Abstract
Dopamine and its receptors appear in the developing brain early in the embryonic period and dopamine receptor activation influences proliferation and differentiation of neuroepithelial precursor cells. Since dopamine D1 and D2 receptor activation produces opposing effects on precursor cell activity, dopamine's overall effects may correlate with relative numbers and activity of each receptor subtype on the precursor cells. Dopamine receptor expression and activity in individual precursor cells in the intact brain are difficult to ascertain. Therefore, cell lines with known receptor expression profiles can be useful tools to study dopamine's influence on neuroepithelial cells. We report characterization of dopamine receptor expression and activity profiles in three mouse striatal precursor cell lines and suggest that these cell lines can be valuable tools to study dopamine's effects on striatal precursor cell proliferation and differentiation.
Keywords: Ganglionic eminence, Dopamine, Cell cycle, Striatum, D1 receptor, D2 receptor
Introduction
Dopamine and its receptors are involved in the regulation of mood, motivation and motor function in the mature central nervous system (CNS). However, dopamine is present in the brain early in development and functional dopamine receptors are expressed in the CNS prior to the onset of synaptogenesis [Jung and Bennett, 1996; Diaz et al., 1997; Shearman et al., 1997; Ohtani et al., 2003], suggesting a role for dopamine in brain development that may be independent of its role at the synapse in the mature CNS [Lauder, 1988, 1993; Levitt et al., 1997; Nguyen et al., 2001; Ohtani et al., 2003]. Neurotransmitters such as dopamine can influence brain development by modulating neurogenesis or neuronal and glial cell differentiation [LoTurco et al., 1995; Schmidt et al., 1996; Lavdas et al., 1997; Bongarzone et al., 1998; Haydar et al., 2000; Popovik and Haynes, 2000; Demarque et al., 2002; Luk et al., 2003; Luk and Sadikot, 2004; Popolo et al., 2004]. Recent studies show that dopamine receptor activation influences the cell cycle of neuroepithelial cells in the lateral ganglionic eminence and the cerebral wall [Zhang and Lidow, 2002; Ohtani et al., 2003; Popolo et al., 2004; Zhang et al., 2005]. Activation of dopamine D1 receptor (D1R) decreases incorporation of the S-phase marker bromodeoxyuridine (BrdU) by neuroepithelial cells whereas activation of the D2 receptor (D2R) increases it [Ohtani et al., 2003; Popolo et al., 2004]. The molecular mechanisms mediating the effects of dopamine receptor activation on cell proliferation or differentiation are only now beginning to be understood [Zhang and Lidow, 2002; Zhang et al., 2005].
Availability of precursor cell lines expressing D1R and D2R could facilitate investigation of the cell and molecular mechanisms of dopamine's action on precursor cells, neurons and glia in the developing brain. We characterized D1R and D2R expression and activity in a mouse striatal precursor cell line STHdh+/Hdh+ [Trettel et al., 2000] that was established by retroviral transduction of the temperature-sensitive mutant tsA58 of the SV40 large T antigen so that the cells remain proliferative at 33°C and lose the SV40 tsA58 oncoprotein and cease proliferation at 39°C [Cattaneo and Conti, 1998; Trettel et al., 2000]. We found that the STHdh+/Hdh+ cell line expressed predominantly D2R mRNA and only low levels of D1R mRNA. Therefore, we stably transfected the STHdh+/Hdh+ cell line with mouse D1R constructs to generate a new cell line that would express functional D1R in addition to the endogenous D2R. We examined the effects of activation of D1R and D2R on cell proliferation and differentiation in these cell lines and our data show that the cell lines can be useful in vitro tools to examine the effects of dopamine receptor activation on the activity of striatal progenitor cells.
Materials and Methods
Generation of Cell Lines Expressing Mouse D1R-EGFP or EGFP Transgenes
Our initial experiments showed that the STHdh+/Hdh+ cell line expressed D2R mRNA and only low levels of D1R mRNA (see below). Therefore, we stably transfected the STHdh+/Hdh+ line with a mouse D1R-EGFP construct or an EGFP reporter construct to generate two new cell lines, as follows.
Plasmids
We cloned mouse dopamine receptor cDNA from Mouse Brain QUICK-Clone cDNA (BD Biosciences/Clontech, San Jose, Calif., USA) by PCR using the following primers: 5′-CGCTCGAGCGCCACCGAAGATGGCTCCTAACACTT-3′ and 5′-CGGGATCCGGTTGAATGCTGCTGTCCGCTGT-3′. The primers were designed to eliminate the stop codon. EGFP-tagged D1R construct (pD1R-EGFP) was designed by in-frame cloning of XhoI and BamHI fragment DNA from PCR products into pEGFP-N3 vector (BD Biosciences/Clontech) (fig. 1).
Fig. 1.
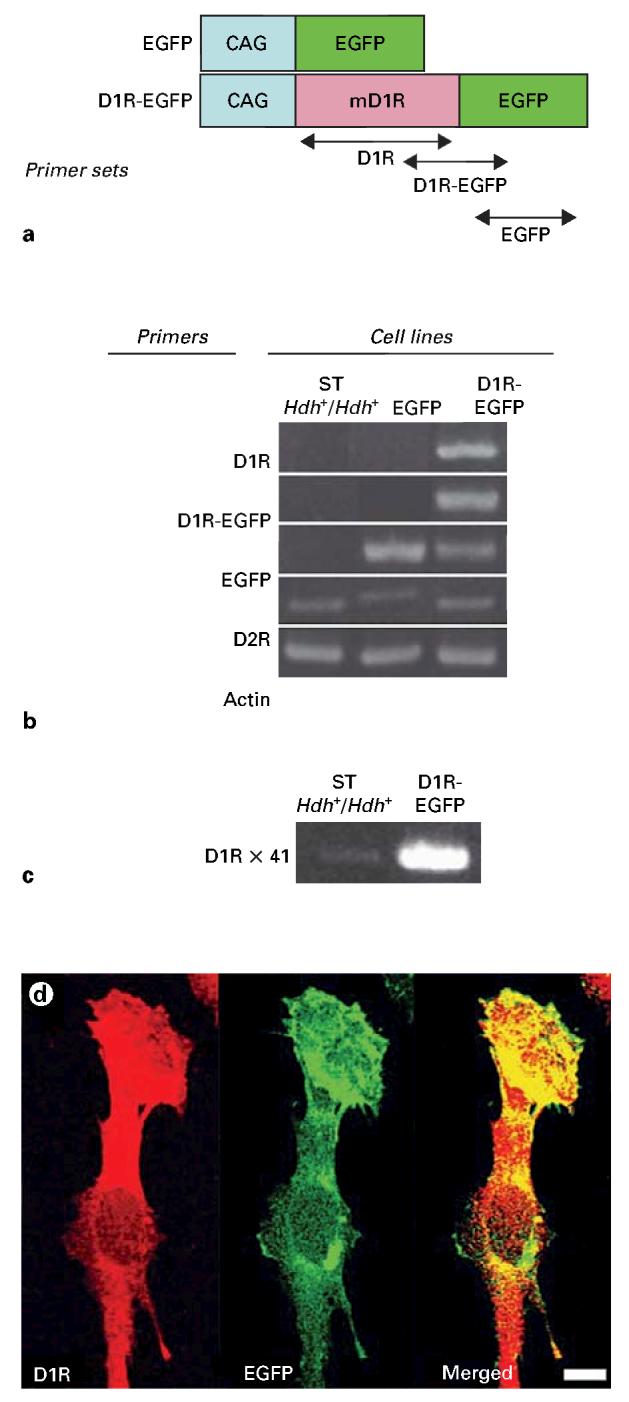
Dopamine receptor expression profiling of striatal progenitor cell lines. a STHdh+/Hdh+ line, established from E14 mouse striatal cells [Trettel et al., 2000], was transfected with EGFP reporter or mouse D1R-EGFP construct to generate EGFP and D1R-EGFP cell lines, respectively. b PCR analysis of the three cell lines shows robust D1R and D1R-EGFP mRNA expression in the D1R-EGFP line, EGFP mRNA expression in the EGFP and D1R-EGFP lines and D2R mRNA expression in all three lines. Actin mRNA levels show equal sample loading in each lane. c When the PCR cycle number was increased to 41 to detect low abundance transcripts, low levels of D1R mRNA could be detected in the STHdh+/Hdh+ line compared to robust expression of this mRNA in the D1R-EGFP line. d Immunocytochemical localization of D1R and expression of EGFP fluorescence show considerable overlap between the two in cells from the D1R-EGFP line indicating that the D1R-EGFP fusion protein is expressed in the same subcellular compartments as the endogenous D1R protein. Scale bar = 10 μm.
Stable Transfection
STHdh+/Hdh+ cells remain proliferative at 33°C and cease proliferation at 39°C [Trettel et al., 2000]. We transfected STHdh+/Hdh+ cells growing at 33°C with either pD1R-EGFP or pEGFP-N3 and pPUR (fig. 1a) (BD Biosciences/Clontech) using LipofectAMINE™ 2000 (Invitrogen, Rockville, Md., USA), according to the manufacturer's instructions. Transfected colonies were selected by growing in medium containing 0.8 mg/ml Geneticin, and 40 μg/ml puromycin (Invitrogen) followed by cell sorting (FACSVantage™; BD Biosciences, Franklin Lakes, N.J., USA). Stable transfection was also confirmed by observing EGFP expression in the cells or colonies under a fluorescence microscope. Thus, we generated two new cell lines: the D1R-EGFP line expressing wild-type mouse D1R-EGFP and the EGFP line expressing only the EGFP reporter sequence. We used these two lines along with the original STHdh+/Hdh+ line in the experiments described here.
Cell Culture
Cells were grown at 33°C in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% nonessential amino acids, 2 mM L-glutamine, 1% penicillin-streptomycin, 0.8 mg/ml Geneticin, and 40 μg/ml puromycin (Invitrogen). The cells remain proliferative under these conditions [Cattaneo and Conti, 1998; Trettel et al., 2000]. In other experiments involving cell differentiation, the cells were cultured at 37°C in 8-well culture chambers (BD Biosciences) at a density of 1 × 104 cells per well in a differentiation medium [Neurobasal/B27, 2 mM L-glutamine, 1% penicillin-streptomycin (Invitrogen), and 0.5% D-glucose (Sigma, St. Louis, Mo., USA)]. The medium was changed every other day. For immunoblot analysis, 5 × 105 cells were cultured in a 100-mm cell culture dish (BD Biosciences).
Application of Drugs
Dopamine (10 μM), D1R agonist SKF81297 (1 μM), D1R antagonist Schering 23390 (10, 20 μM), D2R agonist quinpirole (10 μM) or forskolin (25, 50 μM) (all chemicals from Sigma-RBI) were added to the culture medium directly. D1R antagonist was applied 30 min prior to agonist application in experiments designed to test specificity of D1R activation. For analysis of cell proliferation, BrdU (10 mM; Sigma) was added to the medium 1 h prior to fixation.
RT-PCR for D1R and D2R
We isolated total RNA from cells by using RNAqueous-4PCR (Ambion, Austin, Tex., USA) and reverse transcribed 2 μg of total RNA to synthesize cDNA. The following primer sets were used: for D1R: 5′-AGCAGGACGTATGCCATTTC-3′, 5′-CCACACAAACACATCGAAGG-3′; for D1R-EGFP: 5′-AAGGCATTCTCGACCCTCTT-3′, 5′-TGGTGCAGATGAACTTCAGG-3′; for EGFP: 5′-ACGTAAACGGCCACAAGTTC-3′, 5′-TGTTCTGCTGGTAGTGGTCG-3′; for D2R: 5′-ACCACGGCCTACATAGCAAC-3′, 5′-GTGAAGGCGCTGTAGAGGAC-3′, and for actin: 5′-CACGGCATTGTAACCAACTG-3′, 5′-TCTCAGCTGTGGTGGTGAAG-3′.
Cyclic Adenosine Monophosphate Assay
We examined cyclic adenosine monophosphate (cAMP) accumulation following activation of the dopamine receptors in the different cell lines. For these experiments, we cultured 4 × 104 cells in 24-well plates for 2 days at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% non-essential amino acids, 2 mM L-glutamine, 1% penicillin-streptomycin, 0.8 mg/ml Geneticin, and 40 μg/ml puromycin (Invitrogen). Then, the cells were washed with PBS twice and dopaminergic drugs were applied for 15 min. Cellular cAMP was measured by cAMP Biotrak EIA system (Amersham Bioscience, Piscataway, N.J., USA) according to the manufacturer's protocol. The optical density of each well was measured using VICTOR V (PerkinElmer, Boston, Mass., USA) at 450 nm.
Immunoblot Analysis
Protein extracts were prepared from cultured cells grown at 37°C as described above. Cells were lysed with buffer containing 2% SDS and 62.5 mM Tris-HCl (pH 6.8). The total cell lysates were sonicated and denatured. Protein concentration was measured by using BCA protein assay kit (PIERCE, Rockford, Ill., USA). Total protein extract (10-20 μg) was resolved on SDS-PAGE and transferred to nitrocellulose membranes (BioRad, Hercules, Calif., USA). The blots were incubated in buffer containing 5% skim milk in TBS-T (50 mM Tris-HCl, 150 mM NaCl, pH 7.4, 0.05% Tween R20). The blots were incubated overnight at 4°C with primary antibodies: phospho-CREB (1:1,000), CREB (1:1,000) (both from Upsate, Charlottesville, Virgin Islands, USA), CNPase (1:200; Chemicon, Temecula, Calif., USA), glial fibrillary acidic protein (GFAP; 1:300), and actin (1:1,000) (both from Sigma) in TBS containing 1.5% BSA. Then the blots were incubated with horseradish peroxidase-conjugated donkey antimouse (1:2,000), or antirabbit (1:2,000) (both from Amersham Bioscience) in TBS containing 1.5% BSA. Immunoreactivity was detected using SuperSignal West Pico Chemiluminescent Substrate (PIERCE), and exposed to autoradiographic film (Eastman Kodak, Rochester, N.Y., USA). Optical density of the bands on the film was measured using a computerized image analysis system (MCID Elite, GE Healthcare, Wisc., USA).
Immunocytochemistry
Cultured cells, grown at 37°C as described above, were washed with PBS and fixed with 4% formaldehyde in 0.1 M phosphate buffer, pH 7.2, then washed with PBS three times. Cells were incubated with blocking buffer containing 10% donkey serum and 0.1% Triton X-100 in PBS. For BrdU staining, cells were treated with 2 N HCl for 1 h and washed again with PBS before blocking in 10% donkey serum and 0.5% Tween 20 in PBS. Cells were incubated with primary antibodies: βIII-tubulin (1:500; Covance, Berkeley, Calif., USA), BrdU (1:75; BD Biosciences). Immunoreactivity was detected using Alexa 568-conjugated donkey antimouse antibody (Invitrogen). Cell nuclei were stained with 1 μg/ml Hoechst 33342 (Invitrogen) in PBS. Images were acquired on a Nikon ECLIPSE E400 microscope (Nikon, Melville, N.Y., USA) fitted with FITC, RITC and UV fluorescence attachments. Images were captured using an RT Slider CCD camera and SPOT RT software (Diagnostic Instruments, Sterling Heights, Mich., USA).
For quantitative analysis of βIII-tubulin- or BrdU-labeled cells, each type of culture was examined under a × 20 objective to calculate the number of each type of labeled cells. Microscope fields were chosen randomly from each well and BrdU-, βIII-tubulin- and Hoechst 33342-labeled cells within defined areas of each field were counted. Cell counts were expressed as ratios of BrdU- or βIII-tubulin-labeled cells to Hoechst 33342-labeled cells. We counted cells in 2–3 fields from each of 10 cultures per treatment group. Statistical significance of the overall differences among the experimental groups was analyzed by one-way analysis of variance (ANOVA) and if significant differences were found then to determine more specifically the source of the significant differences, we compared the mean values between each pair of groups using Tukey's multiple comparisons test (GraphPad Software Inc., San Diego, Calif., USA).
Results
Dopamine Receptor mRNA and Protein Expression
Our initial studies examined the expression of D1R and D2R mRNA in the STHdh+/Hdh+ line by RT-PCR. We found that this line expressed D2R mRNA (fig. 1b) and very low levels of D1R mRNA that was detectable only when the PCR cycle number was increased to 41 (fig. 1c). Like the ST Hdh+/Hdh+ line, our EGFP line expressed D2R mRNA. Our D1R-EGFP line, which was stably transfected with mouse D1R-EGFP transgene, expressed both D2R and D1R mRNAs as well as mRNA for the D1R-EGFP fusion sequence. Thus, the STHdh+/Hdh+ and EGFP lines expressed mainly D2R mRNA while the D1R-EGFP line expressed both D1R and D2R mRNAs. The EGFP and D1R-EGFP lines expressed EGFP mRNA as well (fig. 1b). Cells from the D1R-EGFP line showed punctate EGFP labeling (fig. 1d), presumably indicating the distribution of the D1R-EGFP fusion protein, in keeping with the expected intracellular distribution of D1R in neuroepithelial cells [Zhang and Lidow, 2002]. Immunostaining of D1R in the D1R-EGFP cells showed considerable overlap between D1R immunoreactivity and the EGFP signal (fig. 1d) confirming that the D1R-EGFP line overexpressed the D1R protein and that the distribution of EGFP fluorescence overlapped with that of the D1R protein.
D1R and D2R Function
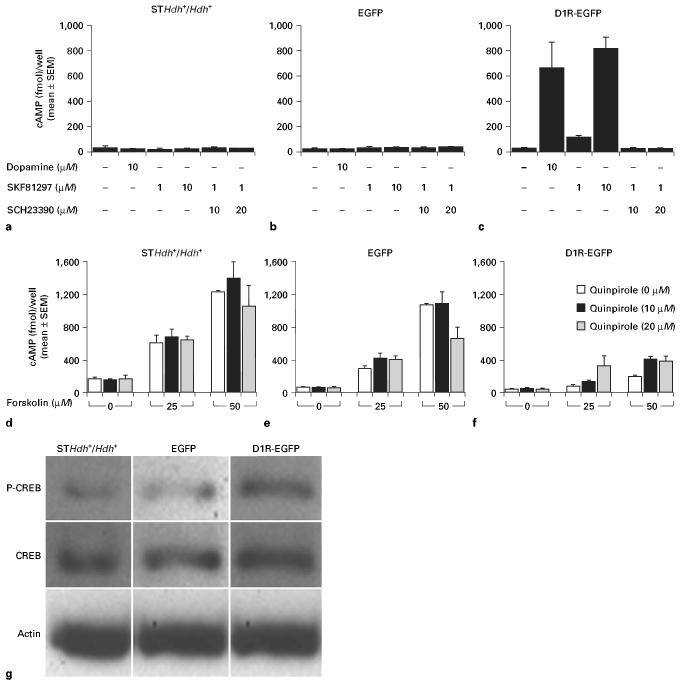
We examined whether D1R and D2R are functional in our cell lines by using cAMP accumulation assays (fig. 2 a–f). D1R is coupled to Gs proteins, which activate adenylate cyclase, which in turn converts ATP to cAMP [Monsma et al., 1990; Schinelli et al., 1994; Robinson and Caron, 1996]. Therefore, D1R activation increases intracellular cAMP levels, which can be measured directly using cAMP Biotrak EIA system. On the other hand, D2R is coupled to Gi proteins, which inhibit adenylate cyclase and decrease intracellular cAMP levels [Monsma et al., 1990; Schinelli et al., 1994; Robinson and Caron, 1996]. Therefore, an indirect method to assay D2R-induced suppression of adenylate cyclase and cAMP synthesis was used. In this method, we initially exposed the cells to forskolin, a direct activator of adenylate cyclase, to induce intracellular cAMP synthesis. In a parallel set of experiments, we exposed the cells to forskolin plus quinpirole, a selective D2R agonist. If the cells expressed functional D2R, then activation of these receptors by quinpirole would suppress the forskolin-induced cAMP synthesis.
Fig. 2.
Analysis of dopamine receptor function by cAMP (a–f) and CREB phosphorylation (g) assays in the STHdh+/Hdh+, EGFP and D1R-EGFP lines. Dopamine and the D1R agonist SKF81297 did not increase cAMP levels in the STHdh+/Hdh+ or EGFP lines (a, b). Both dopamine and SKF81297 increased cAMP levels in the D1R-EGFP line in a dose-dependent manner (c), in comparison to the cAMP level under control conditions (i.e. no drugs). When D1R activity was blocked by the specific D1R antagonist Schering 23390 (SCH23390), prior to exposing the cell lines to the D1R agonist SKF81297, the increase in cAMP levels produced by SKF81297 alone in the D1R-EGFP line was blocked confirming the specificity of the D1R induced by the D1R-EGFP transgene. We examined the function of D2R by analyzing decreases in forskolin-induced cAMP levels following activation of D2R (d–f), because D2R activation decreases intracellular cAMP levels. Cells were exposed to forskolin (0, 25 or 50 μM) to stimulate cAMP synthesis and then exposed to the D2R agonist quinpirole (0, 10 or 20 μM). Forskolin increased the cAMP levels in all three lines in a dose-dependent manner (open bars). Quinpirole (20 μM) decreased the forskolin-induced cAMP levels (at forskolin concentrations of 50 μM) in the STHdh+/Hdh+ and EGFP lines (d, e), confirming that D2R was functional in these two cell lines. Quinpirole did not decrease cAMP levels induced by forskolin in the D1R-EGFP line (f), suggesting that D2R was not functional or that its function in this line could not be ascertained by the assay. Phosphorylation of CREB following activation of D1R by its agonist SKF81297 was assayed by Western blot (g). Phospho-CREB (P-CREB) expression was robust in the D1R-EGFP line and low in the STHdh+/Hdh+ and EGFP lines. CREB (unphosphorylated) expression did not show significant changes. Actin levels indicate equal loading of samples in the different lanes.
In the experiments to assay D1R function, we exposed all three cell lines to dopamine or the D1R agonist SKF81297 (1 or 10 μM) for 15 min and measured cAMP accumulation. We found that cAMP levels increased only in the D1R-EGFP line, indicating that the D1R-EGFP line expressed functional D1R. We did not see increases in the cAMP levels in the STHdh+/Hdh+ or EGFP lines at the concentrations of dopamine or SKF81297 used in this assay (fig. 2a, b), presumably because of the low level of expression of D1R in these two lines (fig. 1b). Therefore, the cAMP data are consistent with the D1R mRNA expression profiles for all three cell lines (fig. 1b). To further assess the specificity of D1R, we exposed the cells to the D1R antagonist Schering 23390 (10 or 20 μM) for 30 min and then to the D1R agonist SKF81297 (1 μM). In these experiments, we did not see an increase in cAMP levels, not even in the D1R-EGFP line, which is confirmation that the transgene-induced D1R was not only functional but also specific (fig. 2c).
We examined D2R function by using a combination of forskolin and the D2R agonist, quinpirole. Forskolin produced a dose-dependent increase in cAMP levels in all three cell lines (compare the open bars in figure 2 d–f) with the D1R-EGFP line showing lower responses than the other lines. The reduced response in the D1R-EGFP line may indicate changes in cyclase activity in this line compared to the other lines and although this difference may have bearing on the forskolin dosage needed in the assays of D2R function for this cell line, unfortunately we were unable to pursue these findings further in the present study.
Next, we exposed the cell lines to forskolin (0, 25 or 50 μM) plus quinpirole (0, 10 or 25 μM). We found that quinpirole did not alter the forskolin-induced cAMP level when forskolin was used at a concentration of 25 μM in the STHdh+/Hdh+ or EGFP lines (fig. 2d, e). However, 20 μM quinpirole (and not 10 μM quinpirole) decreased the cAMP level in the STHdh+/Hdh+ and EGFP lines when forskolin was used at a concentration of 50 μM (fig. 2d, e). Quinpirole did not decrease the forskolin-induced cAMP levels in the D1R-EGFP line (fig. 2f). In fact, in the D1R-EGFP line, 20 μM quinpirole increased the forskolin-induced cAMP levels compared to the levels in the control (0 μM quinpirole) or 10 μM quinpirole groups (fig. 2f). These data suggest that D2R is functional in STHdh+/Hdh+ and EGFP lines, consistent with D2R mRNA expression profiles of these two lines. However, although the D1R-EGFP line expressed D2R mRNA (fig. 1b), the cAMP assays did not confirm D2R function in this line. As mentioned earlier, a higher dose of quinpirole and a more detailed analysis of forskolin-induced cAMP levels in this cell line would be required to further characterize the D2R responses in this cell line.
Next, we examined the intracellular signaling cascade induced by the activation of D1R. Activation of the D1R stimulates adenylate cyclase, increases intracellular cAMP levels and results in phosphorylation of CREB. We found that activation of D1R by SKF81297 (1 μM) resulted in CREB phosphorylation in the D1R-EGFP line (fig. 2g). D1R activation induced considerably lower levels of CREB phosphorylation in the STHdh+/Hdh+ or EGFP lines compared to the D1R-EGFP line (fig. 2g), consistent with low levels of D1R mRNA expression (fig. 1b) and lack of D1R agonist-induced cAMP synthesis (fig. 2a) in the STHdh+/Hdh+ or EGFP lines.
Cell Proliferation
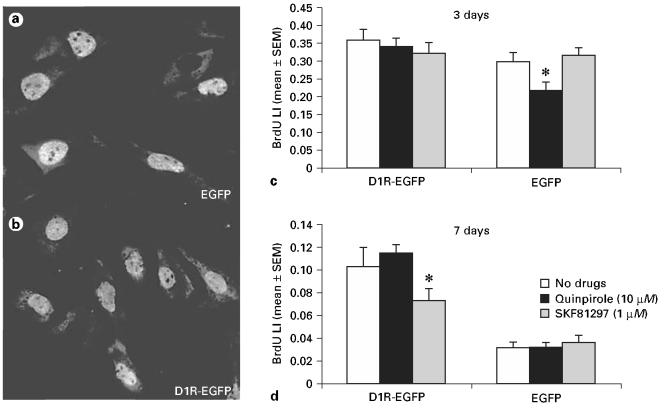
We examined the effects of D1R and D2R activation on BrdU incorporation in our cell lines. Since STHdh+/Hdh+ and EGFP lines showed similar receptor mRNA expression and functional profiles, we used the EGFP line as control (instead of using both STHdh+/Hdh+ and EGFP lines as controls). We exposed the D1R-EGFP and EGFP lines to the D1R agonist SKF81297 (1 μM) or the D2R agonist quinpirole (10 μM) for 3 or 7 days and to BrdU for 1 h prior to fixation. The concentrations of SKF81297 and quinpirole used here produced significant changes in the cAMP assays earlier. Figure 3a and B show examples of BrdU-labeled cells from the EGFP and D1R-EGFP lines, respectively. We found that the BrdU labeling index (BrdU-labeled cells ÷ total cells per microscope field) decreased significantly from 3 to 7 days in culture with or without addition of the drugs (compare the open bars in figure 3c to those in figure 3d), suggesting a gradual decline in cell proliferation during growth of the cells in the differentiation medium at 37°C. We found that neither the D1R agonist SKF81297 (1 μM) nor the D2R agonist quinpirole (10 μM) produced significant changes in the BrdU labeling index in the D1R-EGFP line at 3 days (fig. 3c; ANOVA, p > 0.05). On the other hand, we found significant differences in the BrdU labeling index in the EGFP line at 3 days (fig. 3c; ANOVA; p < 0.05). Tukey's multiple comparisons test indicated that the difference was due to a significant decrease in the BrdU labeling index in the quinpirole-treated group compared to the SKF81297-treated group (p < 0.05). Following exposure to the receptor agonists for 7 days, there was a significant difference in the BrdU labeling index in the D1R-EGFP line (fig. 3d; ANOVA, p < 0.05) but not in the EGFP line. The difference in the D1R-EGFP line was due to a significant decrease in the BrdU labeling index following exposure to SKF81297 compared to exposure to quinpirole or the control condition (Tukey's multiple comparisons test, p < 0.05). We also found that the BrdU labeling index was significantly higher in the D1R-EGFP line compared to the EGFP line at 7 days even without exposure to the dopaminergic drugs (fig. 3d).
Fig. 3.
D1R-EGFP and EGFP lines were exposed to the D1R agonist SKF81297 or D2R agonist quinpirole for 3 or 7 days in culture and to the S-phase marker BrdU for the final 1 h. BrdU immunohistochemistry was performed on the cells and BrdU labeling index (BrdU LI) was compared between D1R-EGFP and EGFP cell lines under the different experimental conditions to examine the effects of receptor activation on cell proliferation. Examples of BrdU-labeled cells are shown in a and b. BrdU LI under the different experimental conditions is shown following 3 days (c) or 7 days (d) in culture. Asterisks indicate statistically significant (p < 0.05) differences (ANOVA followed by Tukey's multiple comparisons test).
Cell Differentiation
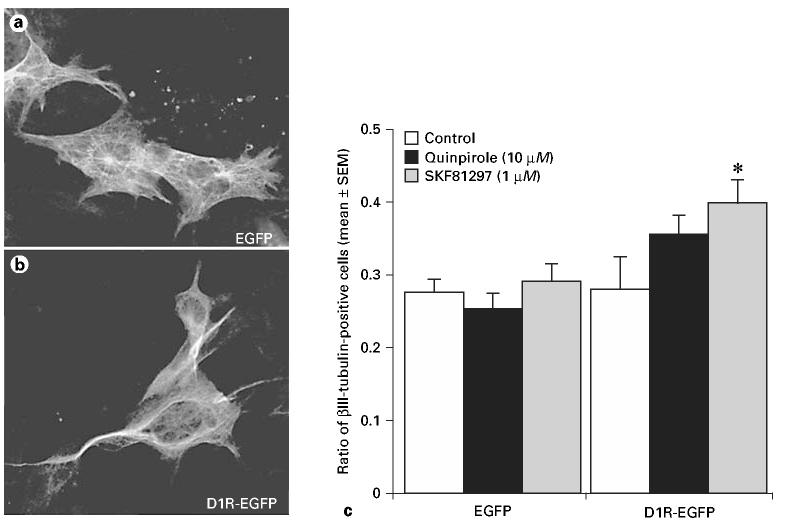
To determine if dopamine receptor activation influences differentiation of cells into neurons or nonneuronal cells, we examined the proportion of βIII-tubulin-labeled cells in the D1R-EGFP or EGFP lines following exposure to the D1R agonist SKF81297 or the D2R agonist quinpirole for 7 days. Examples of βIII-tubulin-labeled cells are shown in figure 4a, b. We found that the proportion of βIII-tubulin-labeled cells was higher in the D1R-EGFP line compared to the EGFP line overall – under control as well as drug-exposed conditions (fig. 4c). Exposure to D1R or D2R agonists did not alter the proportion of βIII-tubulin-labeled cells significantly in the EGFP line (ANOVA, p > 0.05). However, the D1R-EGFP line showed significant changes in the proportion of βIII-tubulin-labeled cells following exposure to the receptor agonists (fig. 4c; ANOVA, p < 0.05). The difference was due to an increase in the proportion of βIII-tubulin-labeled cells following exposure to SKF81297 (fig. 4c; Tukey's multiple comparisons test, p < 0.05).
Fig. 4.
Effects of D1R or D2R activation on differentiation of cells from the EGFP or D1R-EGFP lines into βIII-tubulin-positive presumptive neurons. βIII-tubulin immunocytochemistry was performed following exposure of the cells to the D1R agonist SKF81297 (1 μM) or the D2R agonist quinpirole (10 μM) for 7 days. Examples of βIII-tubulin-labeled cells are shown in a and b. βIII-Tubulin labeling index (βIII-tubulin-positive cells ÷ total cells per microscope field) was compared. c SKF81297 produced statistically significant (p < 0.05) increases in βIII-tubulin labeling index in the D1R-EGFP line (asterisk; ANOVA followed by Tukey's multiple comparisons test).
Next, we examined if dopamine receptor activation promotes glial cell differentiation by analyzing the expression of the oligodendrocyte marker CNPase and the astrocytic marker GFAP by Western blot. We found that CNPase expression level was not altered by activation of D1R or D2R in either the EGFP or the D1R-EGFP line (fig. 5a). We did not find GFAP expression in any of the cell lines under any of the culture conditions (fig. 5b), indicating that the cells did not differentiate into astrocytes.
Fig. 5.
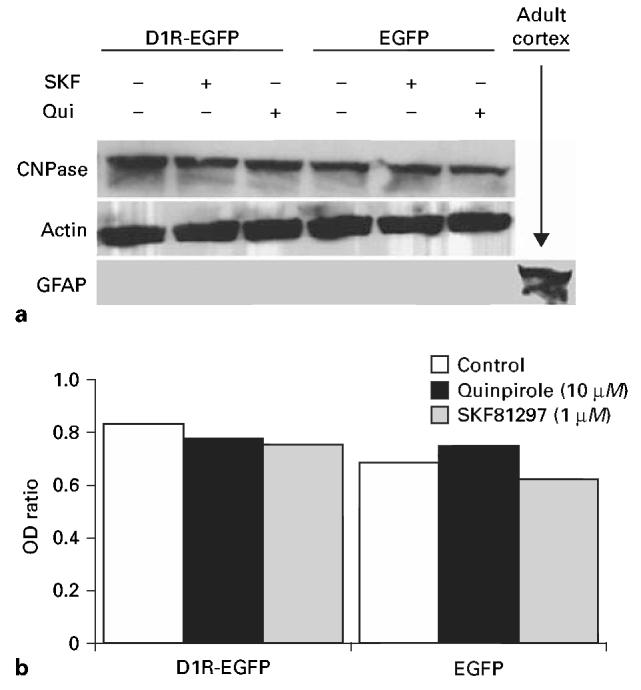
Effects of D1R or D2R activation on glial cell differentiation was analyzed in EGFP and D1R-EGFP lines. The cells were exposed to the D1R agonist SKF81297 or D2R agonist quinpirole for 7 days and the expression levels of oligodendrocyte marker CNPase or astrocytic marker GFAP were analyzed by Western blot (a). We did not see GFAP expression under any of the experimental conditions. Lysate from adult mouse brain was used as positive control for GFAP. Although CNPase was expressed in all the experimental conditions analyzed here, its expression level quantified by optical density (OD) values (ratio of CNPase to total protein, indicated by actin expression) did not show significant differences among the different experimental groups (b).
Discussion
We analyzed dopamine receptor expression and function in three striatal progenitor cell lines and examined the effects of dopamine receptor activation on cell proliferation and differentiation in these cell lines. The STHdh+/Hdh+ line, created from E14 mouse striatum [Trettel et al., 2000], expresses functional D2R, whereas the D1R-EGFP line, created for the present study by modifying the STHdh+/Hdh+ line, expresses functional D1R. The EGFP line that was also created for the present study from the STHdh+/Hdh+ line expresses functional D2R and in that regard resembles the STHdh+/Hdh+ line. Cells of the EGFP line do not express green fluorescence, making this line less valuable as a source of striatal progenitor cells that can be identified under a microscope based on endogenous green fluorescence alone, without additional labeling protocols. Nevertheless, for the present study, the EGFP line served as a valuable control.
It is intriguing that although D2R mRNA is expressed at high levels in the D1R-EGFP line, D2R does not appear to be functional based on the outcome of the cAMP accumulation assays. A number of explanations exist for the loss of D2R function in the D1R-EGFP cell line such as artifact of transfection with the D1R construct, expression of functional D1R, or D1-D2 heterodimer formation with loss of D2R function [So et al., 2005], or internalization of D2R. We are unable to distinguish among these possibilities at the present time. It is also possible that the D2R is functional in this line and that a higher concentration of the D2R agonist quinpirole than that used in the cAMP assays (i.e. higher than 20 μM) is necessary to activate D2R. We were unable to verify this possibility in the present study.
Although the data do not conclusively explain the cause of possible inactivation of D2R in the D1R-EGFP line, this line could be useful to analyze the effects of D1R activation. The D1R-EGFP line (with functional D1R) and the STHdh+/Hdh+ line (with functional D2R) can be complimentary tools to independently evaluate the effects of activation of D1R and D2R in striatal precursor cells.
One aim of the present study was to determine if the effects of activation of D1R and D2R on proliferation and differentiation of the cell lines were similar to the effects in striatal precursor cells in embryonic mice in vivo and in primary neuronal cultures derived from the embryonic mouse striatum in vitro [Ohtani et al., 2003; Popolo et al., 2004]. In the present study, we found that activation of D1R decreased the BrdU labeling index following 7 days in culture, suggesting that D1R activation decreased cell proliferation. These findings are in agreement with the effects of D1R activation in the lateral ganglionic eminence, embryonic source of striatal neurons, and the neuroepithelium of the cerebral cortex in vivo [Ohtani et al., 2003; Popolo et al., 2004] as well as in primary cultures of cells derived from the embryonic striatum and neocortex in vitro [Zhang and Lidow, 2002; Popolo et al., 2004; Zhang et al., 2005]. Activation of D2R decreased the BrdU labeling index in the EGFP line in the present study. This effect is opposite to the effect produced by D2R activation in the lateral ganglionic eminence in vivo and in primary cultures of cells derived from the embryonic mouse striatum in vitro [Ohtani et al., 2003; Popolo et al., 2004]. The present data do not clarify the reasons for the differences. One possibility is that D2R expression without concomitant D1R expression, which is the case in the EGFP line, leads to a different effect of D2R activation on cell proliferation compared to the effects when D2R and D1R expression are concomitant, as is likely to be the case in the cells of the lateral ganglionic eminence [Ohtani et al., 2003]. This interpretation suggests that the cell lines used here may have unveiled interesting interactions between D1R and D2R with respect to mediation of the effects of dopamine on cell proliferation. That is, the two receptors may influence each other's actions such that different effects are produced when a cell expresses both receptors vis-à-vis only one receptor [So et al., 2005]. Therefore, the cell lines may be especially valuable, as the D1R-EGFP, EGFP and STHdh+/Hdh+ lines may uniquely represent precursor cells in different parts of the embryonic CNS that may have unique dopamine receptor expression patterns (i.e. D2R only, D1R only). As mentioned earlier, whether neuroepithelial cells in some regions of the intact brain express only D1R or only D2R is unknown because it is difficult to ascertain dopamine receptor expression/activity patterns at the single cell level in vivo. However, in the case of postmitotic neurons of the striatum, recent reports show that striatal medium spiny neurons express both D1R and D2R [Aizman et al., 2000], although it is possible that striatonigral projection neurons display a predominantly D1R response and striatopallidal neurons a predominantly D2R response [Surmeier et al., 1996].
Our data on the effects of D1R and D2R activation on differentiation of the cells indicate that activation of D1R leads to an increase in the relative numbers of βIII-tubulin-positive presumptive neurons. Activation of D2R under the same conditions did not alter the proportion of βIII-tubulin-positive cells. These data are in agreement with earlier data, which showed that D1R activation led to an increase in the relative proportion of NeuN-positive cells in primary cultures of embryonic mouse striatal cells [Popolo et al., 2004] or an increase in neuronal cells identified by morphological features in cultures from E17 rat striatum [Schmidt et al., 1996]. Moreover, in these earlier studies, D2R activation did not produce significant effects of neuronal numbers.
We found that activation of D1R or D2R did not alter the expression of CNPase, an enzyme associated with myelin ensheathment of axons and virtually exclusively expressed in oligodendrocytes and Schwann cells. Our attempts to count the number of CNPase-positive cells by immunohistochemistry did not succeed due to high background staining. Therefore, we are unable to correlate the Western blot data on CNPase expression with cell counts. We found that the cell lines did not differentiate into GFAP-positive astrocytes under any of the conditions. This is in agreement with findings from the ST14A line, an immortalized striatal progenitor line derived from embryonic rats [Cattaneo and Conti, 1998; Ehrlich et al., 2001]. Thus, the cell lines used in the present study appear to differentiate into presumptive neurons and oligodendrocytes, not astrocytes, and a larger proportion of the cells express βIII-tubulin following activation of D1R.
In summary, we report characterization of dopamine receptor expression and activity profiles of three striatal progenitor cells lines, which may serve as useful models to study the effects of dopamine receptor activation on neuroepithelial cell proliferation and differentiation. Although other cell lines of striatal origin are available [Wainwright et al., 1995; Cattaneo and Conti, 1998], advantages of the cell lines described here are that the lines are of mouse origin, we have characterized the proliferative response of these cell lines to dopamine receptor activation and finally that the STHdh+/Hdh+ line expresses functional D2R only whereas the D1R-EGFP line expresses functional D1R. However, our data do not conclusively establish the cellular mechanism that may have resulted in the loss of D2R function in the D1R-EGFP line. Nonetheless, the cell lines can serve as experimental tools to characterize the role of D1R and D2R in neuroepithelial precursor cell biology.
Acknowledgements
This study was supported by USPHS grants NS 43426, HD 05515, and a grant from the National Alliance for Autism Research. We thank Drs. Jiang-Fan Chen and Liqun Yu of Boston University School of Medicine, Boston, Mass., USA, for providing us with the mouse dopamine D1R sequence and for their advice regarding preparation of primers for PCR. We gratefully acknowledge excellent technical assistance by Igor Bagayev of the MGH Confocal Microscopy Facility.
References
- Aizman O, Brismar H, Uhlen P, Zettergren E, Levey AI, Forssberg H, Greengard P, Aperia A. Anatomical and physiological evidence for D1 and D2 dopamine receptor colocalization in neostriatal neurons. Nat Neurosci. 2000;3:226–230. doi: 10.1038/72929. [DOI] [PubMed] [Google Scholar]
- Bongarzone ER, Howard SG, Schonmann V, Campagnoni AT. Identification of the dopamine D3 receptor in oligodendrocyte precursors: potential role in regulating differentiation and myelin formation. J Neurosci. 1998;18:5344–5353. doi: 10.1523/JNEUROSCI.18-14-05344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo E, Conti L. Generation and characterization of embryonic striatal conditionally immortalized ST14A cells. J Neurosci Res. 1998;53:223–234. doi: 10.1002/(SICI)1097-4547(19980715)53:2<223::AID-JNR11>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Demarque M, Represa A, Becq H, Khalilov I, BenAri Y, Aniksztejn L. Paracrine intercellular communication by a Ca2+ - and SNARE-independent release of GABA and glutamate prior to synapse formation. Neuron. 2002;36:1051–1061. doi: 10.1016/s0896-6273(02)01053-x. [DOI] [PubMed] [Google Scholar]
- Diaz J, Ridray S, Mignon V, Griffon N, Schwartz JC, Sokoloff P. Selective expression of dopamine D3 receptor mRNA in proliferative zones during embryonic development of the rat brain. J Neurosci. 1997;17:4282–4292. doi: 10.1523/JNEUROSCI.17-11-04282.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich ME, Conti L, Toselli M, Taglietti L, Fiorillo E, Taglietti V, Ivkovic S, Guinea B, Tranberg A, Sipione S, Rigamonti D, Cattaneo E. ST14A cells have properties of a medium-size spiny neuron. Exp Neurol. 2001;167:215–226. doi: 10.1006/exnr.2000.7551. [DOI] [PubMed] [Google Scholar]
- Haydar TF, Wang F, Schwartz ML, Rakic P. Differential modulation of proliferation in the neocortical ventricular and subventricular zones. J Neurosci. 2000;20:5764–5774. doi: 10.1523/JNEUROSCI.20-15-05764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung AB, Bennett JP., Jr Development of striatal dopaminergic function. 1. Pre- and postnatal development of mRNAs and binding sites for striatal D1 (D1a) and D2 (D2a) receptors. Brain Res Dev Brain Res. 1996;94:109–120. doi: 10.1016/0165-3806(96)00033-8. [DOI] [PubMed] [Google Scholar]
- Lauder JM. Neurotransmitters as morphogens. Prog Brain Res. 1988;73:365–387. doi: 10.1016/S0079-6123(08)60516-6. [DOI] [PubMed] [Google Scholar]
- Lauder JM. Neurotransmitters as growth regulatory signals: role of receptors and second messengers. Trends Neurosci. 1993;16:233–240. doi: 10.1016/0166-2236(93)90162-f. [DOI] [PubMed] [Google Scholar]
- Lavdas AA, Blue ME, Lincoln J, Parnavelas JG. Serotonin promotes the differentiation of glutamate neurons in organotypic slice cultures of the developing cerebral cortex. J Neurosci. 1997;17:7872–7880. doi: 10.1523/JNEUROSCI.17-20-07872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Harvey JA, Friedman E, Simansky K, Murphy EH. New evidence for neurotransmitter influences on brain development. Trends Neurosci. 1997;20:269–274. doi: 10.1016/s0166-2236(96)01028-4. [DOI] [PubMed] [Google Scholar]
- LoTurco JJ, Owens DF, Heath MJS, Davis MBE, Kriegstein AR. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron. 1995;15:1287–1298. doi: 10.1016/0896-6273(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Luk KC, Kennedy TE, Sadikot AF. Glutamate promotes proliferation of striatal neuronal progenitors by an NMDA receptor-mediated mechanism. J Neurosci. 2003;23:2239–2250. doi: 10.1523/JNEUROSCI.23-06-02239.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Sadikot AF. Glutamate and regulation of proliferation in the developing mammalian telencephalon. Dev Neurosci. 2004;26:218–228. doi: 10.1159/000082139. [DOI] [PubMed] [Google Scholar]
- Monsma FJ, Mahan LC, McVittie LD, Gerfen CR, Sibley DR. Molecular cloning and expression of a D1 dopamine receptor linked to adenylyl cyclase activation. Proc Natl Acad Sci USA. 1990;87:6723–6727. doi: 10.1073/pnas.87.17.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Rigo JM, Rocher V, Belachew S, Malgrange B, Rogister B, Leprince P, Moonen G. Neurotransmitters as early signals for central nervous system development. Cell Tissue Res. 2001;305:187–202. doi: 10.1007/s004410000343. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Goto T, Waeber C, Bhide PG. Dopamine modulates cell cycle in the lateral ganglionic eminence. J Neurosci. 2003;23:2840–2850. doi: 10.1523/JNEUROSCI.23-07-02840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo M, McCarthy DM, Bhide PG. Influence of dopamine on precursor cell proliferation and differentiation in the embryonic mouse telencephalon. Dev Neurosci. 2004;26:229–244. doi: 10.1159/000082140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovik E, Haynes LW. Survival and mitogenesis of neuroepithelial cells are influenced by noradrenergic but not cholinergic innervation in cultured embryonic rat neopallium. Brain Res. 2000;853:227–235. doi: 10.1016/s0006-8993(99)02242-8. [DOI] [PubMed] [Google Scholar]
- Robinson SW, Caron MG. Chimeric D2/D3 dopamine receptors efficiently inhibit adenylyl cyclase in HEk 293 cells. J Neurochem. 1996;67:212–219. doi: 10.1046/j.1471-4159.1996.67010212.x. [DOI] [PubMed] [Google Scholar]
- Schinelli S, Paolillo M, Corona Gl. Opposing actions of D1- and D2-dopamine receptors on arachidonic acid release and cyclic AMP production in striatal neurons. J Neurochem. 1994;62:944–949. doi: 10.1046/j.1471-4159.1994.62030944.x. [DOI] [PubMed] [Google Scholar]
- Schmidt U, Beyer C, Oestreicher AB, Reisert I, Schilling K, Pilgrim C. Activation of dopaminergic D1 receptors promotes morphogenesis of developing striatal neurons. Neuroscience. 1996;74:453–460. doi: 10.1016/0306-4522(96)00201-1. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Zeitzer J, Weaver DR. Widespread expression of functional D1-dopamine receptors in fetal rat brain. Brain Res Dev Brain Res. 1997;102:105–115. doi: 10.1016/s0165-3806(97)00091-6. [DOI] [PubMed] [Google Scholar]
- So CH, Varghese G, Curley KJ, Kong MM, Alijaniaram M, Ji X, Nguyen T, O'Dowd BF, George SR. D1 and D2 dopamine receptors form heterooligomers and cointernalize after selective activation of either receptor. Mol Pharmacol. 2005;68:568–578. doi: 10.1124/mol.105.012229. [DOI] [PubMed] [Google Scholar]
- Surmeier DJ, Song WJ, Yan Z. Coordinated expression of dopamine receptors in neostriatal medium spiny neurons. J Neurosci. 1996;16:6579–6591. doi: 10.1523/JNEUROSCI.16-20-06579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- Wainwright MS, Perry BD, Won LA, O'Malley KL, Wang WY, Ehrlich ME, Heller A. Immortalized murine striatal neuronal cell lines expressing dopamine receptors and cholinergic properties. J Neurosci. 1995;15:676–688. doi: 10.1523/JNEUROSCI.15-01-00676.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Bai J, Undie AS, Bergson C, Lidow MS. D1 dopamine receptor regulation of the levels of the cell-cycle-controlling proteins, cyclin D, P27 and Raf-1, in cerebral cortical precursor cells is mediated through cAMP-independent pathways. Cereb Cortex. 2005;15:74–84. doi: 10.1093/cercor/bhh110. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lidow MS. D1 dopamine receptor regulation of cell cycle in FGF- and EGF-supported primary cultures of embryonic cerebral cortical precursor cells. Int J Dev Neurosci. 2002;20:593–606. doi: 10.1016/s0736-5748(02)00104-1. [DOI] [PubMed] [Google Scholar]