Abstract
This work reports a genetic analysis of the expression of nitrobenzene dioxygenase (NBDO) in Comamonas sp. strain JS765 and 2-nitrotoluene dioxygenase (2NTDO) in Acidovorax sp. strain JS42. Strains JS765 and JS42 possess identical LysR-type regulatory proteins, NbzR and NtdR, respectively. NbzR/NtdR is homologous to NahR, the positive salicylate-responsive transcriptional activator of the naphthalene degradation genes in Pseudomonas putida G7. The genes encoding NBDO and 2NTDO in each strain are cotranscribed, and transcription starts at the same site within identical promoter regions for each operon. Results from a lacZ reporter gene fusion demonstrated that expression of NBDO and 2NTDO is induced by multiple aromatic compounds, including an array of nitroaromatic compounds (nitrobenzene, 2-, 3-, and 4-nitrotoluene, 2,4- and 2,6-dinitrotoluene, and aminodinitrotoluenes), as well as salicylate and anthranilate. The nitroaromatic compounds appear to be the actual effector molecules. Analysis of β-galactosidase and 2NTDO activities with strain JS42 demonstrated that NtdR was required for induction by all of the inducing compounds, high basal-level expression of 2NTDO, and complementation of a JS42 ntdR null mutant. Complementation with the closely related regulators NagR (from Ralstonia sp. strain U2) and NahR restored only induction by the archetype inducers, salicylate or salicylate and anthranilate, respectively, and did not restore the high basal level of expression of 2NTDO. The mechanism of 2NTDO gene regulation in JS42, and presumably that of NBDO gene regulation in JS765, appear similar to that of NahR-regulated genes in Pseudomonas putida G7. However, NbzR and NtdR appear to have evolved a broader specificity in JS42 and JS765, allowing for recognition of nitroaromatic compounds while retaining the ability to respond to salicylate and anthranilate. NtdR is also the first example of a nitroarene-responsive LysR-type transcriptional activator.
Most nitroaromatic compounds are man-made. Examples include nitrobenzene and mono- and dinitrotoluenes, which are used in the manufacturing of dyes, pigments, polymers, and explosives (15). Use and disposal of these and other nitroaromatic compounds have led to extensive environmental contamination. In addition, many nitrophenolic compounds are regularly dispersed as agricultural pesticides. Although this class of chemicals was introduced into the environment relatively recently, bacteria that are able to utilize many nitroaromatic compounds as carbon, nitrogen, and energy sources have been isolated. Examples include bacteria that can grow with nitrobenzene, mononitrotoluenes, and dinitrotoluenes (13, 14, 25-27, 36, 50). Multicomponent dioxygenases are used by many of these bacteria to catalyze the initial step in degradation. All of the nitroarene dioxygenases identified to date fall within the naphthalene dioxygenase family of Rieske nonheme iron oxygenases (12, 31). Interestingly, the identified nitroarene dioxygenases are most similar to the naphthalene dioxygenase from Ralstonia sp. strain U2, a strain that converts naphthalene to central metabolites via gentisate rather than using the meta cleavage of catechol used by Pseudomonas putida G7 (9). Thus, it has been suggested that nitroarene dioxygenases have evolved from a naphthalene dioxygenase system similar to that in strain U2 (9, 17, 22, 31).
Comamonas sp. strain JS765 and Acidovorax (formerly Pseudomonas) sp. strain JS42 were isolated from nitrobenzene-contaminated samples from New Jersey and Mississippi, respectively (14, 27). However, strain JS765 was selected for growth with nitrobenzene, while strain JS42 was selected for growth with 2-nitrotoluene. Metabolism of these compounds is initiated by a nitroarene dioxygenase in each strain. Previous experiments suggested that nitrobenzene dioxygenase (NBDO)activity in strain JS765 was induced during growth with nitrobenzene, while 2-nitrotoluene dioxygenase (2NTDO) activity in strain JS42 appeared constitutive (14, 27). We have cloned and characterized the nbz genes encoding NBDO from strain JS765 and the ntd genes encoding 2NTDO from strain JS42 (22, 29). We now report the identification of LysR-type regulators (NbzR and NtdR) that are similar to NagR from strain U2 (56) and the well-studied activator of naphthalene degradation genes, NahR from P. putida G7 (3, 16, 42-46). The purpose of this study was to determine if these regulators play a role in the expression of NBDO and 2NTDO, to identify the specific inducing compounds, and to determine whether the mechanism of regulation is similar to that for NahR, thus providing further information on the evolution of nitroarene dioxygenases. Based on results presented here, strains JS765 and JS42 have evolved regulatory systems that respond to the presence of nitroaromatic compounds.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown at 37°C in Luria-Bertani (LB) (4) broth or agar unless otherwise indicated. E. coli DH5α was used for cloning and plasmid propagation. All other bacterial strains were grown at 30°C in minimal-salts medium (MSB) (49) containing succinate (10 mM) and Balch's vitamins (without thiamine) (11). For growth with aromatic compounds, strains were grown at 30°C on MSB agar containing Balch's vitamins (without thiamine) and the aromatic compound supplied as a vapor as previously described (26). For plasmid selection and maintenance, antibiotics were added to growth media at the following concentrations: for E. coli, ampicillin, 150 μg/ml; kanamycin, 100 μg/ml; chloramphenicol, 34 μg/ml; tetracycline, 15 μg/ml; and gentamicin, 15 μg/ml; for strains JS765 and JS42, kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml; tetracycline, 20 μg/ml; and gentamicin, 15 μg/ml; for strain G7, gentamicin, 100 μg/ml. Cell densities were determined by measuring the turbidity at 660 nm.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Comamonas sp. | ||
| JS765 | Wild type, nitrobenzene degrader | 27 |
| JS765-1 | JS765 containing nbzAa-lacZ, Gmr | This study |
| Acidovorax sp. | ||
| JS42 | Wild type, 2NT degrader | 14 |
| JS42-1 | JS42 containing nbzAa-lacZ, Gmr | This study |
| JS42-1R | ntdR mutant of JS42-1; Kmr | This study |
| E. coli | ||
| DH5α | Cloning host | 39 |
| HB101 | Cloning host | 21 |
| S17-1 | Cloning strain for plasmid mobilization, thi | 47 |
| BW19851 | Host strain for R6K replicons | 23, 47 |
| GS162 | MG1655 derivative, ΔlacZY, phe, thi | M. L. Urbanowski |
| GS162R | GS162, rifampin-resistant mutant | This study |
| GS162R-1 | GS162R containing nbzAa-lacZ, Gmr | This study |
| P. putida | ||
| G7 | Wild type, naphthalene degrader | 6 |
| G7-1 | G7 containing nbzAa-lacZ, Gmr | This study |
| Ralstonia sp. | ||
| U2 | Wild type, naphthalene degrader | 9 |
| Plasmids | ||
| pMW24-RPOS | lacZ transcriptional fusion vector, Apr | 53 |
| pUTminiTn5-Gm | Delivery plasmid for mini-Tn5-Gm, Apr Gmr | 53 |
| pRK2013 | tra+ helper plasmid for triparental matings, Kmr | 8 |
| pJS31 | pGEM7Z(+) containing dntR, Apr | 51 |
| pHP45Ω-Km | Contains Kmr cassette for insertional inactivation | 7 |
| pRK415 | Broad-host-range vector, Tcr | 19 |
| pEX1.8 | Broad-host-range expression vector, Apr | 35 |
| pBBR1MCS | Broad-host-range vector, Cmr | 20 |
| pMS104 | pRK415 carrying nahR, Tcr | 42 |
| pUC19 | Cloning vector, Apr | 54 |
| pDTG925 | pUC18 containing nbzYRAaAbAcAd genes from JS765 | 22 |
| pDTG928 | pMW24-RPOS containing nbzAa promoter fused to lacZ | This study |
| pDTG931 | pUTminiTn5-Gm carrying nbzAa-lacZ from pDTG928, Apr Gmr | This study |
| pDTG935 | pEX1.8 containing nbzR, Apr | This study |
| pDTG957 | pRK415 containing dntR::Kmr, Kmr Tcr | This study |
| pDTG800 | pUC18 containing ntdAaAbAcAd genes and partial ntdR from JS42, Apr | 29 |
| pDTG864 | pUC19 containing 5′ end of ntdR sequence, Apr | This study |
| pDTG865 | pUC19 containing 3′ end of ntdR sequence from pDTG800, Apr | This study |
| pDTG866 | pUC19 containing complete ntdR sequence, Apr | This study |
| pNtd1 | pBBR1MCS containing 0.9-kb nbzR fragment, Cmr | This study |
| pNag1 | pBBR1MCS containing 0.9-kb nagR fragment, Cmr | This study |
| pNah1 | pBBR1MCS containing 3.0-kb nahR fragment, Cmr | This study |
DNA manipulations.
Standard methods were used to manipulate plasmids and DNA fragments (39). Restriction endonucleases and DNA modification enzymes were purchased from New England Biolabs (Beverly, Mass.). Plasmids were isolated using spin miniprep kits (Qiagen, Chatsworth, Calif.), and DNA fragments were purified using a Qiaquick PCR purification kit (Qiagen). Chromosomal DNA was prepared using a Puregene DNA isolation kit (Gentra systems, Minneapolis, Minn.). DNA fragments were excised and purified from agarose gels using Qiaquick gel extraction kits (Qiagen). PCR was performed with an Expand Long Template PCR system (Roche, Indianapolis, Ind.). DNA was sequenced at the University of Iowa DNA core facility by standard automated-sequencing technology. Strain JS42 was identified as an Acidovorax sp. by partial 16S rRNA gene sequence comparison to the currently available 16S rRNA gene sequences. Identification of strain JS42 was done by MIDI Labs (Newark, Del.).
Cloning of ntdR.
A clone carrying the 5′ end of ntdR, including the promoter region, was identified previously (29). The 3′ end of ntdR was obtained as follows. Southern blot analysis (39) was used to identify the size of the SacI fragment carrying the 3′ end of the gene using dntR as a probe (data not shown). A partial genomic library was made by digesting JS42 DNA with SacI and ligating fragments (1.5 to 2.7 kb) into SacI-digested pUC19. The library was introduced into DH5α by transformation. Colony blot screenings were done using standard protocols with dntR labeled with [α-32P]dCTP as a probe to identify a transformant carrying ntdR (39). The plasmid from this transformant, designated pDTG864, contained the remaining sequence of ntdR within a 1.8-kb SacI fragment. A 0.6-kb SacI/NruI fragment containing the 5′ end of ntdR was subcloned from pDTG800 into SacI/HincII-digested pUC19 to form pDTG865. The 1.8-kb SacI-fragment containing the remaining sequence of ntdR was ligated into SacI-digested pDTG865 to form pDTG866, which contains the complete ntdR gene. The complete sequence of ntdR was obtained from pDTG866.
Primer extension analysis.
Primer extension analysis of the nbz and ntd operons was performed as previously described (52). RNA was prepared from JS765 and JS42 using the Trizol reagent (Life Technologies, Grand Island, N.Y.). RNA was extracted from cultures grown to a turbidity of 0.25. The initial turbidity was 0.05. The extension primer was 5′-GCCGCAGCGGCCCGACATGCA-3′. The primer was 5′ end labeled using [γ-32-P]dATP and a KinaseMax kit (Ambion, Austin, Tex.). The 32P-labeled primer was annealed to 10 to 20 μg of JS765 or JS42 RNA and extended using a First-Strand cDNA synthesis kit (Amersham, Piscataway, N.J.). DNA sequences were obtained using pDTG925 as the template and the same primer used for primer extension. Sequencing was carried out with [α-35S]dATP and a Sequenase, version 2.0, DNA sequencing kit (U.S. Biochemicals, Cleveland, Ohio). DNA fragments were resolved on 8 M urea-8% polyacrylamide gels (39).
RT-PCR.
Total RNA was isolated from JS765 and JS42 grown with succinate alone or in the presence of 500 μM salicylate, nitrobenzene (JS765), or 2-nitrotoluene (JS42) by using an RNeasy total RNA mini kit (Qiagen). Purified RNA was treated with RNase-free DNase I (Qiagen) to remove contaminating DNA. Reverse transcriptase (RT) PCR was carried out with an Access RT-PCR kit (Promega). The following primer pairs were used: P760F (5′-TGCCTAGCGATGCGGAAATG-3′) and P420R (5′-TCTCGGACATGTTCTGCAAC-3′) for the nbzAaAb region and P760F and PprobeR (5′-ACGTGGTAGCCGTCACCTAC-3′) for the nbzAaAbAc region. Control PCRs were performed without addition of RT with primer set 1. Primers for amplification of nbzAbAd (data not shown) were the following: P2100F (5′-ACGATGTCGAGCCTTTCGAG-3′) and P460R (5′-TTGTTGGCCCAGTTCTGAGG-3′).
Construction of the nbzAa-lacZ reporter and introduction into JS765, JS42, and G7.
The nbzAa-lacZ fusion was constructed using the method described by Whiteley et al. (53). A 241-bp DNA fragment encompassing the entire nbzAa promoter region including 135 bp upstream of the putative LysR-type binding site was generated by PCR using the primers NBDOPRO1 (5′-GGGGTACCCCTTTAAGTGAATTGCTGACGGCAGG-3′) and NBDOREV (5′-GCTCTAGAGCGCAAGCTCTTTTTTCAGTTGTCTC-3′). After restriction digestion of the introduced sites (underlined), this 241-bp fragment was ligated to KpnI-XbaI-digested pMW24-RPOS to generate pDTG928. The nbzAa-lacZ fragment from NotI-digested pDTG928 was ligated to NotI-digested pUTminiTn5-Gm to yield pDTG931. pDTG931 is a mobilizable plasmid containing the nbzAa-lacZ transcriptional fusion and a gentamicin resistance marker within mini-Tn5. Introduction of pDTG931 into JS765, JS42, and P. putida G7 was done by mating with E. coli S17-1(pDTG931) or with E. coli HB101(pRK2013) as a helper. Gentamicin-resistant colonies were selected and screened for blue color formation on agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (0.5%). Single Tn5 insertions in the resulting transconjugants (JS765-1, JS42-1, and G7-1) were confirmed by PCR and Southern blot analysis with a probe specific for the gentamicin resistance gene (data not shown). PCR confirmation was done with primers specific for nbzAa-lacZ: NBDOPRO1 and LacZ-rev (5′-GTTGTAAAACGACGGCCAGTGAA-3′). Strains JS765-1, JS42-1, and G7-1 were still able to grow with nitrobenzene, 2-nitrotoluene (2NT), or naphthalene, respectively, similar to their wild-type counterparts. Strains JS765-1, JS42-1, and G7-1 were used for analysis of lacZ expression.
Construction of E. coli carrying the nbzAa-lacZ fusion.
E. coli GS162 was chosen as an alternative recipient strain for determination of nbzAa-lacZ expression. A spontaneous rifampin-resistant mutant of GS162, designated GS162R, was obtained after selection on LB agar plates containing 50 μg of rifampin/ml. pDTG931 was introduced into strain BW19851 by transformation. BW19851(pDTG931) was mated with GS162R as described above. GS162R transconjugants containing insertions of nbzAa-lacZ were selected on MSB agar plates containing 10 mM glucose, 20 μg of gentamicin/ml, 20 μg of rifampin/ml, 1 mM thiamine, 1 mM phenylalanine, and X-Gal (0.5%). An ampicillin-sensitive transconjugant (GS162R-1) was selected, and integration of nbzAa-lacZ into the chromosome of GS162R was verified by PCR with primers specific for nbzAa-lacZ. For expression of NbzR in GS162R-1, nbzR was PCR amplified using primers NbzR1A (5′-CGGAATTCATGGATCTGCGCGACATCG-3′) and NbzR2A (5′-CCCAAGCTTTTATGCTTCAGAGAAAAG-3′). This fragment was digested with EcoRI (underlined) and HindIII (underlined) and ligated to EcoRI/HindIII-digested pEX1.8 to form pDTG935. The NbzR expression plasmid, pDTG935, was introduced into GS162R-1 by transformation.
Disruption of NtdR in JS42-1 and complementation with NtdR, NagR, and NahR.
A 7.0-kb pJS31 BglII fragment containing dntR, from Burkholderia sp. strain DNT (51), was cloned into BamHI-digested pRK415. This plasmid was digested with BamHI (unique site in dntR) and ligated with a 2.4-kb BamHI fragment containing the kanamycin resistance gene from pHP45Ω-Km. The resulting plasmid containing dntR::Km was designated pDTG957 and used for disruption of ntdR, since the nucleotide sequence of dntR was determined to be 97% identical to that of ntdR. pDTG957 was introduced into JS42-1 by mating as described above. We selected kanamycin-resistant colonies and identified a tetracycline-sensitive strain (JS42-1R), which indicated loss of the plasmid. This mutant contained a disrupted regulatory gene in place of ntdR, as shown by PCR (primers, NbzR1A and NbzR2A) and Southern analysis with a probe specific for ntdR (data not shown). For complementation, a 0.9-kb DNA fragment containing ntdR/nbzR was generated by PCR amplification with pDTG925 as the template using primers: NbzRcomp1A (5′-GGGGTACCATGGATCTGCGCGACATCGAC-3′) and NbzRcomp2 (5′-GGGGTACCTTATGCTTCAGAGAAAAGCTC-3′). This fragment was digested with KpnI (underlined) and ligated to KpnI-digested pBBR1MCS to form pNtd1. Introduction of pNtd1 into JS42-1R was done by mating with S17-1(pNtd1) as described above, and chloramphenicol-resistant colonies were selected. The presence of pNtd1 in JS42-1R was confirmed by plasmid purification and transformation of DH5α. A 0.9-kb DNA fragment containing nagR was generated by PCR amplification (primers, NbzRcomp1A and NbzRcomp2) with Ralstonia sp. strain U2 genomic DNA as the template. This fragment was digested with KpnI and ligated to KpnI-digested pBBR1MCS to form pNag1. To generate pNah1, a 3.0-kb PstI fragment containing nahR was excised from pMS104 and ligated to PstI-digested pBBR1MCS. pNag1 and pNah1 were introduced into JS42-1R as described for pNtd1.
Analysis of lacZ expression.
JS765-1, JS42-1, JS42-1R, and G7-1 containing nbzAa-lacZ were grown in MSB supplemented with 10 mM succinate, appropriate antibiotic(s), vitamins, and potential inducing compounds: nitrobenzene, 2NT, 3-nitrotoluene (3NT), 4-nitrotoluene (4NT), salicylate, and anthranilate at 500 μM; 2,4-dinitrotoluene (2,4-DNT), 2,6-dinitrotoluene (2,6-DNT), catechol, 3-methylcatechol, 2-amino-4,6-dinitrotoluene (2ADNT), and 4-amino-2,6dinitrotoluene (4ADNT) at 100 μM. All aromatic compounds were added from methanol stock solutions except salicylate, which was added from an aqueous solution. Cultures were inoculated to an initial turbidity of 0.03 to 0.06, and β-galactosidase was measured when the turbidity reached between 0.2 and 0.3 as outlined by Miller (24). Additional control experiments with 0.5% (final concentration) methanol and 100 to 200 μM (final concentration) nitrite added did not result in detectable induction of β-galactosidase activity (data not shown). GS162R-1, GS162R-1(pDTG935), and GS162R-1(pNag1) were grown in LB with appropriate antibiotic(s) and nitrobenzene, 2NT, catechol, 3-methylcatechol, salicylate, or anthranilate at a final concentration of 500 μM. Cultures were inoculated to an initial turbidity of 0.05 to 0.07, and β-galactosidase activity was measured when the turbidity reached between 0.4 and 0.6.
Analysis of nitroarene dioxygenase activity.
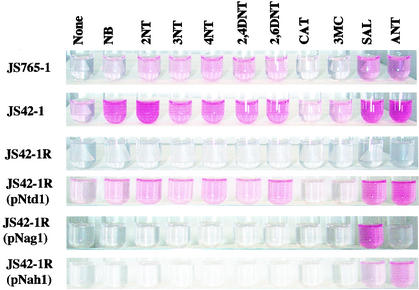
NBDO and 2NTDO activities in strains of JS765 and JS42, respectively, were assayed by monitoring the formation of nitrite as previously described (11). Whole cells of JS765 and JS42 that were used in β-galactosidase assays were also used in nitrite formation assays. Specifically, cells were resuspended to a turbidity of 0.3 to 0.4 in Z-buffer (24) and incubated with 3NT (JS765) or 2NT (JS42) at 30°C with shaking at 220 rpm. After 20 to 40 min, 0.4 ml of 1% (wt/vol) sulfanilamide in 1.5 N HCl was added. After mixing, 0.4 ml of 0.02% (wt/vol) N-(1-naphthyl)ethylenediamine in 1.5 N HCl was added. The formation of a pink-colored complex was documented after 15 min. The assays were done three separate times with similar results obtained, and a representative example is depicted in Fig. 3.
FIG. 3.
Analysis of nitrite release from 3NT by JS765-1 and from 2NT by variants of JS42 after growth in the presence of potential inducing compounds (abbreviations are as in Table 3). Dioxygenase activity is indicated by the formation of a pink-colored complex, using a nitrite assay as described in Materials and Methods.
Chemicals.
Nitrobenzene was from Mallinckrodt Inc. (Paris, Ky.). Salicylate was from Fisher Scientific (Pittsburgh, Pa.). 2-Amino-4,6-dinitrotoluene and 4-amino-2,6-dinitrotoluene were a gift from Jim C. Spain (Tyndall Air Force Base, Panama City, Fla.) All other aromatic compounds were from Aldrich Chemical Inc. (Milwaukee, Wis.).
Nucleotide sequence accession numbers.
The nucleotide sequences of nbzY and nbzR are available in GenBank under accession no. AY223675, that of ntdR is available under accession no. AY223676, and that of dntR is available under accession no. AY223677. The nucleotide sequence of the partial 16S rRNA gene from Acidovorax sp. strain JS42 is available under accession no. AY228545.
RESULTS
Identification of genes encoding LysR-type regulators associated with nitroarene dioxygenase gene clusters.
The sequence further upstream of the nitrobenzene dioxygenase (nbzAaAbAcAd) genes from strain JS765 on plasmid pDTG925 revealed two additional open reading frames (ORFs). One ORF was designated nbzY, a gene that appears to encode a protein similar to methyl-accepting chemotaxis proteins from E. coli and similar to the partial sequence of NagY from strain U2 (56). The second ORF shares homology with the LysR family of transcriptional regulators and was designated nbzR. nbzR is transcribed divergently from nbzAa, which encodes the reductase component of NBDO (Fig. 1A). Additional sequencing upstream of the ntd gene cluster encoding 2NTDO also revealed a divergently transcribed gene encoding a putative regulatory protein, NtdR. The nucleotide sequence of ntdR is identical to that of nbzR. The genes encoding 2,4-dinitrotoluene dioxygenase from the 2,4-DNT degrader Burkholderia sp. strain DNT were previously identified within pJS31 (51). Further sequencing upstream of the 2,4-dinitrotoluene dioxygenase genes within pJS31 identified dntR. An additional putative nitroarene dioxygenase regulator (ORF12) has been identified in another 2,4-DNT degrader, B. cepacia R34 (17). A comparison of the amino acid sequence identities of the putative nitroarene dioxygenase regulators is shown in Table 2. All of the nitroarene dioxygenase regulatory proteins have high levels of sequence identity to NahR from P. putida G7. NahR responds to salicylate and activates expression of the naphthalene degradation genes of the nah and sal operons in G7 (42, 44-46). As shown in Table 2, all of the nitroarene dioxygenase regulators are more similar to NagR than NahR. nagR is transcribed divergently from the nag gene cluster encoding the naphthalene dioxygenase in Ralstonia sp. strain U2 (56). Strain U2 grows on naphthalene through a gentisate pathway (9), unlike strain P. putida G7, in which naphthalene is degraded through the meta cleavage of catechol (55). NbzR, NtdR, DntR, ORF12, and NagR all consist of 301 amino acids and contain an N-terminal helix-turn-helix DNA-binding motif typical of LysR-type regulators (41) and only differ from each other on average in five amino acids.
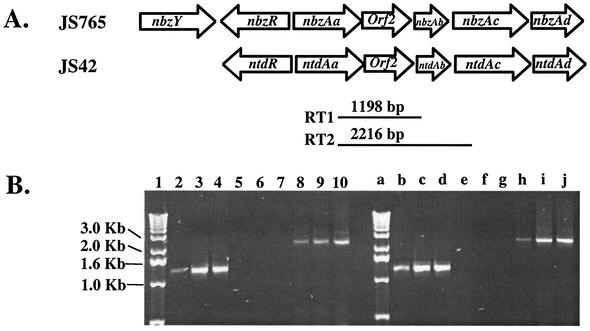
FIG. 1.
(A) The identical genetic organization of the nbz genes encoding NBDO from strain JS765 and ntd genes encoding 2NTDO from strain JS42. Arrows indicate the direction of transcription. The locations of primer sets and the amplified DNA fragments for RT-PCR are designated RT1 and RT2. (B) RT-PCR amplification of the nbz (lanes 2 to 10) and ntd (lanes b to j) gene clusters. Lanes 2, 5, and 8, RT-PCR products from total RNA from succinate-grown JS765; lanes 3, 6, and 9, RT-PCR products from total RNA from JS765 grown with succinate plus nitrobenzene; lanes 4, 7, and 10, RT-PCR products from total RNA from JS765 grown on succinate plus salicylate; lanes b, e, and h, RT-PCR products from total RNA from succinate-grown JS42; lanes c, f, and i, RT-PCR products from total RNA from JS42 grown with succinate plus 2NT; lanes d, g, and j, RT-PCR products from total RNA from JS42 grown with succinate plus salicylate. Samples in lanes 5 to 7 and e to g were without RT. Lanes 2 to 7 and b to g were amplifications performed with the RT1 primer set, and lanes 8 to 10 and h to j were with the RT2 primer set. Lanes 1 and a were loaded with a 1-kb ladder (Invitrogen, Carlsbad, Calif.).
TABLE 2.
Amino acid identities between LysR-type regulatory proteinsa
| Protein | % Identity with:
|
|||||
|---|---|---|---|---|---|---|
| NbzR | NtdR | DntR | Orf12 | NagR | NahR | |
| NbzR | 100 | |||||
| NtdR | 100 | 100 | ||||
| DntR | 97 | 97 | 100 | |||
| ORF12 | 98 | 98 | 99 | 100 | ||
| NagR | 98 | 98 | 99 | 99 | 100 | |
| NahR | 61 | 61 | 61 | 61 | 61 | 100 |
NbzR from JS765 (accession no. AY223675), NtdR from JS42 (accession no. AY223676), DntR from Burkholderia sp. strain DNT (accession no. AY223677), ORF12 from Burkholderia cepacia R34 (accession no. AF169302), NagR from strain U2 (accession no. AF036940), and NahR from P. putida G7 (accession no. J04233).
RT-PCR and primer extension analysis of the nbz and ntd genes.
To begin to understand the regulation of the nbzAaAbAcAd and ntdAaAbAcAd genes, which encode NBDO and 2NTDO, respectively, we determined the transcriptional architecture of the genes. As shown in Fig. 1, RT-PCR was used to determine whether the genes in the nbz and ntd clusters are cotranscribed. We obtained RT-PCR products from primers specific for the regions encoding the reductase to ferredoxin (Aa to Ab genes) and reductase to oxygenase α-subunit (Aa to Ac genes) in each cluster. In addition, a RT-PCR product was obtained with primers specific for the ferredoxin-to-oxygenase β-subunit (Ab to Ad genes) (data not shown). RT-PCR products were obtained with RNA prepared from JS765 and JS42 grown under a variety of conditions. However, more product was obtained from RNA extracted from JS765 or JS42 grown in the presence of salicylate, nitrobenzene, or 2NT than with succinate alone (Fig. 1B). The RT-PCR results demonstrate that the genes in the nbz and ntd clusters are each in an operon.
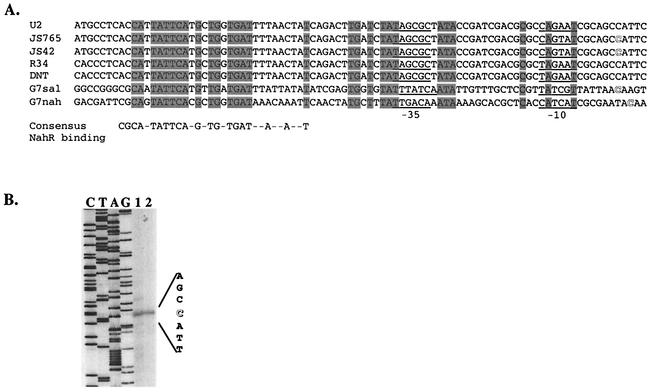
A comparison of the promoter regions of the nbz and ntd operons to those of other nitroarene dioxygenase and naphthalene dioxygenase gene clusters revealed a high degree of similarity in the nucleotide sequence (Fig. 2). In fact, the nucleotide sequences of the nbz and ntd promoter regions are identical. As shown in Fig. 2, many of the promoter elements previously identified in the nah and sal operons in P. putida G7 are conserved in the nitroarene dioxygenase promoters, including the NahR binding site (T-N11-A inverted repeat at −60 bp upstream) (16). Thus, we hypothesized that the start site of transcription of the nbz and ntd operons would be similar to those of the nah and sal operons, which are regulated by NahR in P. putida G7. To address this, we determined the transcriptional start sites for the nbz and ntd operons. Primer extension products were obtained from JS765 and JS42 grown in the presence of salicylate (Fig. 2B). The start sites of the major primer extension product from both strains were identical and mapped to a position 135 bp upstream of the predicted reductase gene translation start site in each operon. Primer extension products obtained from JS765 grown with succinate plus nitrobenzene and JS42 grown with succinate plus 2NT were identical to those obtained from cells grown with succinate plus salicylate (data not shown). The transcriptional start sites of the nbz/ntd operons correspond to the start sites of the nah and sal operons from P. putida G7 (Fig. 2).
FIG. 2.
(A). Alignment of the nitroarene and naphthalene dioxygenase gene promoters. The consensus NahR binding sequence (16) is shown below the alignment. Conserved nucleotides are shaded. The −35 and −10 sites are underlined. Identified transcription start sites are in outlined font. (B) Primer extension analysis of the nbz and ntd transcripts from JS765 and JS42 grown with succinate plus salicylate, respectively. The sequencing ladder is shown at the left, and primer extension products from strain JS765 (lane 1) and from strain JS42 (lane 2) are at the right. The transcription start site is shown in outlined font.
Activity of the nbzAa-lacZ fusion in wild-type JS765 and JS42.
To examine the transcriptional regulation of the nbz and ntd operons, we constructed a promoter reporter fusion. Specifically, the promoter region upstream of the reductase gene (nbzAa) from JS765 was fused to a promoterless lacZ gene located within a mini-Tn5 to form nbzAa-lacZ, as described in Materials and Methods. As mentioned above, the nucleotide sequences of the promoter regions of the nbz and ntd operons are identical, and therefore, nbzAa-lacZ could be used in both JS765 and JS42 to monitor nbz/ntd promoter activity.
β-Galactosidase activity in JS765-1 was analyzed in cells grown in minimal medium with succinate provided as the carbon source and various aromatic compounds added as potential inducers. As shown in Table 3, we observed a significant increase in β-galactosidase activity in JS765-1 after growth in the presence of many of the aromatic compounds over the basal level of activity (succinate alone). All of the nitroaromatic compounds caused a substantial increase in β-galactosidase activity (7- to 28-fold). Nitrobenzene, the growth substrate for JS765-1, was the weakest inducer (sevenfold). 2NT was the strongest inducer (28-fold), even though JS765-1 is unable to grow with 2NT as a carbon source. In addition, 3NT and 4NT were good inducers, and both compounds allow for growth of JS765 (22). The two isomers of dinitrotoluene also caused an increase in β-galactosidase activity, even though they do not support growth of JS765. Interestingly, addition of the reduction products of 2,4,6-trinitrotoluene, 2ADNT, and 4ADNT (48) resulted in a significant increase in β-galactosidase activity. Addition of catechol or 3-methylcatechol, which are intermediates in nitrobenzene and 2NT metabolism, respectively (27, 29), did not result in an increase in β-galactosidase activity, indicating that the nitroaromatic compounds function as the actual effector molecules. Salicylate and anthranilate, the known inducers of the nah and sal operons in P. putida G7 (1, 42), also increased β-galactosidase activity in JS765-1, indicating that NbzR is able to recognize inducers of the naphthalene degradation genes. However, JS765 is unable to grow with naphthalene or salicylate as a carbon source.
TABLE 3.
Activity of the nbzAa-lacZ fusion in strains of JS765 and JS42
| Compound addedd | β-Galactosidase activitya (fold inductionb) in strain:
|
|||||
|---|---|---|---|---|---|---|
| JS765-1 | JS42-1 | JS42-1Rc | JS42-1R (pNtd1) | JS42-1R (pNag1) | JS42-1R (pNah1) | |
| None | 8 ± 3 | 62 ± 19 | 4 ± 1 | 217 ± 50 | 7 ± 2 | 12 ± 4 |
| NB | 56 ± 18 (7) | 225 ± 63 (4) | 5 ± 1 (1) | 410 ± 84 (2) | 8 ± 1 (1) | 15 ± 2 (1) |
| 2NT | 228 ± 82 (28) | 452 ± 101 (7) | 4 ± 1 (1) | 632 ± 79 (3) | 11 ± 3 (2) | 21 ± 6 (2) |
| CAT | 11 ± 4 (1) | 77 ± 4 (1) | 5 ± 1 (1) | 248 ± 35 (1) | 9 ± 2 (1) | 20 ± 2 (2) |
| 3MC | 17 ± 5 (2) | 61 ± 5 (1) | 5 ± 1 (1) | 231 ± 58 (1) | 9 ± 2 (1) | 15 ± 4 (1) |
| 3NT | 112 ± 24 (14) | 385 ± 70 (6) | 5 ± 1 (1) | 644 ± 65 (3) | 12 ± 4 (2) | 16 ± 5 (1) |
| 4NT | 200 ± 28 (25) | 323 ± 82 (5) | 4 ± 1 (1) | 651 ± 104 (3) | 12 ± 4 (2) | 16 ± 3 (1) |
| 2,4-DNT | 110 ± 15 (14) | 369 ± 104 (6) | 4 ± 1 (1) | 547 ± 33 (3) | 9 ± 2 (1) | 15 ± 2 (1) |
| 2,6-DNT | 190 ± 15 (24) | 365 ± 140 (6) | 4 ± 1 (1) | 585 ± 58 (3) | 10 ± 3 (1) | 13 ± 3 (1) |
| 2ADNT | 97 ± 11 (9) | 433 ± 35 (7) | 4 ± 1 (1) | 772 ± 82 (4) | 11 ± 3 (2) | 15 ± 4 (1) |
| 4ADNT | 83 ± 12 (8) | 409 ± 22 (7) | 4 ± 1 (1) | 576 ± 29 (3) | 12 ± 4 (2) | 17 ± 4 (1) |
| SAL | 225 ± 61 (28) | 691 ± 180 (11) | 5 ± 1 (1) | 1087 ± 164 (5) | 342 ± 92 (49) | 504 ± 20 (42) |
| ANT | 144 ± 33 (18) | 568 ± 89 (9) | 5 ± 1 (1) | 714 ± 95 (3) | 10 ± 2 (1) | 390 ± 42 (32) |
Expressed as Miller units ± standard deviation.
Fold induction = Miller units (compound added)/Miller units (succinate alone), same strain.
Strain JS42-1R contained the vector pBBR1MCS.
NB, nitrobenzene; 2NT, 2-nitrotoluene; 3NT, 3-nitrotoluene; 4NT, 4-nitrotoluene; 2,4-DNT, 2,4-dinitrotoluene; 2,6-DNT, 2,6-dinitrotoluene; 2ADNT, 2-amino-4,6-dinitrotoluene; 4ADNT, 4-amino-2,6-dinitrotoluene; CAT, catechol; 3MC, 3-methylcatechol; SAL, salicylate; ANT, anthranilate.
Analysis of β-galactosidase activity in strain JS42-1 produced similar results (Table 3). However, the basal level of activity in JS42-1 was significantly higher than in JS765-1 (approximately eightfold). All of the nitroaromatic compounds tested caused a significant increase in β-galactosidase activity (four- to sevenfold), with nitrobenzene again the weakest inducer. JS42 is unable to grow with 3NT and 4NT as carbon sources, and only weak growth has been observed with nitrobenzene (14, 22). The metabolites catechol and 3-methylcatechol did not increase activity, suggesting that metabolism of the nitroaromatic compounds is also not required for induction in JS42. Salicylate and anthranilate were also good inducers, suggesting that NtdR can also respond to these compounds in JS42-1. JS42 is also unable to grow with naphthalene or salicylate.
Activity of the nbzAa-lacZ fusion in JS42-1R and complementation with NtdR, NagR, and NahR.
To determine whether NtdR is directly involved in expression of the ntd genes, NtdR was disrupted in JS42-1. JS42-1R was unable to grow with 2NT as a carbon source, indicating that NtdR is required for 2NT metabolism. β-Galactosidase activity was analyzed in JS42-1R under the same growth conditions as JS42-1 (Table 3). No significant increase in β-galactosidase activity was observed after growth in the presence of the aromatic compounds. In addition, the basal level of activity was substantially lower in JS42-1R than in JS42-1 (approximately 16-fold). When NtdR was added in trans, induction by all of the nitroaromatic compounds, as well as salicylate and anthranilate, was restored, and the ability to grow with 2NT was also restored. However, the basal level of β-galactosidase activity was approximately threefold higher in JS42-1R(pNtd1) than in JS42-1 (Table 3). Since NtdR differs from NagR by only five amino acids and is 61% identical to NahR, we tested whether NagR and NahR could restore induction in JS42-1R. Neither NagR nor NahR could complement for induction by any of the nitroaromatic compounds, but they did restore induction by salicylate, indicating that NagR and NahR are expressed in JS42-1R. NahR, but not NagR, was able to restore induction by anthranilate (Table 3). Finally, the basal level of activity in JS42-1R was only slightly increased when NagR or NahR was present. However, the ability to grow with 2NT was restored in both strains.
Analysis of nitroarene dioxygenase activity.
To verify results from lacZ fusion studies, we examined NBDO and 2NTDO activity directly using a nitrite assay. As depicted in Fig. 3, dioxygenase activity correlates well with β-galactosidase activity in all of the strains. An increase in dioxygenase activity (increase in pink color indicating nitrite formation) was observed in JS765-1 and JS42-1 after growth in the presence of the nitroaromatic compounds, as well as salicylate and anthranilate. Growth in the presence of catechol and 3-methylcatechol resulted in dioxygenase activity similar to that seen after growth with succinate alone. A low basal level of dioxygenase activity and no induction were observed in JS42-1R. An increase in dioxygenase activity was observed only with salicylate in JS42-1R(pNag1), a result identical to that seen with the lacZ fusion. Dioxygenase activity levels in JS42-1R(pNah1) were similar to results of the lacZ fusion studies, with only salicylate and anthranilate functioning as inducers.
Activity of the nbzAa-lacZ fusion in P. putida G7 and E. coli.
As a control experiment, we tested whether the native NahR is able to regulate expression of nbzAa-lacZ in a P. putida G7 background. P. putida G7-1, which contains a chromosomal insertion of nbzAa-lacZ, demonstrated an increase in β-galactosidase activity in the presence of salicylate and anthranilate (Table 4). However, there was no significant increase in β-galactosidase activity in the presence of nitrobenzene or 2NT. Therefore, the nbz promoter alone is not sufficient for induction by nitroaromatic compounds. However, this result confirms that NahR can recognize nbzAa-lacZ and respond to the presence of salicylate and anthranilate.
TABLE 4.
Activity of the nbzAa-lacZ fusion in P. putida G7 and E. coli GS162
| Compound addedc | β-Galactosidase activitya (fold inductionb) in strain:
|
|||
|---|---|---|---|---|
| G7-1 | GS162R-1 | GS162R-1 (pDTG935) | GS162R-1 (pNag1) | |
| None | 4 ± 1 | 2 ± 0.2 | 16 ± 2 | 1.4 ± 0.6 |
| NB | 6 ± 1 (1) | 2 ± 0.1 (1) | 13 ± 3 (1) | 1.0 ± 0.4 (0.7) |
| 2NT | 8 ± 2 (1) | 2 ± 0.3 (1) | 17 ± 3 (1) | 2.2 ± 0.8 (1) |
| CAT | 5 ± 1 (1) | 2 ± 0.2 (1) | 12 ± 2 (1) | 1.0 ± 0.4 (0.7) |
| 3MC | 4 ± 1 (1) | 2 ± 0.3 (1) | 9 ± 1 (1) | 1.0 ± 0.4 (0.7) |
| SAL | 107 ± 15 (27) | 2 ± 0.2 (1) | 396 ± 96 (25) | 85 ± 24 (60) |
| ANT | 88 ± 22 (18) | 2 ± 0.2 (1) | 88 ± 31 (5) | 1.1 ± 0.2 (0.8) |
Expressed as Miller units ± standard deviation.
Fold induction = Miller units (compound added)/Miller units (succinate alone).
Compound abbreviations are as in Table 3.
To attempt to determine the actual effector compounds, E. coli GS162R, which is unable to oxidize nitroaromatic compounds, was used in expression studies. A strain of GS162R harboring a chromosomal copy of nbzAa-lacZ (GS162R-1) was constructed. nbzR and nagR were then expressed separately in GS162R-1 in order to examine the inducer profile of each regulatory protein. GS162R-1 containing the vector only had a low basal level of β-galactosidase activity, and no induction occurred when cells were grown in the presence of any of the compounds tested (Table 4). When NbzR was expressed in GS162R-1, the basal level of β-galactosidase activity increased significantly. Unexpectedly, only salicylate and anthranilate resulted in an increase in β-galactosidase activity, and neither nitrobenzene nor 2NT functioned as an inducer (Table 4). Similarly, when NagR was expressed in GS162R-1, only salicylate induced β-galactosidase activity, which is consistent with results obtained with JS42-1R(pNag1). Also, the basal level of activity did not increase when NagR was expressed in trans. To determine whether the lack of induction by nitrobenzene and 2NT in GS162R-1(pDTG935) was a specific characteristic of E. coli, we conducted similar experiments with other bacteria, including P. putida PRS2000 (28, 49) and Ralstonia eutropha JMP289 (5). In each case, when NbzR was provided in trans, the results obtained were similar to obtained with GS162R-1(pDTG935), where only salicylate and anthranilate acted as inducers (data not shown).
DISCUSSION
To date, all bacteria that utilize a nitroarene dioxygenase to initiate aerobic growth with nitroaromatic compounds have a nahR homolog transcribed divergently from the nitroarene dioxygenase gene cluster. This suggests that regulation of nitroarene dioxygenase genes, including the nbz and ntd genes, may be similar to the regulation of expression of the naphthalene degradation (nah and sal) genes in P. putida G7. Primer extension and RT-PCR analyses support this hypothesis.
Results from β-galactosidase and nitrite assays with JS765-1 and JS42-1 revealed that nbz and ntd expression is induced by multiple aromatic compounds. The same series of aromatic compounds induced expression in both strains, which was not unexpected, since the identified regulatory elements are identical in JS765 and JS42. However, the basal level of expression of the nbz operon in JS765-1 is much lower than that of the ntd operon in JS42-1, which suggests some fundamental differences between the strains. The difference in the basal level of expression between JS765-1 and JS42-1 corresponds well to the previous observation that NBDO activity was inducible, while 2NTDO activity was constitutive (14, 27). However, this study revealed that the ntd genes could be induced approximately fourfold over the basal level of expression. Even compounds that do not serve as growth substrates for these strains induced expression, including 2,4-DNT, 2,6-DNT, 2ADNT, 4ADNT, salicylate, and anthranilate. It is also important to note that NBDO is able to release nitrite from all of the nitroaromatic compounds tested except for 2ADNT (18, 22). 2NTDO can oxidize nitrobenzene, 2NT, 3NT, and 4NT but not 2,4-DNT (30). 2NTDO can also oxidize 4ADNT, with the release of nitrite, but not 2ADNT (K.-S. Lee and R. E. Parales, unpublished data), while 2,6-DNT oxidation has not been examined. These observations, along with the lack of induction by catechol and 3-methylcatechol, suggest that metabolism is not required for induction and that the nitroaromatic compounds function as the actual effector molecules. Based on the nbzAa-lacZ fusion results, 2NT appeared to be the strongest effector and nitrobenzene the weakest effector in both JS765-1 and JS42-1. It has been suggested by Cebolla et al. that specific contacts between NahR and compounds with substituents at positions C-1 and C-2 of the aromatic ring, such as salicylate and anthranilate, are requirements for effector interaction. However, wild-type NahR was shown to accept a wide variety of salicylate structural analogs as effectors. Even those with substituents at the C-3, C-4, and C-5 positions were tolerated (3). Based on homology to NahR, NbzR/NtdR might also favor aromatic compounds with C-1 and C-2 substituents, because 2NT and salicylate appeared to be the strongest inducers. However, it appears that unlike wild-type NahR, NbzR/NtdR can respond to nitroaromatic compounds as effectors and tolerate changes in the position of single or multiple substituents.
As shown in Table 3 and Fig. 3, disruption of NtdR resulted in a decrease in the basal level of expression and a loss of induction of the 2NTDO genes. When NtdR was expressed in trans, the basal level of expression was significantly increased and induction by all of the compounds was restored. Therefore, NtdR must play a direct role in responding to nitroaromatic compounds, as well as salicylate and anthranilate. In addition, it is apparent that NtdR is required for the high basal level of activity in JS42. Binding of NahR to DNA and interaction with the α-subunit of RNA polymerase (RNAP) have been demonstrated in the absence of added inducer (16, 34, 44). Similarly, in the absence of inducer, NtdR may bind the recognition site upstream of the −35 site and interact with RNAP, increasing transcription from the ntd promoter. The increase in the basal level of β-galactosidase activity in JS42-1R(pNtd1) over that in JS42-1 is probably due to increased expression of ntdR from the multicopy plasmid (Table 3). Finally, NtdR is required for growth with 2NT, providing further evidence that NtdR functions as an activator that is directly involved in regulating expression of the 2NTDO genes.
Complementation of JS42-1R with NahR did not restore induction by the nitroaromatic compounds, and NagR only restored induction with salicylate (Table 3). This result is consistent with results of naphthalene (nag) gene induction in Ralstonia sp. strain U2. In this organism, only salicylate, but not anthranilate or nitroaromatic compounds, serve as an inducer (R. M. Jones and P. A. Williams, personal communication). In addition, the basal level of β-galactosidase activity was only slightly increased for JS42-1R(pNag1) or JS42-1R(pNah1) (Table 3). Although 2NT did not function as an inducer in JS42-1R(pNag1) or JS42-1R(pNah1), the slight increase in the basal level of expression of 2NTDO in JS42-1R appeared to be sufficient to allow for growth with 2NT.
As shown in Table 5, there are only five amino acids that differ between NtdR and NagR. Functional domains have been previously identified within the NahR protein. These include the N-terminal portion containing the helix-turn-helix motif (residues 23 to 45), which is important for DNA binding, the central coinducer response-recognition regions (residues 95 to 173 and 196 to 206), and a C-terminal portion (residues 227 to 253) believed to be involved in both inducer response and multimerization, as NahR is believed to function as a tetramer (3, 16, 43). Interestingly, single-amino-acid changes at position 169 of NahR resulted in proteins that were able to respond to benzoate, an aromatic compound lacking a C-2 substituent (3). Also, when the proline at position 227 was changed to a serine, NahR no longer responded to salicylate as an inducer (43). Residue 227 is a serine in NtdR, but NtdR responds to salicylate. Considering the numerous differences in the coinducer response regions between NahR and NtdR, this is not surprising. Also, proteins with semiconstitutive phenotypes that display a significant increase in the basal level of expression of the nah and sal operons have been shown to result from single mutations in NahR (3, 16). Similar results have also been observed in other systems. For example, XylR, a member of the NtrC family of transcriptional activators, has a broad effector profile, and single-amino-acid changes in this regulator resulted in altered effector specificity, including the ability to respond to nitrotoluenes (10, 38). Also, the regulators of TOL meta-cleavage pathway genes, XylS and XylS1, differ in only five amino acids but have different effector profiles and display different sigma factor dependence (37). Finally, the ability of bacterial strains to grow with multiple hydroxylated aromatic substrates has been linked to enhanced effector responses, such as those caused by single-amino-acid changes in DmpR, a regulator of phenol degradation genes (40). Therefore, it seems reasonable that the few differences between NtdR and NagR could drastically alter the effector profile as well as the observed basal level of expression.
TABLE 5.
Comparison of the amino acid differences between NbzR/NtdR and NagR
The attempt to examine the effector profile of NtdR by expression in E. coli produced interesting results (Table 4). The lack of induction by nitroaromatic compounds in E. coli cannot be due to the inability of these compounds to be transported into the cell cytoplasm, because whole-cell biotransformations with E. coli strains expressing cloned dioxygenase genes demonstrated that nitrobenzene, mononitrotoluenes, and dinitrotoluenes are able to enter the cell (22, 30). In addition, functional analysis of NahR in E. coli has been successful (3). The differences in the effector profiles of NtdR in E. coli and JS42 could be due to differences in the RNA polymerases in the two strains and the interaction of the specific RNAP with NtdR. This hypothesis is supported by the results demonstrating differences in basal and induced levels of activity between JS765-1 and JS42-1 and the presence of a minor primer extension product with JS765 (Fig. 2B), even though all of the identified regulatory elements are identical in these two strains (Table 3). Alternatively, differences in mRNA or regulator stability could affect activity in the different strains. The data presented here suggest that JS42 and JS765 are unique in their ability to utilize NtdR/NbzR to sense nitroaromatic compounds. Whether this is due to differences in RNAP or the presence of some other unidentified regulatory factor(s) remains to be determined.
This is the first clear demonstration of a nitroarene-responsive activator involved in the expression of nitroarene degradation genes. However, there is another report of a regulator responding to nitrobenzene as an effector. P. putida HS12 grows with nitrobenzene through a partial reductive pathway where nitrobenzene is converted to 2-aminophenol (33). All of the nitrobenzene degradation genes in P. putida HS12 are encoded on two catabolic plasmids. A negative regulator (NbzR) of the aminophenol degradation operon has been identified in P. putida HS12. However, it is unclear whether NbzR in P. putida HS12 responds directly to nitrobenzene or to hydroxylaminobenzene, because in a P. putida strain containing only NbzR and the aminophenol degradation operon, nitrobenzene induction was not observed. This study also suggested that a secondary trans-acting factor may be required for the induction of the aminophenol degradation genes (32). Similarly, the requirement for an additional trans-acting factor in the expression of the JS765 and JS42 nitroarene dioxygenases cannot be ruled out.
Most bacteria that are able to grow using nitroaromatic compounds have been isolated from sites contaminated with one or more of these compounds. Thus, due to selective pressure, bacteria appear to have evolved the necessary enzymes and pathways required for nitroaromatic compound degradation. It has been suggested that the next step in the evolution process would be to coordinate the expression and activities of these enzymes in response to novel man-made compounds, in this case nitroaromatic compounds (2). Based on the data presented here, strains JS765 and JS42 not only have evolved the necessary catabolic enzymes for the degradation of nitroaromatic compounds but are also able to regulate expression of the genes encoding the initial enzyme. However, JS765 and JS42 appear to be at an intermediate step in the evolution process, because although NbzR/NtdR can recognize multiple nitroaromatic compounds, it still responds to salicylate and anthranilate, molecules that are not produced during the degradation of nitrobenzene or 2NT. The identified regulatory systems provide an additional evolutionary link between nitroarene dioxygenases and the naphthalene dioxygenase from U2, since apparently only five amino acid substitutions are required to change NagR from a salicylate-only responsive regulator to a nitroarene-responsive regulator with broad specificity (NtdR). Additional characterization of NtdR, including DNA-binding studies and site-directed mutagenesis, will be required in order to understand the fundamental differences in NtdR function within JS42 and heterologous hosts.
Acknowledgments
We thank Mark Urbanowski and George Stauffer for providing E. coli GS162, Jim Spain for providing pJS31, Peter Williams for providing Ralstonia sp. strain U2, and Marvin Whiteley for helpful discussions. We also thank Rheinallt Jones and Peter Williams for sharing data prior to publication.
This research was supported in part by funds from the Strategic Environmental Research and Development Program, project CU1212 (R.E.P.), and the U.S. Army Research Office under grant DAAD19-99-1-0285 (D.T.G). D.J.L. has been supported by a National Science Foundation research training grant (DBI9602247) and a National Institutes of Health traineeship in Biotechnology (T32GM8365).
REFERENCES
- 1.Barnsley, E. A. 1975. The induction of the enzymes of naphthalene metabolism in pseudomonads by salicylate and 2-aminobenzoate. J. Gen. Microbiol. 88:193-196. [DOI] [PubMed] [Google Scholar]
- 2.Cases, I., and V. de Lorenzo. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cebolla, A., C. Sousa, and V. de Lorenzo. 1997. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J. Biol. Chem. 272:3986-3992. [DOI] [PubMed] [Google Scholar]
- 4.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 5.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn, N., and I. C. Gunsalus. 1973. Transmissable plasmids coding early enzymes of naphthalene oxidation in Pseudomonas putida. J. Bacteriol. 114:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 8.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuenmayor, S. L., M. Wild, A. L. Boyles, and P. A. Williams. 1998. A gene cluster encoding steps in the conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garmendia, J., D. Devos, A. Valencia, and V. de Lorenzo. 2001. A la carte transcriptional regulators: unlocking responses of the prokaryotic enhancer-binding protein XylR to non-natural effectors. Mol. Microbiol. 42:47-59. [DOI] [PubMed] [Google Scholar]
- 11.Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.). 1994. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 12.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 13.Haigler, B. E., and J. C. Spain. 1993. Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl. Environ. Microbiol. 59:2239-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haigler, B. E., W. H. Wallace, and J. C. Spain. 1994. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl. Environ. Microbiol. 60:3466-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartter, D. R. 1985. The use and importance of nitroaromatic chemicals in the chemical industry, p. 1-14. In D. E. Rickert (ed.), Toxicity of nitroaromatic compounds. Chemical Industry Institute of Toxicology series. Hemisphere, Washington, D.C.
- 16.Huang, J., and M. A. Schell. 1991. In vivo interactions of the NahR transcriptional activator with its target sequences. J. Biol. Chem. 266:10830-10838. [PubMed] [Google Scholar]
- 17.Johnson, G. R., R. K. Jain, and J. C. Spain. 2002. Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol. 184:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, G. R., B. F. Smets, and J. C. Spain. 2001. Oxidative transformation of aminodinitrotoluene isomers by multicomponent dioxygenases. Appl. Environ. Microbiol. 67:5460-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 20.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. A broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 21.Lacks, S., and J. R. Greenberg. 1977. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J. Mol. Biol. 114:153-168. [DOI] [PubMed] [Google Scholar]
- 22.Lessner, D. J., G. R. Johnson, R. E. Parales, J. C. Spain, and D. T. Gibson. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol. 68:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metcalf, W. W., W. Jiang, and B. L. Wanner. 1994. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6Kv origin plasmids at different copy numbers. Gene 138:1-7. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1975. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Nishino, S. F., G. C. Paoli, and J. C. Spain. 2000. Aerobic degradation of dinitrotoluenes and the pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol. 66:2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishino, S. F., and J. C. Spain. 1993. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol. 59:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishino, S. F., and J. C. Spain. 1995. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl. Environ. Microbiol. 61:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ornston, L. N., and D. Parke. 1976. Properties of an inducible uptake system for β-ketoadipate in Pseudomonas putida. J. Bacteriol. 125:475-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parales, J. V., A. Kumar, R. E. Parales, and D. T. Gibson. 1996. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181:57-61. [DOI] [PubMed] [Google Scholar]
- 30.Parales, J. V., R. E. Parales, S. M. Resnick, and D. T. Gibson. 1998. Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the C-terminal region of the α subunit of the oxygenase component. J. Bacteriol. 180:1194-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parales, R. E. 2000. Molecular biology of nitroarene degradation, p. 63-89. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. CRC Press, Boca Raton, Fla.
- 32.Park, H.-S., and H.-S. Kim. 2001. Genetic and structural organization of the aminophenol catabolic operon and its implication for evolutionary process. J. Bacteriol. 183:5074-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, H.-S., and H.-S. Kim. 2000. Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J. Bacteriol. 182:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, W., C. O. Jeon, and E. L. Madsen. 2002. Interaction of NahR, a LysR-type transcriptional regulator, with the α subunit of RNA polymerase in the naphthalene degrading bacterium Pseudomonas putida. NCIB 9816-4. FEMS Microbiol. Lett. 213:159-165. [DOI] [PubMed] [Google Scholar]
- 35.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhys-Williams, W., S. C. Taylor, and P. A. Williams. 1993. A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J. Gen. Microbiol. 139:1967-1972. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz, R., and J. L. Ramos. 2002. Residues 137 and 153 at the N terminus of the XylS protein influence the effector profile of this transcriptional regulator and the σ factor used by RNA polymerase to stimulate transcription from its cognate promoter. J. Biol. Chem. 277:7282-7286. [DOI] [PubMed] [Google Scholar]
- 38.Salto, R., A. Delgado, C. Michan, S. Marques, and J. L. Ramos. 1998. Modulation of the function of the signal receptor domain of XylR, a member of a family of prokaryotic enhancer-like positive regulators. J. Bacteriol. 180:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Sarand, I., E. Skarfstad, M. Forsman, M. Romantschuk, and V. Shingler. 2001. Role of the DmpR-mediated regulatory circuit in bacterial biodegradation properties in methylphenol-amended soils. Appl. Environ. Microbiol. 67:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 42.Schell, M. A. 1985. Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene 36:301-309. [DOI] [PubMed] [Google Scholar]
- 43.Schell, M. A., P. H. Brown, and S. Raju. 1990. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcription activator. J. Biol. Chem. 265:3844-3850. [PubMed] [Google Scholar]
- 44.Schell, M. A., and E. F. Poser. 1989. Demonstration, characterization, and mutational analysis of NahR protein binding to nah and sal promoters. J. Bacteriol. 171:837-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schell, M. A., and M. Sukordhaman. 1989. Evidence that the transcription activator encoded by the Pseudomonas putida nahR gene is evolutionarily related to the transcription activators encoded by the Rhizobium nodD genes. J. Bacteriol. 171:1952-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schell, M. A., and P. E. Wender. 1986. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J. Bacteriol. 166:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon, R., U. Priefer, and A. Puhler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:37-45. [Google Scholar]
- 48.Spain, J. C. 1995. Biodegradation of nitroaromatic compounds. Annu. Rev. Microbiol. 49:523-555. [DOI] [PubMed] [Google Scholar]
- 49.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads; a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 50.Suen, W.-C., B. E. Haigler, and J. C. Spain. 1996. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J. Bacteriol. 178:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suen, W.-C., and J. C. Spain. 1993. Cloning and characterization of Pseudomonas sp. strain DNT genes for 2,4-dinitrotoluene degradation. J. Bacteriol. 175:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 55.Yen, K.-M., and I. C. Gunsalus. 1982. Plasmid gene organization: naphthalene/salicylate oxidation. Proc. Natl. Acad. Sci. USA 79:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, N.-Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]