Abstract
A monooxygenase encoded by the mtmOIV gene from the mithramycin gene cluster of Streptomyces argillaceus was purified 21-fold by a three-step purification procedure. This monooxygenase catalyzes the oxidative cleavage of the fourth ring of premithramycin B. The enzyme was dependent on NADPH and flavin adenine dinucleotide for activity with optimal pH at 9.5, and the Km values for NADPH and premithramycin B were 269.22 and 23.35 μM, respectively. The reaction catalyzed by MtmOIV yields two possible isomers of the same basic shortened aliphatic chain molecule. One of the reaction products showed important biological activity, thus highlighting the importance of the cleavage of the fourth ring of the aglycon for biological activity.
Mithramycin is a member of the aureolic acid group of antitumor compounds (8, 12). It shows antibacterial activity against gram-positive bacteria and also a remarkable cytotoxicity against a variety of tumor cell culture lines. Mithramycin has been clinically used for the treatment of certain tumors, such as disseminated embryonic cell carcinoma, as well as for Paget's bone disease (12) and is also used for control of hypercalcemia in patients with malignant disease (1). The biosynthetic gene cluster for mithramycin has been fully characterized (2-6, 9), and it has been found that although mithramycin is a tricyclic aromatic polyketide, its biosynthesis proceeds through tetracyclic intermediates. Many of these intermediates have been isolated from mutants blocked at different steps in the biosynthesis (2-6). Insertional inactivation of one of the genes of the mithramycin cluster, mtmOIV, generated a non-mithramycin-producing mutant that accumulated a fully glycosylated tetracyclic intermediate, premithramycin B (6). This gene codes for an oxygenase responsible for the oxidative cleavage of the fourth ring of premithramycin B, and after this oxidation step, the final mithramycin molecule is produced by a ketoreduction of the 4′ position at the generated aliphatic chain (6). Since only mithramycin, and not the biosynthetic intermediates, possesses antibiotic and antitumor properties, the opening of the fourth ring appears as a very important step in the biosynthesis of this antitumor compound.
Purification and characterization of the MtmOIV oxygenase.
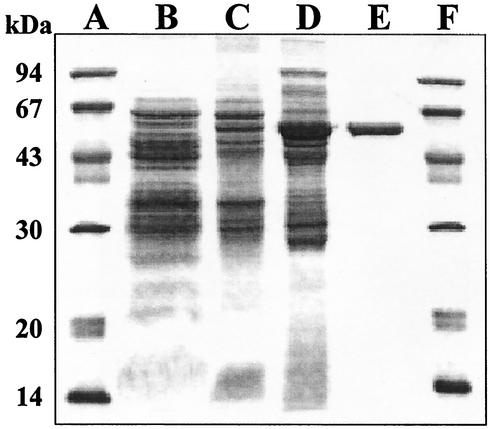
The MtmOIV oxygenase was purified from Streptomyces albus LP04, a strain that overexpresses MtmOIV. Enzyme activity was monitored at 30°C by using premithramycin B as a substrate to determine NADPH oxidation at 340 nm. The enzyme was purified about 21-fold with a three-step purification procedure (including ammonium sulfate precipitation and two chromatographic steps) using affinity chromatography on the immobilized reactive dye Blue 4 and anionic exchange on Mono Q HR 5/5 (Table 1; Fig. 1). The enzyme was purified as a single band, as observed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), migrating at 56.1 kDa (Fig. 1). The enzyme was eluted from the Blue 4 column at 40 mM NaCl and from the Mono Q HR 5/5 column at 230 mM NaCl. Gel filtration with Superose-6 indicated an apparent molecular mass of the native enzyme of 56.2 kDa. Together with the SDS-PAGE results, this suggested that the native enzyme is a monomer.
TABLE 1.
Purification of MTMOIV enzyme
| Purification step | Total protein (mg) | Total activity (μmoles/min) | Specific activity (nmoles/min/mg of protein) | Purifi- cation (fold) |
|---|---|---|---|---|
| Cell-free extract | 120 | 0.088 | 0.7 | 1 |
| (NH4)2SO4 | 43.2 | 0.124 | 2.9 | 4.1 |
| Sepharose Blue 4 | 4.7 | 0.030 | 6.4 | 9.1 |
| Mono Q | 0.34 | 0.005 | 14.7 | 21 |
FIG. 1.
SDS-PAGE analysis of the different steps in the purification of the MtmOIV monooxygenase. Lanes A and F, molecular mass markers; lane B, cell extract; lane C, ammonium sulfate precipitation; lane D, active fractions eluted from the Blue 4 column; lane E, active fractions eluted from the Mono Q column. Protein (15 μg) was loaded on lanes B, C, and D. Pure MtmOIV (3 μg) was loaded on lane E.
The purified enzyme was very unstable, losing most of its activity within a few days even when kept at a low temperature. The enzyme was not stabilized with 20% glycerol, although stability increased at 40% glycerol. Other stabilizing agents, such as 1 M MgCl2, the nonionic detergent Triton X-100 at 1% vol/vol, or albumin at 10 mg/ml, were also not very effective. The best results were obtained when 1 M ammonium sulfate was used; the activity of the enzyme increased about three times compared with the results obtained using other stabilizing agents. Nevertheless, even under these conditions, the enzyme was active only for a few weeks when kept at −70°C.
NADPH was more efficient than NADH as a cofactor for the oxidoreductase reaction, and the oxidation was also dependent on the presence of flavin adenine dinucleotide. pH dependence for the reaction showed a typical bell-shaped curve, with an optimum value in the alkaline region at pH 9.5. Steady-state initial velocity studies of the reaction were performed by using different concentrations of NADPH at different fixed concentrations of premithramycin B. To obtain estimates of the kinetic constants, the full data set was then fitted by performing nonlinear regression analysis with the equation of Alberty. The parameters obtained were as follows: KmNADPH, 269.2 μM; Kmpremithramycin B, 23.3 μM; and Vmax, 12.1 nmol min−1 mg protein−1.
Five intermediates of the mithramycin biosynthetic pathway were used as potential substrates for the MtmOIV oxygenase: premithramycinone, premithramycin A1, 9-demethyl-premithramycin A2, premithramycin A2, and premithramycin A3. Premithramycin B was used as a control. Several other tetracyclic aromatic polyketide antibiotics were also tested: chromocyclomycin, tetracenomycin C, tetracycline, and daunorubicin. Reactions analyzed by monitoring the oxidation of NADPH produced positive results when premithramycin A2, premithramycin A3, and chromocyclomycin were used as substrates. Nevertheless, when the reaction was analyzed by high-pressure liquid chromatography (HPLC), no transformation of any of the substrates mentioned above except premithramycin B was detected.
Isolation and analysis of the reaction products.
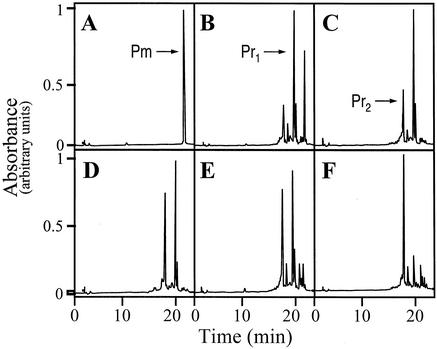
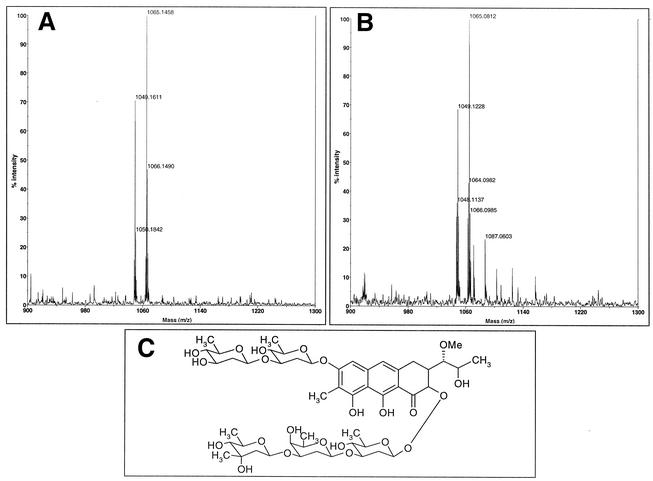
Discontinuous reactions were performed for the isolation of the oxygenase reaction product. After incubation, the reaction mixture was extracted twice with 1 vol of ethyl acetate after decreasing the pH by the addition of 100 μl of 0.2 N HCl. After evaporation of the solvent, the residue was dissolved in 50 μl of methanol and the product of the reaction was monitored and purified by analytical HPLC as described previously (6). At short reaction times (t = 5 min), a major product (Pr1) appeared with an HPLC retention time of 19.6 min (Fig. 2) and with an absorption spectrum different from that of premithramycin B and very similar to that of mithramycin; the absorption peaks of this molecule were displaced towards shorter wavelengths, as expected from the opening of the fourth ring. After 15 min of reaction, a second peak (Pr2) appeared in a different region of the chromatogram (17.7 min) whose spectrum showed some slight differences when compared with the spectrum obtained for the first product of the reaction. At longer times of reaction, the first product was progressively transformed into the second one (Fig. 2). Both HPLC-isolated products were incubated in the presence and in the absence of the enzyme under reaction conditions and it was found that Pr1 was transformed into Pr2 when the enzyme was present, but no conversion of Pr2 into Pr1 was observed. Individual peaks remained stable in the absence of the enzyme. Attempts to obtain milligram amounts of the reaction product for its structural elucidation were unsuccessful, due to the high instability of the enzyme. Matrix-assisted laser desorption ionization-time of flight (mass spectrometry) analysis showed, surprisingly, identical molecular mass values for both products (1,048.1 and 1,064.1 for the Na+ and K+ adducts, respectively) (Fig. 3). This finding strongly suggests that Pr1 and Pr2 are isomers for the same molecule. The biological activity of the reaction products was determined by bioassays performed with Micrococcus luteus ATCC 10240. Product Pr1 presented an important antibiotic activity, in contrast to premithramycin B, which showed no activity at all. The antibiotic activity of this product was about 75% of that shown by an equivalent amount of mithramycin. In contrast, product Pr2 did not show any biological activity when assayed against M. luteus. That Pr2 was obtained by isomerization, together with the resulting changes in protonation state at the phenolic O8 responsible for the binding of the molecule to the DNA duplex, could explain this loss of activity. Other possible candidates for this chemical change (involved in the loss of activity) are the phenolic O9 or the carbonilic O1 of the aglycon, which are essential for mithramycin dimerization, as they couple with stoichiometric amounts of Mg2+ (11). Interestingly, and considering that there is no biological activity in any of the mithramycin biosynthetic intermediates, the opening of the fourth ring becomes essential for the generation of the bioactivity of the molecule. This observation seems to be related to the way in which mithramycin exercises its biological effect. Opening of the fourth ring can be essential for the generation of the dimer and its interaction with DNA; structural studies have shown that 3′-OH and 4′-OH hydroxyl protons on the hydrophilic side chain can form hydrogen bonds with the backbone phosphates of the DNA in this way (11).
FIG. 2.
Time course for the reaction catalyzed by MtmOIV. The results of analytical HPLC after the indicated reaction times are shown as follows: panel A, 0 min; panel B, 5 min; panel C, 15 min; panel D, 30 min; panel E, 1 h; panel F, 4 h. Chromatographic peaks corresponding to premithramycin B (Pm) and the first (Pr1) and second (Pr2) reaction products are indicated by arrows.
FIG. 3.
Mass spectrum analysis of the MtmOIV monooxygenase reaction product. (A) First reaction product (Pr1). Molecular masses of 1,049.1 and 1,065.1 corresponded to the respective adducts of the product with Na+ and K+. (B) Second reaction product (Pr2). Molecular masses of 1,049.1 and 1,065.08 corresponded to the respective adducts of the product with Na+ and K+. Both products are probably isomers for the same molecule. (C) Proposed structure for the short-chain product obtained by the activity of the MtmOIV monooxygenase.
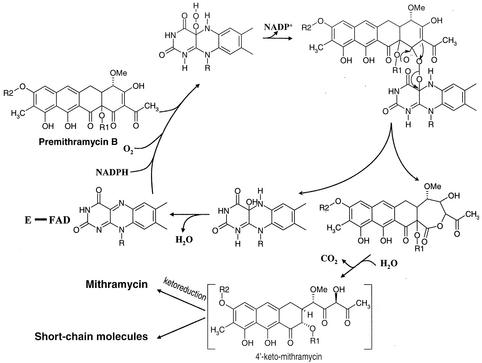
Considering all the preceding data, it is possible to suggest a Baeyer-Villiger-based mechanism for the oxidative cleavage of the fourth ring of premithramycin B by MtmOIV monooxygenase (Fig. 4). Binding of NADPH, O2, and premithramycin B by the enzyme is required. Although to know the order of substrate addition would require additional kinetic data, information from other Baeyer-Villiger monooxygenase studies suggests an ordered mechanism (10) in which NADPH reduces flavin adenine dinucleotide and the subsequent addition of an oxygen molecule generates a flavin hydroperoxide. The distal oxygen of this hydroperoxide would react as a nucleophilic oxygen atom attacking the electrophilic carbonyl ketone carbon in the cosubstrate. The result would be the oxygen transfer and ring expansion to form a seven-membered cyclic product. Subsequent hydrolysis and loss as CO2 of one carbon unit, presumably spontaneous, would generate the product 4′-keto-mithramycin. This would not be the final product of the pathway. To generate mithramycin, a further reduction to alcohol of the keto group at position 4′ of the newly generated hydrophilic side chain would be necessary. Coordination of the activities of the MtmOIV monooxygenase and the ketoreductase seems to be essential for the stability of the final mithramycin molecule, as the presence of the nonreduced keto group drives great instability at the hydrophilic chain, probably by tautomerism. This conclusion is reinforced when considering the structure of the products obtained in the MtmOIV reaction (Fig. 4), lacking aliphatic carbons as deduced from mass spectrum analysis. Some other short-chain mithramycin (7) and chromomycin (Nuria Menéndez, unpublished results) intermediates have been isolated. In both cases, these compounds were accumulated by mutants lacking the 4′ ketoreductase. The chemical pathways leading to the formation of these shortened-chain intermediates remain unknown. Their elucidation could contribute to explaining the significance of the ketoreduction in the stability of the compounds once the fourth ring has been opened.
FIG. 4.
Proposed Baeyer-Villiger-based oxidation mechanism for the oxidative opening of the fourth ring of premithramycin B by MtmOIV.
Acknowledgments
This research was supported by a grant of the Plan Regional de Investigación del Principado de Asturias (GE-MED01-05).
We wish to thank Felipe Lombó for discussions and for helping us in the MALDI-TOF analyses.
D.R. and L.M.Q. have equally contributed to this work.
REFERENCES
- 1.Dewick, P. M. 1997. Medicinal natural products: a biosynthetic approach, p. 85. John Wiley & Sons, Chichester, N.Y.
- 2.Fernández, E., U. Weiβbach, C. Sánchez Reillo, A. F. Braña, C. Méndez, J. Rohr, and J. A. Salas. 1998. Identification of two genes from Streptomyces argillaceus encoding two glycosyltransferases involved in the transfer of a disaccharide during the biosynthesis of the antitumour drug mithramycin. J. Bacteriol. 180:4929-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernández-Lozano, M. J., L. Remsing, L. M. Quirós, A. F. Braña, E. Fernández, C. Sánchez C. Méndez, J. Rohr, and J. A. Salas. 2000. Characterization of two polyketide methyltransferases involved in the biosynthesis of the antitumour drug mithramycin by Streptomyces argillaceus. J. Biol. Chem. 275:3065-3074. [DOI] [PubMed] [Google Scholar]
- 4.Lombó, F., G. Blanco, E. F. Fernández, C. Méndez, and J. A. Salas. 1996. Characterization of Streptomyces argillaceus genes encoding a polyketide synthase involved in the biosynthesis of the antitumour mithramycin. Gene 172:87-91. [DOI] [PubMed] [Google Scholar]
- 5.Lombó, F., K. Siems, A. F. Braña, C. Méndez, K. Bindseil, and J. A. Salas. 1997. Cloning and insertional inactivation of Streptomyces argillaceus genes involved in earliest steps of sugar biosynthesis of the antitumour mithramycin. J. Bacteriol. 179:3354-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prado, L., E. Fernández, U. Weissbach, G. Blanco, L. M. Quirós, A. F. Braña, C. Méndez, J. Rohr, and J. A. Salas. 1999. Oxidative cleavage of premithramycin B is one of the last steps in the biosynthesis of the antitumor drug mithramycin. Chem. Biol. 6:19-30. [DOI] [PubMed] [Google Scholar]
- 7.Remsing, L. L., A. M. Gonzalez, M. Nur-e-Alam, M. J. Fernandez-Lozano, A. F. Braña, U. Riz, M. A. Oliveira, C. Méndez, J. A. Salas, and J. Rohr. Mithramycin SK, a novel antitumour drug with improved therapeutic index, mithramycin SA, and demycarosyl-mithramycin SK: three new products generated in the mithramycin producer Streptomyces argillaceus through combinatorial biosynthesis. J. Am. Chem. Soc., in press. [DOI] [PMC free article] [PubMed]
- 8.Rohr, J., C. Méndez, and J. A. Salas. 1999. The biosynthesis of aureolic acid group antibiotics. Bioorg. Chem. 27:41-54. [Google Scholar]
- 9.Rohr, J., U. Weissbach, C. Beninga, E. Künzel, K. Siems, K. Bindseil, L. Prado, F. Lombó, A. F. Braña, C. Méndez, and J. A. Salas. 1998. The structure of premithramycinone and demethylpremythramycinone, early intermediates of the aureolic acid group antibiotic mithramycin. Chem. Commun. 3:437-438. [Google Scholar]
- 10.Ryerson, C. C., D. P. Ballou, and C. Walsh. 1982. Mechanistic studies on cyclohexanone oxygenase. Biochemistry 21:2644-2655. [DOI] [PubMed] [Google Scholar]
- 11.Sastry, M., and D. J. Patel. 1993. Solution structure of the mithramycin dimer-DNA complex. Biochemistry 32:6588-6604. [DOI] [PubMed] [Google Scholar]
- 12.Skarbek, J. D., and M. K. Speedie. 1981. Antitumor antibiotics of the aureolic acid group: chromomycin A3, mithramycin A, and olivomycin A, p. 191-235. In A. Aszalos (ed.), Antitumor compounds of natural origin: chemistry and biochemistry, 8th ed., vol. 1. CRC Press, Boca Raton, Fla.