmid pECD738 expressed CusA as a C-terminal fusion protein with Strep-TagII (amino acid sequence SAWSHPNFEK). The cloned CusA possessed also eight additional residues at the N terminus (GDRGPEF). A similar approach was used to create the CusF expression vector plasmid pECD735.
Site-directed mutagenesis.
The Quick-Change site-directed mutagenesis system (Stratagene, La Jolla, Calif.) was used to alter individual amino acid residues in CusA and CusF. The plasmids pECD735 (cusF) and pECD738 (cusA) that were used as template were isolated from a dam+ strain of E. coli. The primer pairs used to introduce the point mutation were antiparallel and overlapping. PCR products were treated with DpnI to digest the dam-methylated template plasmid. The purified PCR product containing the point mutation was directly transformed into E. coli. The correct mutations were verified by DNA sequence analysis.
Protein expression and purification.
Proteins CusA and CusF were purified with Strep-TagII technology. For overexpression the recombinant plasmids pECD735 (CusF) and pECD738 (CusA) were always transformed freshly into E. coli strain BL21(pLYS) (Stratagene). Gene expression was induced by addition of anhydrotetracycline to a final concentration of 200 μg/liter of culture. CusF was purified from periplasmic extract (21). Alternatively, cells were suspended in buffer W (100 mM Tris-HCl, pH 8.0) containing DNase I (10 mg/liter), protease inhibitor phenylmethylsulfonyl fluoride (1 mM), and avidin (0.02 U) and lysed with a French press (1,260 lb/in2, cell type 20K; Amicon, SLM Instruments, Inc., Rochester, N.Y.). After centrifugation (23,400 × g, 15 min, 4°C) the supernatant was applied to a streptactin-agarose column (1 ml/400 ml of culture) equilibrated with the same buffer. The column was washed extensively with buffer W containing 1 M NaCl. The CusF protein was then eluted in aliquots of 1 ml of buffer E (buffer W containing 2.5 mM desthiobiotin). For purification of CusA, the same steps were used. However, CusA was solubilized from the membrane fraction of the cells with N-lauroylsarcosine (20 g/liter), and all buffers used to purify the solubilized protein contained N-lauroylsarcosine (1 g/liter). Polyclonal antibodies against both proteins were raised in rabbits.
Immunoblotting.
Protein concentrations were determined by the Bradford method (9). To detect CusF, E. coli was grown in LB broth to 100 Klett units and incubated with different concentrations of CuCl2 for 30 min. Cells were harvested by centrifugation, and a periplasmic extract was prepared as described above. Protein samples were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels and blotted (SemiDry-Blot; Biometra, Göttingen, Germany) onto a polyvinylidene difluoride membrane as previously described (8). The membrane was subsequently incubated with the CusF-specific antibody (diluted 1:1,000) and a monoclonal anti-rabbit immunoglobulin G antibody conjugated with horseradish peroxidase (diluted 1:50,000; Sigma-Aldrich, Deisenhofen, Germany). For detection, the polyvinylidene difluoride membrane was incubated 1:1 with solution 1 (0.1 M Tris-HCl [pH 8.5], 0.4 mM p-coumaric acid, 2.5 mM Luminol) and solution 2 (0.1 mM Tris-HCl [pH 8.5], 5.4 M H2O2) for 1 min. The membrane was exposed for 5 s to 5 min to ECL Hyperfilm (Amersham Pharmacia Biotech, Uppsala, Sweden). Strep-TagII epitopes were detected with a streptavidin-alkaline phosphatase conjugate.
Analysis of copper binding.
A CusF-containing solution (0.15 mM protein in 100 mM Tris-HCl buffer, pH 8.0) was incubated for 30 min at 0°C with 0.5 mM CuCl2. Unbound copper was removed by dialysis against 10 mM Tris-HCl buffer (pH 7.0). The protocol that was used for copper analysis was adapted from Environmental Protection Agency method 6020A. Up to 0.5 ml of the CusF sample (1 μM protein) was placed in a 5-ml Teflon digestion bomb, and 1 ml of concentrated trace-metal-grade nitric acid was added. The bomb was capped tightly and placed in water in a microwave-safe dish. The samples were heated for 5 min at 560 W and allowed to cool, and then the pressure was vented. The samples were heated an additional 5 min at 100% power, allowed to cool, quantitatively transferred to a volumetric flask, and brought to volume with Milli-Q water. The samples were finally analyzed by inductively coupled plasma-mass spectrometry with an external standard calibration curve. Terbium was added online as an internal standard. The UV-visible (UV-vis) spectrum of CusF in the absence or presence of copper was recorded on a Uvikon 922A spectrometer (Kontron Instruments, Zürich, Switzerland) at room temperature.
Assay of protein-protein interactions.
The CytoTrap (Stratagene) yeast two-hybrid system was employed to study protein-protein interactions. The cusF gene was amplified without leader sequence by PCR and cloned into the bait vector pSOS with BamHI and SalI restriction sites introduced by PCR. The resulting plasmid expressed CusF (from amino acid 23) as an N-terminal fusion with hSOS. The genes encoding the soluble part of CusC (from amino acid 21) and CusB (from amino acid 29) were also amplified by PCR and cloned into the target vector pMYR with EcoRI and XhoI restriction sites. The resulting plasmids expressed these two proteins as N-terminal fusions with a myristylation signal targeting the fusion proteins to the cytoplasmic membrane. The plasmids were verified by DNA sequencing and transformed into Saccharomyces cerevisiae strain cdc25H (Stratagene). Interaction studies were carried out as described in the manufacturer's manual.
RESULTS
All Cus structural genes are required for full copper resistance.
The presence of the membrane protein CusA, the central pump of the CusC(F)BA copper efflux complex, is essential for cus-mediated copper resistance, since E. coli strain GR15 (ΔcueO ΔcusA::Cm) exhibits a copper-sensitive phenotype. A ΔcusA deletion did not differ in copper sensitivity from GR10 (ΔcueO ΔcusCFBA::Cm) with all cus structural genes deleted (21, 22). In order to investigate the influence of the other components of the Cus transport complex on copper resistance, cusC, cusF, or cusB was deleted (avoiding polar effects) in E. coli strain GR1 (ΔcueO), which led to strains EC951 (ΔcueO ΔcusC), EC933 (ΔcueO ΔcusF), and EC950 (ΔcueO ΔcusB), respectively. In all three mutant strains, unaltered expression of CusA was confirmed by immunoblotting (data not shown).
Single deletions of all four cus structural genes in strain GR1 (ΔcueO) led to a decrease in copper resistance (Table 1). Although the cusF deletion had the smallest effect on copper resistance, involvement of the product of this previously uncharacterized gene in copper resistance could be clearly demonstrated.
Deletion of cusB led to a complete loss of Cus-mediated copper tolerance (Table 1), comparable to the effect of the ΔcusA deletion. This demonstrated the essential function of the MFP CusB for copper resistance. A deletion of cusC, encoding a putative OMF, led to a strong decrease in copper resistance. The ΔcusC deletion strain was slightly more resistant to copper than was the ΔcusB or the ΔcusA deletion strain (Table 1). This small increase in copper resistance may be due to a complementation of the ΔcusC deletion by a gene for another OMF. To test if TolC might be this OMF, the tolC gene was replaced by a kanamycin resistance cassette. However, copper resistance of a ΔcueO ΔcusC ΔtolC::Kan triple mutant strain compared to that of the ΔcueO ΔcusC double mutant strain was unchanged, showing that TolC cannot partially substitute for a missing CusC protein (Table 1). Taken together, these data indicated that the three subunits of the putative CusCBA transenvelope efflux complex were all essential for cus-mediated copper resistance.
Conserved methionines of CusA are required for copper resistance.
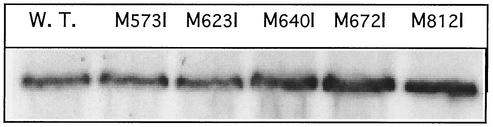
Comparison of CusA with SilA (23) and other putative RND proteins revealed conserved methionine residues in the second periplasmic domain of CusA (M573, M623, M640, M672, and M812). These residues are not present in RND proteins involved in the export of divalent cations, such as CzcA (GenBank accession number P13511 [18, 45]) or CnrA (accession number G47056 [35]) from Ralstonia metallidurans strain CH34 or NccA from R. metallidurans strain 31A (accession number I39580 [59]). Each single methionine residue was mutated to isoleucine by site-directed mutagenesis. The mutant proteins were expressed in E. coli strain GR15 (ΔcueO ΔcusA::Cm). Correct expression of all mutated proteins was controlled by Western blot analysis (shown for some mutant proteins in Fig. 1; other data not shown). Copper resistance of this strain could be partially restored by expression of the CusA wild-type protein in trans, and the effect of the mutant proteins on copper resistance was compared to that of the wild-type protein (Table 1).
FIG. 1.
Expression level of CusA mutant proteins. An immunoblot against CusA of crude extracts (50 μg [dry weight] of cells) from cells of strain GR15 expressing mutated CusA variants is shown.
The three CusA mutant proteins M573I, M623I, and M672I were no longer able to relieve the copper sensitivity of strain GR15 (ΔcueO ΔcusA::Cm) in trans (Table 1). Therefore, these methionine residues were essential for the function of CusA. Expression of CusA M640I in GR15 did not change the level of copper tolerance from that of cells expressing wild-type CusA (Table 1). Only a small difference in resistance between E. coli GR15 strains expressing CusA and those expressing CusA M812I could be detected on copper-containing solid medium (Table 1). In liquid medium, however, expression of M812I led to an intermediate level of copper resistance (data not shown). Although conserved, M640 and M812 appeared not to be essential for CusA function.
Sequence comparison of the second periplasmic domain of CusA and SilA (data not shown) revealed four additional conserved methionines (M738, M755, M792, and M833). Single mutations of these four methionines to isoleucine had no or only a slight influence on copper resistance (Table 1). An amino acid sequence, M875XXM878XXM881, similar to a copper-binding motif described by Zhou and Thiele (67), was found adjacent to the second periplasmic domain (data not shown). A CusA triple mutant (M875I-M878I-M881I) did not differ much in its ability to confer copper resistance from the wild-type protein (Table 1). Thus, some methionine residues were not essential for CusA function although they are conserved and even located within putative copper-binding motifs. However, three methionine residues were indispensable for the function or structural integrity of CusA.
Two amino acid residues (D405 and E412) within or close to transmembrane helix IV of CusA are conserved in the family of metal-transporting RND proteins (64). These two charged amino acid residues were also essential for copper resistance mediated by Cus (Table 1). In RND proteins involved in the transport of monovalent cations, an alanine residue (A399) within this transmembrane helix is also conserved. RND proteins dealing with divalent heavy metal cations contain a D residue at this position (44). Alteration of A399 in CusA to D resulted in a clear effect on copper resistance (Table 1), showing that this amino acid residue was also essential for the function of the copper-transporting RND protein CusA.
CusF is a periplasmic protein.
Involvement of CusF is a unique feature of cus-mediated copper detoxification. Sequence analysis of CusF predicted an N-terminal leader sequence and a potential signal peptidase cleavage site (data not shown), suggesting a periplasmic location of the CusF protein. A C-terminal fusion, CusF′-PhoA, was constructed to study the cellular location of CusF. The alkaline phosphatase activity of the CusF′-PhoA fusion was compared to that of the negative control, PhoA located in the cytoplasm, and to that of the positive control, PhoA in the periplasm. The CusF′-PhoA fusion showed a 30-fold-higher PhoA activity (160.1 ± 37.42 U/mg [dry weight] of cells) than did the negative control, a β-lactamase-PhoA fusion without the β-lactamase leader peptide (5.2 ± 0.76 U/mg [dry weight] of cells). The activity of the CusF′-PhoA fusion was even higher than that of the positive control, a β-lactamase-PhoA fusion containing the β-lactamase leader peptide (56.3 ± 1.78 U/mg [dry weight] of cells). These results confirmed a periplasmic location of CusF.
The CusF protein was overexpressed as a C-terminal Strep-TagII fusion and could be purified from the periplasmic extract of E. coli cells expressing this fusion protein (Fig. 2). The amino-terminal sequence of the purified protein was determined by Edman degradation as ANEHH, corresponding to the predicted sequence of CusF from A22. This indicates that CusF is processed by the leader peptidase after translocation into the periplasm.
FIG. 2.
Purification of CusF. CusF was expressed as a C-terminal Strep-TagII fusion in E. coli BL21(pLYS). Crude extract before (lane 1) and after (lane 2) induction of the E. coli cells with anhydrotetracycline is shown. After the cells were harvested, periplasmic extract (lane 3) was prepared, and CusF was purified by affinity chromatography. Lanes 4 to 6 show elution fractions 2 to 4, respectively, containing the CusF protein. From the different purification steps, protein samples were separated by SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue 250. M, molecular mass markers.
CusF could be detected in periplasmic extracts of E. coli strains W3110 and GR1 (ΔcueO), but only after induction with CuCl2 (data not shown). The size of the detected protein was about 10 kDa. This value correlated well with the predicted size of the leaderless form of CusF. In contrast, CusF was not found in two independent cusF deletion strains, EC952 and EC933 (data not shown). These data clearly indicated that cusF is expressed in E. coli after induction with copper and its product is transported into the periplasm and processed into a mature protein after cleavage of the preprotein between positions 21 and 22.
Interaction of CusF with the proteins of the CusCBA efflux complex.
Besides CusF, most parts of the CusCBA efflux complex are predicted to be located in the periplasm. To investigate whether there is an interaction between CusF and the CusCBA complex, a yeast two-hybrid system was applied. In this system, two proteins that may interact are encoded by a bait and by a target vector, respectively. If these proteins interact, a stress response pathway in the yeast S. cerevisiae is successfully switched on and the cells are able to grow at 37°C.
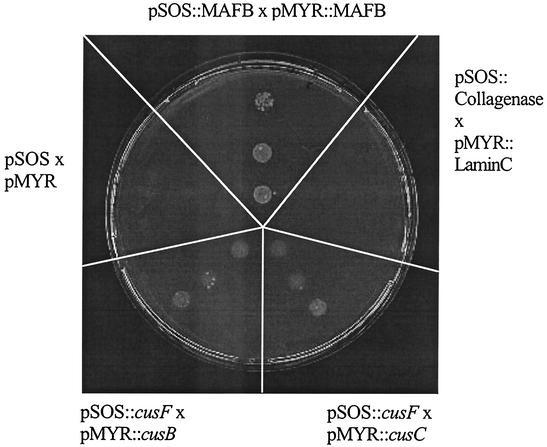
The gene cusF was PCR amplified and cloned into the CytoTrap bait vector pSOS. The genes encoding CusC and CusB were also amplified and each cloned into the CytoTrap target vector pMYR. Since the experiment was done with proteins natively located in the periplasm, only the hydrophilic portions without the leader sequences (CusF′ and CusC′) or membrane anchor (CusB′) were used. The resulting constructs plus controls provided by the manufacturer were transformed into the temperature-sensitive S. cerevisiae strain cdc25H, and growth on galactose-containing agar plates at 37°C was monitored. Yeast strains containing cusF′ in the bait vector and cusB′ or cusC′ in the predator vector were able to grow at elevated temperature, as was the positive control provided by the manufacturer (Fig. 3). This indicated a possible interaction of CusF with CusB and of CusF with CusC.
FIG. 3.
Interaction among CusF, CusB, and CusC. Results of a CytoTrap yeast two-hybrid experiment are shown. Positive control refers to the MAFB-MAFB interaction (top), and negative controls refer to lamin C and collagenase as described in the protocol provided by the manufacturer (upper right) and to the vector plasmids without insert (upper left). The interaction between cusF′ cloned in pSOS and the gene for CusB′ (lower left) and CusC′ (lower right) cloned in pMYR indicates a possible interaction between the periplasmic protein CusF and the MFP CusB or the OMF CusC, respectively. None of the three cus genes contained the coding region for the leader signal peptide (CusF and CusC) or the N-terminal membrane anchor (CusB).
CusF is a pink copper-binding protein.
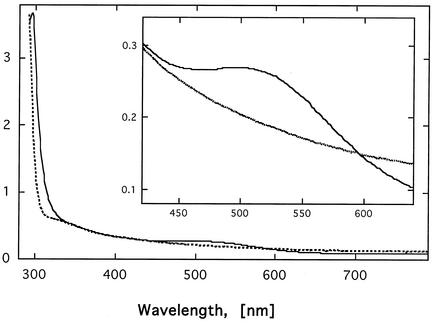
CusF is involved in copper resistance and contains several methionine, aspartate, and histidine residues that might be involved in copper binding. To examine binding of copper, purified CusF was incubated with CuCl2 and excessively bound copper was removed by dialysis. CusF with bound copper was of a pink color (data not shown). The copper content of CusF was analyzed by inductively coupled plasma-mass spectrometry, and a ratio of 0.925 ± 0.115 copper atoms per CusF polypeptide was calculated. The UV-vis spectrum of the copper-containing CusF showed a maximum around 510 nm (Fig. 4). To our knowledge, such an absorption maximum has not been reported previously for a copper protein. Since CusF was clearly identified as a copper-binding protein, CusF may contain a novel type of copper binding site.
FIG. 4.
UV-vis spectrum of CusF. The UV-vis spectrum of CusF (0.75 mM) in the absence (dashed line) and presence (solid line) of copper was recorded on a Uvikon 922A spectrometer at room temperature in 10 mM Tris-HCl, pH 7.0. The inset shows an expansion of the range between 420 and 640 nm to demonstrate the copper-specific shoulder at about 510 nm.
Amino acid residues of CusF involved in copper binding.
Sequence alignment of CusF with homologous proteins deduced from completed bacterial genome sequences showed a conserved methionine-containing motif (M69TM71). Site-directed mutagenesis was performed to replace both methionines with isoleucine. The effect of this substitution was analyzed by trans-complementation of E. coli strain EC933 (ΔcueO ΔcusF) with the mutated cusF gene (Table 1). Expression of the CusF mutant protein was confirmed by immunoblotting (data not shown). The ΔcusF phenotype of strain EC933 could be partially complemented in trans by the cusF wild-type gene but not by the M69I-M71I double mutant (Table 1). The CusF M69I-M71I protein was purified (data not shown), and copper binding was analyzed. The molar ratio between the CusF M69I-M71I polypeptide and bound copper was 0.225 ± 0.085. Since wild-type CusF bound about 1 copper atom per molecule, the MTM motif appears to be involved in binding of copper.
In copper chloride-containing LB medium, the difference among copper resistance of the control strain GR1 (MIC, 3.25 mM), GR1 with an additional ΔcusF deletion (MIC, 2.25 mM), and the same ΔcusF deletion strain complemented in trans by cusF (MIC, 2.75 mM) was rather small (Table 1). Therefore, the growth medium was optimized for a better differentiation between copper-sensitive and copper-resistant bacterial strains. In the resulting LB medium (pH 7.5) with copper sulfate instead of copper chloride, the MIC for the control strain GR1 dropped to 3.0 mM CuSO4. However, the MIC for the ΔcusF strain decreased to 1.25 mM CuSO4 and the MIC for the ΔcusF strain complemented in trans by cusF was 2.25 mM CuSO4 (Table 2). In this improved medium, the effect of mutations in CusF could be studied with enhanced resolution.
TABLE 2.
MICs of copper for E. coli strain GR1 mutant derivatives in LB medium (pH 7.5) containing copper sulfate
| Bacterial strain | Relevant genotype | Complementing gene in trans | MIC (mM) of CuSO4a |
|---|---|---|---|
| GR1(pASK-IBA3) | ΔcueO | None | 3.0 |
| EC933(pASK-IBA3) | ΔcueO ΔcusF | None | 1.25 |
| EC933(pECD735) | ΔcueO ΔcusF | cusF | 2.25 |
| EC933(pECD736) | ΔcueO ΔcusF | cusF (M69I-M71I) | 1.25 |
| EC933(pECD775) | ΔcueO ΔcusF | cusF (M69I) | 1.5 |
| EC933(pECD776) | ΔcueO ΔcusF | cusF (M71I) | 1.5 |
| EC933(pECD737) | ΔcueO ΔcusF | cusF (H25R-H26R-H27R-H57R-H58R) | 1.0b |
| EC933(pECD766) | ΔcueO ΔcusF | cusF (H57R-H58R) | 1.75 |
| EC933(pECD763) | ΔcueO ΔcusF | cusF (H57Q-H58Q) | 2.0 |
| EC933(pECD764) | ΔcueO ΔcusF | cusF (H25Q-H26Q-H27Q) | 2.25 |
| EC933(pECD765) | ΔcueO ΔcusF | cusF (D48N) | 2.25 |
| EC933(pECD762) | ΔcueO ΔcusF | cusF (F73Y) | 2.25 |
The MIC is defined as the lowest concentration at which no growth was observed. Strains were grown overnight and diluted 1:500 into fresh LB medium. After 2 h of growth at 37°C they were plated onto LB agar, carefully adjusted to pH 7.5 with NaOH, containing different concentrations (0.5 to 4.0 mM in steps of 0.25 mM) of CuSO4. LB agar plates for complementation experiments contained anhydrotetracycline (200 μg/liter) to induce expression of complementing genes. Growth was monitored after 16 h at 37°C. All experiments were done at least three times with identical results.
Reduced plating efficiency was also observed on LB agar plates containing anhydrotetracyline without CuSO4.
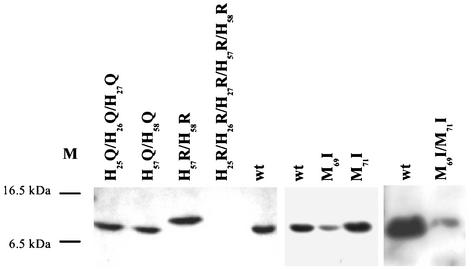
The CusF M69I-M71I protein was not able to complement the copper sensitivity of the ΔcusF deletion, emphasizing an essential function of the methionine residues. Both CusF variants with a single methionine-to-isoleucine mutation mediated slightly increased copper resistance (Table 2). All three mutant proteins appeared in the processed form in the periplasm as shown by Western analysis of periplasmic extracts; however, the expression level of the M69I-M71I double mutant protein and the M69I single mutant protein was below the expression level of the wild-type protein (Fig. 5). The expression level of the M71I single mutant protein was unaltered (Fig. 5). Thus, both methionine residues contributed to copper binding.
FIG. 5.
Expression profile of CusF mutant proteins. Cells of E. coli strain EC933 (ΔcueO ΔcusF) were complemented in trans with the cusF wild-type gene (wt) or with plasmids (Table 2) containing cusF mutant genes. After induction with anhydrotetracyline (200 μg/liter) to induce cusF gene expression and continuing growth, periplasmic extracts were prepared and separated by SDS-polyacrylamide gel electrophoresis (10 μg of protein per lane). The proteins were visualized via streptavidin-alkaline phosphatase conjugates. M, positions of two marker proteins.
A quintuple (H25R-H26R-H27R-H57R-H58R) mutant protein did not complement copper sensitivity (Table 2), because it was not transported into the periplasm (Fig. 5). Thus, these mutations may have resulted in structural deformations. Mutation of only two of the five histidine residues to arginine (H57R-H58R) led to a medium level of complementation (Table 2). However, this mutant protein appeared as an unprocessed form in the periplasm (Fig. 5), which also indicated a structural effect of the mutation. Therefore, both histidine residues were changed to glutamine residues. The resulting mutant protein appeared in the correct size in the periplasm (Fig. 5) and complemented the ΔcusF mutation nearly as well as the wild-type CusF did (Table 2). When the remaining three histidine residues were also changed to glutamine residues, this triple mutant protein was also correctly exported into the periplasm (Fig. 5), and it mediated full complementation to copper resistance (Table 2). Thus, the five histidine residues of CusF are probably not essential for copper binding and/or CusF function. Mutation of two other conserved amino acid residues that may be involved in copper binding, D48 and F73, had also no effect (Table 2).
DISCUSSION
E. coli detoxifies excess copper by the interaction of at least three systems encoded on the bacterial chromosome, CopA, CueO, and Cus. Additional tolerance may be conferred by plasmid-encoded resistance determinants (10). When the effects of single deletions of the three chromosomal systems on copper resistance are compared, inactivation of copA had the strongest effect (22). CopA is a soft-metal-transporting P-type ATPase (53, 54). According to the structural model for a related protein (63), CopA is located in the cytoplasmic membrane. Since CopA pumps Cu(I) into inside-out vesicles (53), it should export Cu(I) from the cytoplasm into the periplasm in vivo. CopA is regulated by CueR, a MerR-like transcriptional activator induced by Cu(I). CueR has been shown elsewhere to regulate two genes, copA and cueO (formerly yacK) (47, 50, 61). CueO is a multicopper oxidase that protects periplasmic enzymes from copper-induced damage (21).
The second copper-induced regulon is the cus determinant that was previously described as a copper and silver resistance system (17, 22, 24, 41). The cusRS genes form a sensor-regulator pair that activates the adjacent but divergently transcribed genes cusCFBA (41), encoding the subunits for a transport complex that traverses both membranes of the gram-negative bacterium. Since Ag(I) is also detoxified (17), the substrate for Cus is Cu(I) rather than Cu(II).
The central transport protein of the Cus protein complex, CusA, is a member of the RND protein superfamily of proton-driven cation symporters and antiporters (57, 64). The presence of CusA is essential for Cus function (22). The first three-dimensional structure of an RND protein, AcrB, shows an extensive periplasmic domain of the protein that is organized as a homotrimer (42). Three vestibules, thought to act as a conduit for substrates, connect these periplasmic domains to the periplasm (42). Thus, access of copper to the RND protein CusA may be possible from the cytoplasm as well as from the periplasm.
Further transenvelope transport of copper should be accomplished by the MFP CusB and the OMF CusC. Although CusC is related to the OMF TolC (33), the latter protein was not able to substitute for a missing CusC protein (Table 1), although cross-complementation of OMFs has been shown in many cases (2, 5, 20, 29, 33, 39, 55, 60, 65).
The organization of the genes for CBA efflux systems agrees with this hypothesis. All described resistance determinants containing a cusA-like gene are usually preceded by a cusB-like gene and are organized as an operon. In the heavy metal resistance determinants czc and cnr of R. metallidurans (35, 45), the sil determinant of S. enterica serovar Typhimurium (23), and cus of E. coli (17, 41), the gene for the outer membrane protein is also part of this operon. However, in other resistance systems of E. coli (acrAB and acrCD), the tolC gene encoding the OMF is located elsewhere on the chromosome (5). The cusBA-like operons on the chromosomes of R. metallidurans (contigs 619 and 709, Department of Energy Joint-Genome Institute) and of P. syringae (orfEF) are lacking the genes for the respective OMFs. These putative efflux pumps need to recruit an OMF from another resistance determinant, as has been shown elsewhere for acrAB and tolC (1).
In contrast to cusC, deletion of cusB led to a complete loss of copper resistance. The importance of the MFP CusB is reminiscent of previous data obtained from studies of the czc and cnr determinants of R. metallidurans. A deletion of czcB led to the nearly complete loss of resistance to cobalt, zinc, and cadmium (55), and a deletion of cnrB led to highly reduced nickel and cobalt resistance (20). As proposed by Andersen et al. (1), the function of MFP such as CusB may be that of an adaptor that brings the putative CusC3 tube close to the catalytic center CusA3. Thus, an RND protein might not be able to bind to an OMF in the absence of its specific MFP adaptor, and the data obtained with CusB fit into this picture.
In this emerging model for the function of a CBA efflux complex, the roles of the MFP adaptor and the OMF tube are more static than dynamic, with the RND protein being the catalytic center. Substrates coming from the periplasm and/or the cytoplasm require a substrate-binding site proximal to the central cavity of the RND protein, formed by the two large periplasmic loops (LPLs) of each monomer (42). Recent findings indicate that these LPLs of RND proteins indeed have a function in substrate recognition. When the LPLs of AcrD were exchanged with the corresponding LPLs from AcrB, the chimeric protein displayed the substrate specificity of AcrB rather than that of AcrD (15). Similar results were obtained with chimeric AcrB-MexB proteins (62), and essential amino acids probably involved in substrate recognition were identified in both LPLs of MexD (37). The LPLs of MexF were also required for correct assembly of this CBA efflux pump (38), assigning just a second essential function to the headpiece of RND proteins.
Sequence analysis of CusA showed no known potential copper-binding motif such as CXnC (46) in loops predicted to be located in the cytoplasm or in the two LPLs of this protein. However, the second periplasmic loop of CusA contains a large number of methionine residues that are conserved within a group of CusA-related proteins. Similar methionine-rich motifs were also identified in copper transport systems from Schizosaccharomyces pombe (67), but not in other RND proteins such as CzcA, CnrA, or NccA, which are described as transporters of divalent heavy metal cations (35, 45, 59). Site-directed mutagenesis of these conserved methionines in CusA showed that M573, M623, and M672 were absolutely necessary to confer copper resistance in E. coli and that M812 is also involved in this process. A mutation of the sequence motif M875VPM878TLM881, which is similar to the copper-binding motifs of Ctr1 and Ctr3 (S. cerevisiae) (12, 31, 49) and Ctr4 (S. pombe) (67), had no influence on copper resistance. Taken together, these data show that methionine residues in the LPLs of CusA may be part of a periplasmic copper binding site or necessary to stabilize the CusCBA efflux complex. This indicates similarity of function between the metal-transporting RND protein CusA and the related RND proteins (AcrB, AcrD, MexB, and MexF) involved in the transport of hydrophobic organic substrates.
The CusCBA protein complex may capture Cu(I) from the cytoplasm and export the cation across both membranes of E. coli, thereby preventing the appearance of Cu(I) in the periplasm. CzcA, the first member of the RND family, also transports divalent heavy metal cations across the cytoplasmic membrane (18, 55). Additionally or alternatively, the Cus protein complex may capture copper from the periplasm for direct export to the outside, as has been suggested elsewhere for RND proteins exporting organic substrates (66). Which access pathways to CusA are used?
Deletion of the structural gene region of the cus determinant or of the cusA gene had no effect on copper resistance in the E. coli wild type under aerobic conditions (22) but impaired silver resistance (17). Deletion of cueO in a ΔcusA single mutant strain diminishes copper resistance from the wild-type level to the most sensitive phenotype observed (22). A ΔcueO ΔcusA double deletion strain is as copper sensitive as the copA::Km ΔcueO ΔcusA triple deletion strain. Deletion of cusA in a copA::Km background has no further effect on copper tolerance (17, 22), probably because protection of the periplasm by CueO is sufficient to maintain copper resistance under aerobic conditions. Although these data do not exclude transenvelope efflux of Cu(I) by Cus, they favor detoxification of copper from the periplasm, as has already been discussed (22, 47). Interestingly, the Cus system contains CusF, a periplasmic protein that is absent in CzcCBA and related protein metal detoxification systems.
The cusF gene is the second gene of the cusCFBA operon and encodes a protein with a predicted size of 110 amino acids and a potential leader sequence (M1 to Q21). The leader sequence was cleaved off after amino acid Q21, and the mature protein was located in the periplasm. In vitro, purified CusF bound one copper per polypeptide, leading to a complex with a pink color corresponding to an absorption maximum of 510 nm in the UV-vis spectrum. Other proteins with a similar color have a binuclear copper center, composed of two copper atoms connected by the sulfhydryl groups of two cysteines. Each of these coppers is also connected to a histidine residue (7, 26, 32). Recently, another purple periplasmic copper-binding protein has been described (28). PcoC is part of a plasmid-borne copper resistance determinant of E. coli (10, 34). Like CusF, PcoC does not contain cysteine residues. At least one ligand of the copper center of PcoC is predicted to be a histidine residue. Participation of a methionine residue in copper binding to PcoC has been discussed elsewhere (28). Since CusF does not contain cysteine residues, CusF might likewise bind copper via histidine and methionine residues. While the five histidines of CusF were not required for full CusF function, two conserved methionines were essential and may be part of a copper site, MXM (67).
Deletion of cusF led to a partial loss of copper tolerance. CusF might have an auxiliary function improving the efficiency of the CusCBA system. CusF could act as a periplasmic copper chaperone shuttling copper to the CusCBA efflux complex and thereby increasing the accessibility of periplasmic copper. This function is reminiscent of periplasmic substrate binding proteins that shuttle their substrates to the MerT transporter or ABC uptake systems (25, 27, 30, 36, 43, 48, 51, 52). Another small periplasmic copper-binding protein, the CopC protein from P. syringae, binds Cu(I) and Cu(II) to different sites, and redox change of the bound copper ion causes its migration from one site of CopC to the other (6). CusF may also bind both Cu(I) and Cu(II) and deliver Cu(I) directly to CusCBA. Maybe, Cu(II) can be transferred to CusCBA only after reduction to Cu(I). These attributes of CusF; the existence of a possible metal binding site in CusA located in the periplasm; and the copper resistance phenotypes of copA, cueO, and cus mutants add to the initial evidence (22) that CBA transport systems may also detoxify periplasmic metal cations by directly exporting cations from the periplasm across the outer membrane. This would be a novel mechanism of metal resistance. Future work will show if this hypothesis can be validated.
Acknowledgments
This work was supported by Hatch project 136713 and NIEHS grant ESO4940 with funds from the EPA to C.R., a DAAD fellowship to S.F., and grant Ni262/3-3 of the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie to D.H.N. Copper analyses were performed by the Analytical Section of the Hazard Identification Core from our NIEHS-supported Superfund Basic Research Program grant (NIH ES-04940)
We acknowledge Michael Kopplin for performing the analyses. The generation of antibodies to CusF was performed by the Institute of Physiological Chemistry of the Martin Luther University Halle-Wittenberg. We acknowledge Beate Fricke for antibody generation. Preliminary genomic sequence data for R. metallidurans were obtained from the DOE Joint-Genome Institute (JGI) at http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html.
REFERENCES
- 1.Andersen, C., C. Hughes, and V. Koronakis. 2001. Protein export and drug efflux through bacterial channel-tunnels. Curr. Opin. Cell Biol. 13:412-416. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, C., E. Koronakis, C. Hughes, and V. Koronakis. 2002. An aspartate ring at the TolC tunnel entrance determines ion selectivity and presents a target for blocking by large cations. Mol. Microbiol. 44:1131-1139. [DOI] [PubMed] [Google Scholar]
- 3.Anraku, Y., and R. B. Gennis. 1987. The aerobic respiratory chain of Escherichia coli. Trends Biochem. Sci. 12:262-266. [Google Scholar]
- 4.Anton, A., C. Große, J. Reißman, T. Pribyl, and D. H. Nies. 1999. CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. J. Bacteriol. 181:6876-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnesano, F., L. Banci, I. Bertini, S. Mangani, and A. R. Thompsett. 2003. A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) to different sites. Proc. Natl. Acad. Sci. USA 100:3814-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, S. M., X. Wang, and Y. Lu. 2000. Ligand replacement study at the His120 site of purple CuA azurin. J. Inorg. Biochem. 78:89-95. [DOI] [PubMed] [Google Scholar]
- 8.Bloss, T., S. Clemens, and D. H. Nies. 2002. Characterisation of the ZAT1p zinc transporter from Arabidopsis thaliana in microbial model organisms and reconstituted proteoliposomes. Planta 214:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Brown, N. L., S. R. Barrett, J. Camakaris, B. T. Lee, and D. A. Rouch. 1995. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17:1153-1166. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, R. A., P. F. Knowles, D. E. Brown, M. A. McGuirl, and D. M. Dooley. 1992. Evidence for copper and 3,4,6-trihydroxyphenylalanine quinone cofactors in an amine oxidase from the Gram-negative bacterium Escherichia coli K12. Biochem. J. 288:337-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dancis, A., D. Haile, D. S. Yuan, and R. D. Klausner. 1994. The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269:25660-25667. [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinh, T., I. T. Paulsen, and M. H. Saier, Jr. 1994. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 176:3825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan, B., G. Grass, C. Rensing, and B. P. Rosen. 2001. Escherichia coli CopA N-terminal Cys(X)(2)Cys motifs are not required for copper resistance or transport. Biochem. Biophys. Res. Commun. 286:414-418. [DOI] [PubMed] [Google Scholar]
- 17.Franke, S., G. Grass, and D. H. Nies. 2001. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147:965-972. [DOI] [PubMed] [Google Scholar]
- 18.Goldberg, M., T. Pribyl, S. Juhnke, and D. H. Nies. 1999. Energetics and topology of CzcA, a cation/proton antiporter of the RND protein family. J. Biol. Chem. 274:26065-26070. [DOI] [PubMed] [Google Scholar]
- 19.Gort, A. S., D. M. Ferber, and J. A. Imlay. 1999. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol. Microbiol. 32:179-191. [DOI] [PubMed] [Google Scholar]
- 20.Grass, G. 2000. Molekulargenetische und biochemische Charakterisierung der cnr Cobalt/Nickel-Resistenz-Determinante aus Ralstonia metallidurans CH34. Dissertation. Martin-Luther-Universität Halle-Wittenberg, Halle/Saale, Germany.
- 21.Grass, G., and C. Rensing. 2001. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286:902-908. [DOI] [PubMed] [Google Scholar]
- 22.Grass, G., and C. Rensing. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, A., K. Matsui, J. F. Lo, and S. Silver. 1999. Molecular basis for resistance to silver in Salmonella. Nat. Med. 5:183-188. [DOI] [PubMed] [Google Scholar]
- 24.Gupta, A., L. T. Phung, D. E. Taylor, and S. Silver. 2001. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology 147:3393-3402. [DOI] [PubMed] [Google Scholar]
- 25.Hantke, K. 2001. Bacterial zinc transporters and regulators. Biometals 14:239-249. [DOI] [PubMed] [Google Scholar]
- 26.Hay, M., J. H. Richards, and Y. Lu. 1996. Construction and characterization of an azurin analog for the purple copper site in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 93:461-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins, C. F. 1992. ABC-transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 28.Huffman, D. L., J. Huyett, F. W. Outten, P. E. Doan, L. A. Finney, B. M. Hoffman, and T. V. O'Halloran. 2002. Spectroscopy of Cu(II)-PcoC and the multicopper oxidase function of PcoA, two essential components of Escherichia coli pco copper resistance operon. Biochemistry 41:10046-10055. [DOI] [PubMed] [Google Scholar]
- 29.Hwang, J., X. Zhong, and P. C. Tai. 1997. Interactions of dedicated export membrane proteins of the colicin V secretion system: CvaA, a member of the membrane fusion protein family interacts with CvaB and TolC. J. Bacteriol. 179:6264-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight, S. A., S. Labbe, L. F. Kwon, D. J. Kosman, and D. J. Thiele. 1996. A widespread transposable element masks expression of a yeast copper transport gene. Genes Dev. 10:1917-1929. [DOI] [PubMed] [Google Scholar]
- 32.Kolczak, U., J. Salgado, G. Siegal, M. Saraste, and G. W. Canters. 1999. Paramagnetic NMR studies of blue and purple copper proteins. Biospectroscopy 5(Suppl.):19-32. [DOI] [PubMed] [Google Scholar]
- 33.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 34.Lee, S. M., G. Grass, C. Rensing, S. R. Barrett, C. J. Yates, J. V. Stoyanov, and N. L. Brown. 2002. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295:616-620. [DOI] [PubMed] [Google Scholar]
- 35.Liesegang, H., K. Lemke, R. A. Siddiqui, and H.-G. Schlegel. 1993. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J. Bacteriol. 175:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makdessi, K., J. R. Andreesen, and A. Pich. 2001. Tungstate uptake by a highly specific ABC transporter in Eubacterium acidaminophilum. J. Biol. Chem. 276:24557-24564. [DOI] [PubMed] [Google Scholar]
- 37.Mao, W. M., M. S. Warren, D. S. Black, T. Satou, T. Murata, T. Nishino, N. Gotoh, and O. Lomovskaya. 2002. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 46:889-901. [DOI] [PubMed] [Google Scholar]
- 38.Maseda, H., M. Kitao, S. Eda, E. Yoshihara, and T. Nakae. 2002. A novel assembly process of the multicomponent xenobiotic efflux pump in Pseudomonas aeruginosa. Mol. Microbiol. 46:677-686. [DOI] [PubMed] [Google Scholar]
- 39.Maseda, H., H. Yoneyama, and T. Nakae. 2000. Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:658-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mills, S. D., C. K. Lim, and D. A. Cooksey. 1994. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol. Gen. Genet. 244:341-351. [DOI] [PubMed] [Google Scholar]
- 41.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182:5864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami, S., R. Nakashima, R. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 43.Navarro, C., L. F. Wu, and M. A. Mandrand Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9:1181-1191. [DOI] [PubMed] [Google Scholar]
- 44.Nies, D. H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev., in press. [DOI] [PubMed]
- 45.Nies, D. H., A. Nies, L. Chu, and S. Silver. 1989. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 86:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nittis, T., G. N. George, and D. R. Winge. 2001. Yeast Sco1, a protein essential for cytochrome c oxidase function, is a Cu(I)-binding protein. J. Biol. Chem. 276:42520-42526. [DOI] [PubMed] [Google Scholar]
- 47.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 48.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 49.Pena, M. M. O., S. Puig, and D. J. Thiele. 2000. Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275:33244-33251. [DOI] [PubMed] [Google Scholar]
- 50.Petersen, C., and L. B. Moller. 2000. Control of copper homeostasis in Escherichia coli by a P-type ATPase, CopA, and a MerR-like transcriptional activator, CopR. Gene 261:289-298. [DOI] [PubMed] [Google Scholar]
- 51.Qian, H., L. Sahlman, P. O. Eriksson, C. Hambraeus, U. Edlund, and I. Sethson. 1998. NMR solution structure of the oxidized form of MerP, a mercuric ion binding protein involved in bacterial mercuric ion resistance. Biochem. USA 37:9316-9322. [DOI] [PubMed] [Google Scholar]
- 52.Rech, S., C. Wolin, and R. P. Gunsalus. 1996. Properties of the periplasmic ModA molybdate-binding protein of Escherichia coli. J. Biol. Chem. 271:2557-2562. [DOI] [PubMed] [Google Scholar]
- 53.Rensing, C., B. Fan, R. Sharma, B. Mitra, and B. P. Rosen. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 97:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rensing, C., M. Ghosh, and B. P. Rosen. 1999. Families of soft-metal-ion-transporting ATPases. J. Bacteriol. 181:5891-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rensing, C., T. Pribyl, and D. H. Nies. 1997. New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 179:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saier, M. H., Jr., R. Tam, A. Reizer, and J. Reizer. 1994. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol. Microbiol. 11:841-847. [DOI] [PubMed] [Google Scholar]
- 57.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 59.Schmidt, T., and H. G. Schlegel. 1994. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J. Bacteriol. 176:7045-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srikumar, R., X. Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoyanov, J. V., J. L. Hobman, and N. L. Brown. 2001. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39:502-511. [DOI] [PubMed] [Google Scholar]
- 62.Tikhonova, E. B., Q. J. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai, K. J., Y. F. Lin, M. D. Wong, H. H. C. Yang, H. L. Fu, and B. P. Rosen. 2002. Membrane topology of the pl258 CadA Cd(II)/Pb(II)/Zn(II)-translocating P-type ATPase. J. Bioenerg. Biomembr. 34:147-156. [DOI] [PubMed] [Google Scholar]
- 64.Tseng, T.-T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 65.Yoneyama, H., A. Ocaktan, M. Tsuda, and T. Nakae. 1997. The role of mex-gene products in antibiotic extrusion in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 233:611-618. [DOI] [PubMed] [Google Scholar]
- 66.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, H., and D. J. Thiele. 2001. Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276:20529-20535. [DOI] [PubMed] [Google Scholar]