Abstract
The cortex peptidoglycan from endospores of Bacillus subtilis is responsible for the maintenance of dormancy. LytH (YunA) has been identified as a novel sporulation-specific component with a role in cortex structure determination. The lytH gene was expressed only during sporulation, under the control of the mother cell-specific sigma factor σK. Spores of a lytH mutant have slightly reduced heat resistance and altered staining when viewed by electron microscopy. Analysis of the peptidoglycan structure of lytH mutant spores shows the loss of muramic acid residues substituted with l-alanine and a corresponding increase in muramic acid residues substituted with tetrapeptide compared to those in the parent strain. In a lytH cwlD mutant, the lack of muramic acid residues substituted with l-alanine and δ-lactam leaves 97% of residues substituted with tetrapeptide. These results suggest that lytH encodes an l-Ala-d-Glu peptidase involved in production of single l-alanine side chains from tetrapeptides in the spore cortex. The lack of di- or tripeptides in a lytH mutant reveals the enzyme is an endopeptidase.
Bacterial endospores are characterized by their extreme dormancy and high-level resistance to a range of stresses, in particular heat (7). The specialized spore structure determines resistance properties, with the dehydrated spore core being crucial for heat resistance. The spore cell wall peptidoglycan, known as the spore cortex, is essential for the maintenance of the dehydrated core, heat resistance, and dormancy (7). The cortex has a unique, spore-specific structure, and recent analysis has begun to identify features important in its role during differentiation and to reveal components responsible for cortex synthesis, modification during sporulation, and hydrolysis during germination (1, 2). Spore cortex is characterized by an extremely low level of cross-linking, which is important for the maintenance of heat resistance (1, 15). A unique feature of the dormant spore cortex is the presence of muramic acid δ-lactam residues. This modification has a specific role during germination, because the putative germination-specific lytic enzymes SleB and CwlJ, which are responsible for cortex degradation, require the presence of muramic acid δ-lactam for substrate recognition, since cortex lacking this modification is unable to be hydrolyzed during germination (2).
During cortex maturation, a sporulation-specific amidase, CwlD, and a recently described polysaccharide deacetylase, PdaA, are responsible for the formation of the characteristic δ-lactam residues of bacterial endospores (1, 8, 15). Although cwlD or pdaA mutants and a cwlD pdaA double mutant can produce resistant endospores, no cortex hydrolysis is observed during germination, and the spores do not outgrow (1, 2, 8, 16, 18). The final characteristic feature of the cortex is that about 25% of the muramic acid side chains are substituted for with single l-alanine residues (1, 17). These may be formed by the action of an l-Ala-d-Glu endopeptidase on nascent stem peptides. The enzyme or enzymes responsible for the single l-Ala side chains have up to now been unknown, as has the role of this peptidoglycan modification. In this paper, we describe a novel peptidoglycan hydrolase, LytH (formerly YunA), which is required for the production of the single l-Ala side chains in the spore cortex.
MATERIALS AND METHODS
Bacterial strains, growth, and preparation of spores.
All Bacillus subtilis strains used in this study are shown in Table 1. Mutations were transformed into the laboratory wild-type background (HR) used for muropeptide analysis (1). Vegetative cells of B. subtilis were grown in nutrient broth (Oxoid) or on nutrient agar plates. Sporulation was initiated in CCY medium, and spores of B. subtilis were prepared as described by Stewart et al. (23). Spores were stored at a concentration of 10 mg (dry weight) ml−1 in distilled water at −20°C.
TABLE 1.
B. Subtilis strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or referencea | |
|---|---|---|---|
| Strains | |||
| B. subtilis 168 | |||
| 1A304 | trpC2 metB5 xin-1 SPβ(s) | BGSCb | |
| HR | trpC2 | Laboratory stock | |
| 1A304 background | |||
| GJH160 | trpC2 metB5 xin-1 SPβ(s) lytH::pMUTIN4 Emr | This study | |
| HR background | |||
| SH243 | trpC2 Pspac-sigE Kanr | Laboratory stock | |
| SH61 | trpC2 Pspac-sigF Cmr | Laboratory stock | |
| SH248 | trpC2 Pspac-sigG Kanr | Laboratory stock | |
| SH132 | trpC2 Pspac-sigK Cmr | Laboratory stock | |
| 1295 | trpC2 spoIIGB::kan | J. Errington, Oxford University | |
| 650 | trpC2 iluB2 leuB16 spoIIAABC::cat | J. Errington, Oxford, University | |
| 618 | trpC2 spoIIIC::cat | 25 | |
| 1296 | trpC2 spoIIIG::kan | J. Errington, Oxford University | |
| GJH161 | trpC2 lytH::pMUTIN4 Emr | This study | |
| GJH162 | trpC2 spoIIGB::kn lytH::pMUTIN4 Emr | 1295 → GJH161 | |
| GJH163 | trpC2 spoIIAABC::cat lytH::pMUTIN4 Emr | 650 → GJH161 | |
| GJH164 | trpC2 spoIIIG::kn lytH::pMUTIN4 Emr | 1296 → GJH161 | |
| GJH165 | trpC2 spoIIIC::cat lytH::pMUTIN4 Emr | 618 → GJH161 | |
| GJH166 | trpC2 Pspac-sigE lytH::pMUTIN4 Emr Kanr | GJH161 → SH243 | |
| GJH167 | trpC2 Pspac-sigF lytH::pMUTIN4 Emr Cmr | GJH161 → SH61 | |
| GJH168 | trpC2 Pspac-sigG lytH::pMUTIN4 Emr Kanr | GJH161 → SH248 | |
| GJH169 | trpC2 Pspac-sigK lytH::pMUTIN4 Emr Cmr | GJH 161 → SH132 | |
| AA107 | trpC2 cwlD::cat | 1 | |
| GJH171 | trpC2 cwlD::cat lytH::pMUTIN4 Emr | GJH160 → AA107 | |
| Plasmids | |||
| pMUTIN4 | Suicide vector in B. subtilis; Emr | 26 | |
| pGJH160 | pMUTIN4 containing 359-bp HindIII-BamHI insert carrying part of lytH gene | This study |
Arrows indicate transformation of recipient strain with donor chromosomal DNA.
Bacillus Genetic Stock Center, Ohio State University, Columbus.
Plasmids were constructed in and prepared from Escherichia coli strain DH5α grown in Luria-Bertani (LB) broth or on LB agar. When appropriate, chromosomal drug resistance markers in E. coli were selected with ampicillin (50 μg ml−1), and those in B. subtilis were selected with chloramphenicol (5 μg ml−1), kanamycin (10 μg ml−1) or erythromycin (1 μg ml−1), and lincomycin (25 μg ml−1). All bacterial cultures were grown at 37°C.
Construction of a lytH mutation. (i) Plasmid construction.
Primers were designed to enable the amplification, by PCR, of a 359-bp fragment of DNA internal to the lytH gene. The forward primer GH18 (5′-GCCGAAGCTTGGCACAGATGAAGACAACA-3′) contained a HindIII site (boldface) followed by 19 nucleotides identical to the DNA sequence between bases 472 and 490 within the lytH gene (underlined). The reverse primer GH19 (5′-CGCGGATCCTGGGCAAAATAATGATACG-3′) contained a BamHI site (boldface) followed by 19 nucleotides identical to residues 806 to 824 (underlined). The 359-bp DNA fragment corresponding to part of the lytH gene was amplified by PCR, purified, digested with HindIII and BamHI, and then ligated into HindIII- and BamHI-restricted pMUTIN4 to create plasmid pGJH160.
(ii) Transformation of E. coli and B. subtilis.
Transformation of E. coli DH5α was performed as described by Hanahan (9). Transformation of B. subtilis 1A304 with pGJH160 was performed by the method of Kunst and Rapoport (10). Disruption of the lytH gene by Campbell-type recombination was confirmed by Southern blot analysis with the insert in pGJH160 as a probe. Hybridization, probe labeling, and detection were done with the Boehringer Mannheim nonradioactive DNA labeling and detection kit.
Multiple mutant crosses.
Chromosomal DNA isolated from strain GJH160 (lytH) was transformed into HR (wild type) to create strain GJH161. The transformation into HR was performed because previous peptidoglycan structural analysis had been carried out in this background (1-3). Chromosomal DNA from strain GJH160 was also transformed into strains carrying various sporulation sigma factor mutations (Table 1). These crosses created strains GJH162, GJH163, GJH164, GJH165, GJH166, GJH167, GJH168, and GJH169, which carry mutations in sigE, sigF, sigG, sigK, Pspac sigE, Pspac sigF, Pspac sigG, and Pspac sigK, respectively. The Pspac sigma factor strains are mutants in which the genes have been separated from their own promoters and placed under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter, so expression of the genes can be controlled. Chromosomal DNA was isolated from strain AA107 (cwlD) and transformed into strain GJH161 (lytH), creating strain GJH171 (lytH cwlD). All strains were verified by the appropriate drug resistance tests and Southern blotting.
Measurement of spore heat resistance.
Spores were resuspended in 1.5 ml of sterile distilled water to a final optical density at 600 nm (OD600) of 2.5. The spore samples were then incubated at 90°C in a water bath, and 0.1-ml aliquots were removed at intervals and diluted into 0.9 ml of ice-cold sterile distilled water. Appropriate dilutions were made in sterile distilled water, and samples were spread on nutrient agar in duplicate. The results obtained were the average from two independent experiments.
Spore protoplast wet density.
The wet density of the B. subtilis spore protoplast was determined by equilibrium density centrifugation on metrizamide (Sigma) gradients as described previously (12). Prior to density determination, spores were decoated for 90 min at 37°C in a mixture of 50 mM Tris-HCl (pH 8.0), 8 M urea, 1% (wt/vol) sodium dodecyl sulfate, and 50 mM dithiothreitol and washed three times with 150 mM NaCl and twice with water by centrifugation (11,000 × g for 5 min at room temperature) and resuspension. The decoated spores were equilibrated for 60 min in 50% (vol/vol) metrizamide prior to loading onto gradients. Results are an average of two different spore preparations, which show variation between each other of <0.001 g/ml.
Analysis of gene expression.
Synchronous sporulation was performed by the resuspension method of Sterlini and Mandelstam (22). Samples were harvested every hour after the initiation of sporulation (t0) for 7 h, and sporulation morphology was monitored by microscopy. Induced expression of sigma factor genes under the control of Pspac was carried out by adding IPTG (1 mM final concentration) to cells growing in LB at an OD600 of 0.25 (24). β-Galactosidase assays, using MUG (methylumbelliferyl β-d-galactoside) as the substrate, were performed as described by Youngman (27), except that cells were permeabilized by incubation with lysozyme (250 μg ml−1) on ice for 20 min. MUG was used at a final concentration of 600 μg ml−1 in dimethyl sulfoxide, and the assay mixture was incubated at 28°C. Fluorescence was measured on a fluorometer (Hoefer). One unit of β-galactosidase activity was defined as the amount of enzyme that releases 1 pmol of methylumbelliferone min−1 ml−1 (25), normalized to a culture OD600 of 1.0.
RP-HPLC analysis of spore peptidoglycan.
Vegetative peptidoglycan extraction, cortex extraction from dormant and germinating spores, muropeptide separation by reverse-phase high-performance liquid chromatography (RP-HPLC), and amino acid and mass spectrometry analyses were performed as previously described (1-3).
Transmission electron microscopy.
Samples for examination were recovered by centrifugation, and the supernatant was removed. The pellet was fixed by resuspension in glutaraldehyde fixative (3% [vol/vol] glutaraldehyde in 0.1 M phosphate buffer). Samples were incubated in fixative for 2 h at room temperature. Following fixing, the samples were washed five times (10 min each) in 0.1 M sodium cacodylate buffer and then postfixed in 2% (wt/vol) aqueous osmium tetroxide. Following a wash in distilled water, samples were incubated in 75% (vol/vol) ethanol (three changes, 5 min each), 95% (vol/vol) ethanol (three changes, 5 min each), and then absolute ethanol (two changes, 10 min each). Further incubation with dried absolute ethanol (two changes, 10 min each) and propylene oxide (two changes, 10 min each) was followed by overnight incubation in a 1:1 mixture of propylene oxide and araldite. Two 3-h incubations in fresh 100% (vol/vol) araldite were followed by embedding in 100% (vol/vol) araldite and polymerization (60°C, 48 h). Sections from the embedded sample were cut into 80-nm sections on a Reicher Ultracut. The sections were then stained with saturated uranyl acetate in 50% (vol/vol) ethanol for 15 min, washed in distilled water, and then stained in Reynolds lead citrate for 5 min. Specimens were viewed at a range of magnifications on a Phillips CM-10 electron microscope operated at 80 kV. At least 50 spores of each strain were examined.
RESULTS
Role and regulation of lytH.
The predicted protein encoded by lytH (yunA) is 349 amino acids (aa) in length with a calculated Mr of 39.5 kDa and an estimated pI of 6.89. A general BLAST search revealed LytH to have the highest homology to peptidase M37 (54% identity, 309 aa) from Bacillus anthracis. This is part of the M23/37 family of zinc metallopeptidases, in which are included the Gly-Gly endopeptidases, such as lysostaphin. Smith et al. (20) had previously identified YunA (LytH) as a lysostaphin homologue. Interestingly, the presence of muropeptides containing glycine has been observed previously in spores of a cwlD mutant (16) and vegetative cell wall peptidoglycan (3). However, the presence of these structures has been shown to be dependent on the medium (3). The lytH gene, which is not located within any potential prophage, is followed by a putative ρ-independent terminator and is preceded by a divergently transcribed gene, yutB, which is itself followed by a putative ρ-independent terminator and encodes a protein similar to lipoic acid synthase.
In order to determine the role of lytH, a mutation was created by the insertion of the suicide plasmid pGJH160 into the B. subtilis chromosome. This led to inactivation of lytH and the creation of a lytH::lacZ transcriptional fusion. No defects in vegetative growth rate or sporulation kinetics were found for GJH161 (lytH) compared to its parent (HR).
Expression analysis.
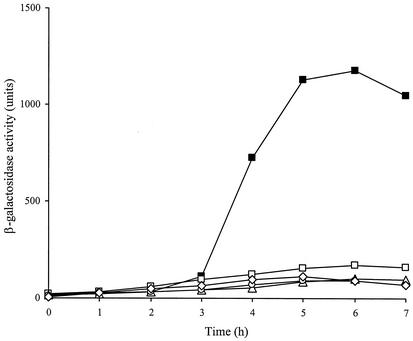
Expression of lytH was monitored during vegetative growth and sporulation. No β-galactosidase activity of strain GJH161 was observed during vegetative growth (data not shown). When analyzed for expression during sporulation, β-galactosidase activity was first detected between t2 and t3 (2 to 3 h after the onset of sporulation, after asymmetric septation) and was maximal around t5 in two separate experiments, suggesting that the operon is regulated by a sporulation-specific sigma factor (Fig. 1).
FIG. 1.
Expression of lytH during sporulation. β-Galactosidase levels were measured following induction of synchronous sporulation in strain GJH161 (lytH; ▪) and various sigma factor mutant background strains: GJH162 (sigE; ○), GJH163 (sigF; ▵), GJH164 (sigG; □), and GJH165 (sigK; ⋄).
Chromosomal DNA carrying the lytH::lacZ fusion was transferred by transformation into strains carrying various spo mutations, and β-galactosidase activity was measured during sporulation (Fig. 1). The absence of expression of lytH::lacZ in strain GJH165 carrying a mutation in the spoIIIC gene (σK) but still containing an intact spoIIIG gene (σG) indicates that σK is most likely required for the in vivo expression of lytH::lacZ. As expected, expression was blocked in strains GJH163, GJH164, and GJH165, because σE, σF, and σG are all required for later sigma factor activity.
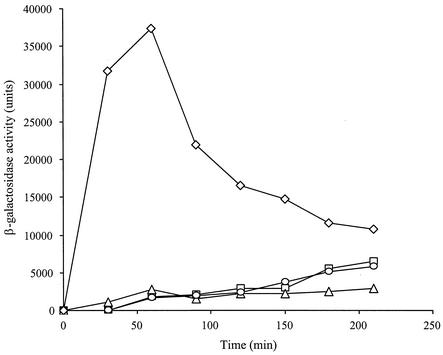
To test the sigma factor specificity of RNA polymerase that could transcribe the lytH gene, the lytH::lacZ fusion was introduced by transformation into strains SH243 (Pspac-sigE), SH61 (Pspac-sigF), SH248 (Pspac-sigG), and SH132 (Pspac-sigK) to produce strains GJH166, GJH167, GJH168, and GJH169, respectively. Expression of β-galactosidase activity was detected on induction of σK, showing that production of σK is directing the expression from the lytH promoter (Fig. 2). Expression of β-galactosidase did not occur following induction of σE, σF, and σG (Fig. 2).
FIG. 2.
Expression of lytH::lacZ in strains carrying various sporulation-specific sigma factors under Pspac control during vegetative growth. Strains GJH166 (Pspac-sigE; □), GJH167 (Pspac-sigF; ▵), GJH168 (Pspac-sigG; ○), and GJH169 (Pspac-sigK; ⋄) were grown in L broth and induced by addition of IPTG to a final concentration of 1 mM.
Role of LytH in endospore heat resistance.
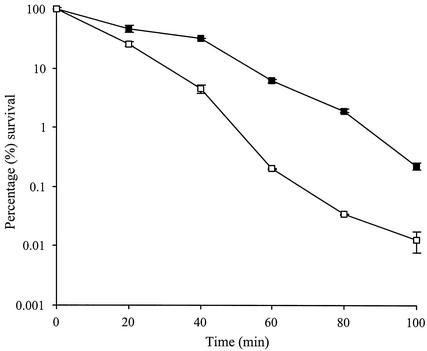
Endospores of strain GJH161 (lytH) were analyzed to determine the role of lytH in spore heat resistance. Spores were heated at 90°C, and the comparison of the percentages of loss of CFU showed that GJH161 had slightly reduced heat resistance compared to the parental strain (Fig. 3). After 100 min, GJH161 showed greater than 10-fold-less survival than HR (wild type).
FIG. 3.
Heat resistance of spores of strains GJH161 (lytH) and HR. Solid squares represent HR (wild type), while open squares represent strain GJH161 (lytH). Error bars show the standard deviation. Results are from two independent experiments. Survival was measured as CFU after heat treatment at 90°C.
Role of lytH in spore water content.
In comparison to a vegetative cell, the protoplast of the spore is relatively dehydrated, even when suspended in water. This dehydration results in metabolic dormancy and is mainly responsible for the heat resistance properties of the spore (4, 13, 15). The spore protoplast wet densities of permeabilized endospores of B. subtilis HR (wild type) and mutant strains were measured. Strains HR, GJH161 (lytH), AA107 (cwlD), and GJH171 (lytH cwlD) had protoplast wet densities of 1.346, 1.344, 1.343, and 1.342 g ml−1, respectively.
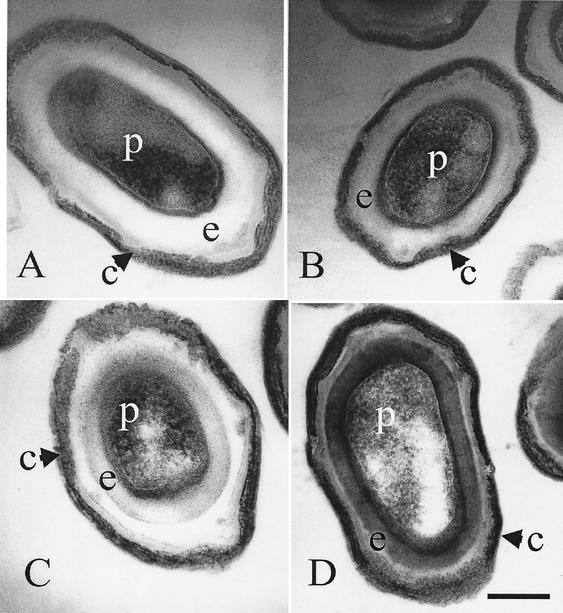
Transmission electron micrograph studies of lytH spores.
As observed previously, micrographs of dormant wild-type spores show a densely stained spore protoplast and an electron-transparent spore cortex (Fig. 4). When the mutant strains were viewed by electron microscopy, it was noted that strain GJH161 (lytH) showed a slightly more electron-dense spore cortex (Fig. 4). In strain AA107 (cwlD), as previously observed by Popham et al. (16), the amount of staining observed in the spore cortex was greater than that in the parent and similar to that seen in a lytH mutant (Fig. 4). However, when dormant endospores of strain GJH171 (lytH cwlD) were observed, the degree of staining to the spore cortex was dramatically greater than that observed in both the lytH and cwlD mutants (Fig. 4).
FIG. 4.
Transmission electron microscopy of dormant endospores of HR (wild type) (A), GJH161 (lytH) (B), AA107 (cwlD) (C), and GJH171 (lytH cwlD) (D). c, spore coats; e, spore cortex peptidoglycan; p, spore protoplast. Bar, 0.25 μm.
Role of LytH in peptidoglycan structure determination.
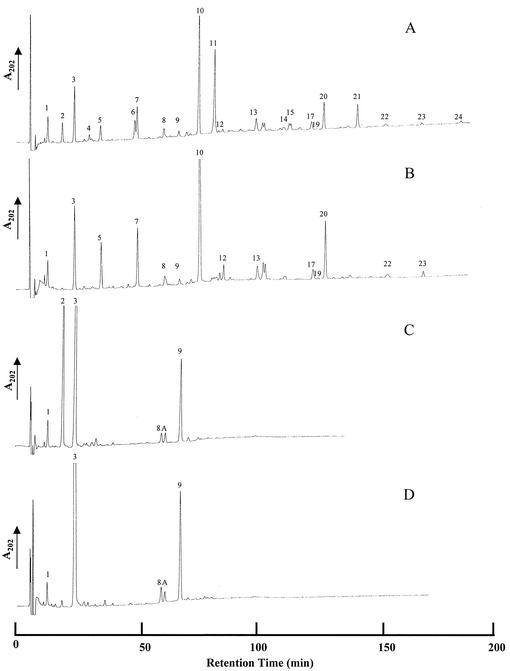
Strain GJH161 (lytH) was analyzed for any changes in the cell wall peptidoglycan structure of vegetative cells, endospores, and during germination by muropeptide HPLC analysis. No change in cell wall peptidoglycan structure of vegetative cells was observed compared to the level in its parent (data not shown). The characteristic appearance of muropeptides during germination also occurred in GJH161 (lytH). Analysis of endospore cell wall peptidoglycan in strain GJH161 (lytH) compared to that in HR showed the loss of a number of peaks that correspond to muropeptides with a single alanine side chain (Fig. 5A and B, peaks 2, 4, 6, 11, 14, 15, 21, and 24; and Tables 2 and 3) (1). Two other strains, AA107 (cwlD) and GJH171 (lytH cwlD), were also analyzed for changes in endospore cell wall peptidoglycan structure. Strain AA107 (cwlD) has previously been shown to have a dramatic alteration in peptidoglycan structure, which was due to the total lack of muramic acid δ-lactam residues in the peptidoglycan (Fig. 5C, peaks 4, 5, 6, 7, 12, 14, 15, 16, 17, and 18) (1). A strain carrying mutations in both lytH and cwlD, GJH171, was also analyzed and showed the loss of a peak corresponding to disaccharide alanine compared to AA107 (cwlD) (Fig. 5D, peak 2). Quantification of the spore muropeptides of strains GJH161 (lytH) and GJH171 (lytH cwlD) (Tables 2 and 3) revealed the percentage of cross-linking index and the muramic acid substitution compared to those in the parental strain. The cross-linking index per diaminopimelic acid and per muramic acid residue of GJH161 (lytH) was found to be similar to that of the wild type. This suggests the mutation in lytH has no effect on peptidoglycan cross-linking. The cross-linking index per diaminopimelic acid residue of GJH171 (lytH cwlD) was also found to be similar to that of the wild type. However, the cross-linking index per muramic acid residue is approximately doubled. A mutation in lytH results in a complete loss of muramic acid residues substituted with l-alanine. The loss of muramic acid residues substituted with alanine corresponds with an increase in muramic acid residues substituted with tetrapeptide in GJH161 (28.8 to 50.5%). In GJH171 (lytH cwlD), the lack of any muramic acid residues substituted with alanine and δ-lactam leaves 97% of residues substituted with tetrapeptide. These results determine that LytH is likely to be an l-Ala-d-Glu peptidase involved in the production of the single l-alanine side chains from tetrapeptides in the spore cortex. The lack of di- or tripeptides in GJH161 suggests the enzyme is an endopeptidase.
FIG. 5.
RP-HPLC muropeptide elution patterns of peptidoglycan from B. subtilis 168 endospores. (A) HR (wild type). (B) GJH161 (lytH). (C) AA107 (cwlD). (D) GJH171 (lytH cwlD). Purified Cellosyl-digested peptidoglycan samples were separated on an octadecylsilane column, and the A202 of the eluate was measured.
TABLE 2.
Muropeptide identities and quantification from peptidoglycan of B. subtilis HR (wild type), GJH161 (lytH), and GJH171 (lytH cwlD)
| Muropeptidea | Identity | Concn (mol%) in:
|
||
|---|---|---|---|---|
| HR (wild type) | GJH161 (lytH) | GJH171 (lytH cwlD) | ||
| 1 | Disaccharide tripeptide | 4.6 | 4.4 | 1.9 |
| 2 | Disaccharide alanine | 6.2 | ||
| 3 | Disaccharide tetrapeptide | 13.2 | 21.2 | 91.4 |
| 4 | Tetrasaccharide alanine with open lactam | 1.2 | ||
| 5 | Tetrasaccharide tetrapeptide with open lactam | 0.9 | 5.3 | |
| 6 | Tetrasaccharide alanine with reduced lactam | 3.1 | ||
| 7 | Tetrasaccharide tetrapeptide with reduced lactam | 4.5 | 10.7 | |
| 8 | Disaccharide tripeptide disaccharide tetrapeptide | 0.9 | 1.0 | 0.6 |
| 9 | Disaccharide tetrapeptide disaccharide tetrapeptide | 0.6 | 1.0 | 5.3 |
| 10 | Tetrasaccharide tetrapeptide | 21.5 | 36.0 | |
| 11 | Tetrasaccharide alanine | 21.7 | ||
| 12 | Hexasaccharide tetrapeptide with 1 reduced lactam | 1.3 | 2.3 | |
| 13 | Disaccharide tetrapeptide tetrasaccharide tetrapeptide | 1.6 | 1.9 | |
| 14 | Hexasaccharide alanine with 1 reduced lactam | 1.3 | ||
| 15 | Hexasaccharide alanine with 1 reduced lactam | 1.3 | ||
| 17 | Hexasaccharide tetrapeptide with 1 reduced lactam | 1.4 | 2.1 | |
| 19 | Tetrasaccharide tetrapeptide tetrasaccharide tetrapeptide | 0.8 | 1.0 | |
| 20 | Hexasaccharide tetrapeptide | 6.7 | 11.5 | |
| 21 | Hexasaccharide alanine | 6.0 | ||
| 22 | Tetrasaccharide tetrapeptide hexasaccharide tetrapeptide | 0.3 | 0.5 | |
| 23 | Octasaccharide tetrapeptide | 0.5 | 1.1 | |
| 24 | Octasaccharide alanine | 0.4 | ||
| A | Disaccharide tripeptide disaccharide tetrapeptide with 1 amidation | 0.8 | ||
Muropeptides are numbered as indicated in Fig. 5.
TABLE 3.
Cross-linking index and muramic acid substitution of B. subtilis spore peptidoglycana
DISCUSSION
The protein encoded by the lytH gene was predicted to be a peptidoglycan hydrolase because of its homology to lysostaphin (20). However lysostaphin is a Gly-Gly endopeptidase, and such a combination of residues has not been identified in B. subtilis peptidoglycan. Thus, the role of LytH was unsure. The lytH gene was found to be expressed specifically during sporulation, under the control of the late mother cell-specific sigma factor, σK. Thus LytH has a likely role during differentiation. Two peptidoglycan hydrolases of B. subtilis also transcribed by EσK RNA polymerase have previously been identified and characterized (11, 14, 21). These are the amidases CwlC and CwlH, which are involved in mother cell lysis.
Minimal differences in the wet density of spore protoplasts were observed in lytH and cwlD mutants compared to the parent, whereas the lytH cwlD double mutant showed a small but reproducible difference. Also the heat resistance of lytH mutant spores was reduced compared to that of the wild type, suggesting a difference in core dehydration and/or cortex structure. The relative dehydration of the spore core is considered the most important factor in the resistance to heat, and the correlation between heat resistance and spore core water content has been shown in a variety of species (5, 13).
Electron microscopy of the wild type and the lytH, cwlD, and lytH cwlD mutants revealed that the lytH and cwlD mutants showed an increase in staining of the cortex compared with that in the wild type. An increase in staining in a cwlD mutant has previously been observed (16). This increase in staining is probably due to the changes in the peptidoglycan architecture of the mutants. This in turn suggests the peptidoglycan of the spore cortex of the mutants may be more permeable to the stain, or a charge alteration allows for greater staining.
In order to determine whether LytH has a role in cortex structural determination, muropeptide analysis was carried out. The spore cortex peptidoglycan structure of a lytH mutant revealed that peaks corresponding to muropeptides containing single l-alanine side chains were no longer present. This determined that LytH was a likely l-Ala-d-Glu endopeptidase involved in the production of the single l-alanine side chains from tetrapeptides in the spore cortex. Alternatively it is possible that LytH is a carboxypeptidase cleaving d-Ala from position 4 of the peptide side chain, which is then processively cleaved by another enzyme or enzymes to give single l-Ala side chains. The role of LytH in single l-Ala side chain production was confirmed in a lytH cwlD mutant. In this strain, 97% of the muropeptides had tetrapeptide side chains due to the absence of δ-lactam as a result of the lack of CwlD (1, 16) and the absence of single l-alanine side chains from the lytH mutation. The lack of l-alanine side chains and an increase in tetrapeptide side chains may bring about a conformational change in the peptidoglycan structure in such a way as to decrease heat resistance. No decrease in heat resistance was observed in a cwlD mutant, which besides having no δ-lactam residues also has increased amounts of tetrapeptides. It must therefore be a lack of the single l-alanine residues that results in the reduced heat resistance. The purpose of the production of such single l-alanine residues could be to reduce the number of muramyl peptides per disaccharide, thus decreasing the amount of cross-links. An increase in the amount of cross-linking may reduce the spores' ability to be resistant to heat, as seen in a dacB mutant (15); however, a significant increase in cross-linking was not observed in a lytH mutant. Another explanation is that single l-alanine residues, the low number of cross-links compared to those in vegetative peptidoglycan, and the absence of amidation on the d-carboxyl group of diaminopimelic acid account for the fact that the cortical peptidoglycan is more negatively charged than vegetative peptidoglycan (6). It has been suggested that this high net negative charge may be essential for the function of the cortex in heat resistance (19). The lytH cwlD mutant exhibits the simplest peptidoglycan profile of any strain so far examined by muropeptide analysis. This is most likely possible as a result of the very specific role of the cortex in the maintenance of spore dormancy. How such an apparently simple structure is able to maintain dehydration is a central unanswered question.
The mechanism of production of muropeptides with single l-alanine side chains has been unknown up to now, but several ideas have been previously put forward (1). The mechanism has now been determined as a likely single-step process due to the activity of a sporulation-specific endopeptidase, LytH. LytH is an apparent l-Ala-d-Glu endopeptidase that cleaves nascent side chain to give a single l-alanine substitution. It likely only acts on non-cross-linked chains, and its activity is not dependent on the presence of δ-lactam residues. This modification occurs at a late stage during the maturation of spore cortex via the activity of a mother cell-derived protein. To date, almost the entire complement of components necessary for peptidoglycan structure dynamics during differentiation has now been identified. However, even though the chemistry of the process has now been established, the functions of components in determining peptidoglycan architecture and how the cortex is able to maintain dormancy have remained elusive.
Acknowledgments
We are grateful to the BBRSC (G.J.H. and A.A.) and the Royal Society (S.J.F.) for funding this research.
REFERENCES
- 1.Atrih, A., P. Zöllner, G. Allmaier, and S. J. Foster. 1996. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J. Bacteriol. 178:6173-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atrih, A., P. Zöllner, G. Allmaier, M. P. Williamson, and S. J. Foster. 1998. Peptidoglycan structural dynamics during germination of Bacillus subtilis 168 endospores. J. Bacteriol. 180:4603-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaman, T. C., J. T. Greenamyre, T. R. Corner, H. S. Pankratz, and P. Gerhardt. 1982. Bacterial spore heat resistance correlated with water content, wet density, and protoplast/sporoplast volume ratio. J. Bacteriol. 150:870-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaman, T. C., and P. Gerhardt. 1986. Heat resistance of bacterial spores correlated with protoplast dehydration, mineralization, and thermal adaptation. Appl. Environ. Microbiol. 52:1242-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchanan, C. E., A. O. Henriques, and P. J. Piggot. 1994. Cell wall changes during bacterial endospore formation, p. 167-186. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 7.Ellar, D. J. 1978. Spore specific structures and their function. Symp. Soc. Gen. Microbiol. 28:295-325. [Google Scholar]
- 8.Fukushima, T., H. Yamamoto, A. Atrih, S. J. Foster, and J. Sekiguchi. 2002. A polysaccharide deacetylase gene (pdaA) is required for germination and for production of muramic δ-lactam residues in the spore cortex of Bacillus subtilis. J. Bacteriol. 184:6007-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda, A., Y. Asami, and J. Sekiguchi. 1993. Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J. Bacteriol. 175:6260-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindsay, J. A., T. C. Beaman, and P. Gerhardt. 1985. Protoplast water content of bacterial spores determined by buoyant density sedimentation. J. Bacteriol. 163:735-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakashio, S., and P. Gerhardt. 1985. Protoplast dehydration correlated with heat resistance of bacterial spores. J. Bacteriol. 162:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nugroho, F. A., H. Yamamoto, Y. Kobayashi, and J. Sekiguchi. 1999. Characterization of a new sigma-K-dependent peptidoglycan hydrolase gene that plays a role in Bacillus subtilis mother cell lysis. J. Bacteriol. 181:6230-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popham, D. L., B. Illades-Aguiar, and P. Setlow. 1995. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J. Bacteriol. 177:4721-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc. Natl. Acad. Sci. USA 93:15405-15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 178:6451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekiguchi, J., K. Akeo, H. Yamamoto, F. K. Khasanov, J. C. Alonso, and A. Kuroda. 1995. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J. Bacteriol. 177:5582-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata, H., H. Murakami, and I. Tani. 1980. Delayed germination of Bacillus cereus T spores after treatment with trichloroacetic acid and their reactivation by heating. Microbiol. Immunol. 24:291-298. [DOI] [PubMed] [Google Scholar]
- 20.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Peptidoglycan hydrolases of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 21.Smith, T. J., and S. J. Foster. 1995. Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J. Bacteriol. 177:3855-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart, G. S. A. B., K. Johnstone, E. Hagelberg, and D. J. Ellar. 1981. Commitment of bacterial spores to germinate. Biochem. J. 198:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun, D., P. Stragier, and P. Setlow. 1989. Identification of a new σ-factor involved in compartmentalized gene expression during sporulation of Bacillus subtilis. Genes Dev. 3:141-149. [DOI] [PubMed] [Google Scholar]
- 25.Turner, S. M., J. Errington, and J. Mandelstam. 1986. Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIIC in spo mutants of Bacillus subtilis: a branched pathway of expression of sporulation operons. J. Gen. Microbiol. 132:2995-3003. [DOI] [PubMed] [Google Scholar]
- 26.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 27.Youngman, P. 1990. Use of transposons and integrational vectors for mutagenesis and construction of gene fusions in Bacillus species, p. 221-266. In C. R. Harwood and S. M. Cutting (ed.), Molecular biology methods for Bacillus. Wiley, New York, N.Y.