Abstract
Recent reports suggested that phosphatase of regenerating liver (PRL)-3 might be involved in colorectal carcinoma metastasis with an unknown mechanism. Here we demonstrated that PRL-3 expression was up-regulated in human liver carcinoma compared with normal liver. PRL-3 was also highly expressed in metastatic melanoma B16-BL6 cells but not in its lowly metastatic parental cell line, B16 cells. B16 cells transfected with PRL-3 cDNA displayed morphological transformation from epithelial-like shape to fibroblast-like shape. PRL-3-overexpressed cells showed much higher migratory ability, which could be reversed by specific anti-sense oligodeoxynucleotide and the phosphatase inhibitors sodium orthovanadate or potassium bisperoxo oxovanadate V. Meanwhile, the expression of the catalytically inactive PRL-3 mutations (D72A or C104S) significantly reduced the cell migratory capability. In addition, PRL-3 transfectants demonstrated altered extracellular matrix adhesive property and up-regulated integrin-mediated cell spreading efficiency. Furthermore, we confirmed that PRL-3 could facilitate lung and liver metastasis of B16 cells in an experimental metastasis model in mice, consistent with accelerated proliferation and growth rate both in vitro and in vivo. Together, these observations provide convincing evidence that PRL-3 truly plays a causal role in tumor metastasis.
Metastasis is the leading cause of death in cancer patients and involves a complex, multistep process including detachment of tumor cells from a primary cancer, invasion of surrounding tissue, entry into the circulatory system, reinvasion, and proliferation at a distant secondary site.1,2 A wide variety of stimuli have been associated with the spread of tumor cells, including cytokines, hormones, growth factors, cell adhesion molecules, and extracellular components. Many of these stimuli transmit signals via a tyrosyl phosphorylation pathways that dictate whether a tumor cell will grow and divide, change shape, migrate, differentiate, or die.3
Protein tyrosine phosphorylation is a major posttranslational modification that cells use to regulate signal transduction. The homeostasis of tyrosine phosphorylation is controlled by protein tyrosine kinases (PTKases) that catalyze tyrosine phosphorylation, and protein tyrosine phosphatase (PTPases) that are responsible for dephosphorylation. PTKases, PTPases, and their corresponding substrates are integrated into elaborate signal-transducing networks. Deregulation of phosphorylation is known to result in neoplastic or nonneoplastic disease.4 Phosphatases are as important as the well-studied PTKases because phosphorylation is a dynamic and reversible process.5 The PTPase superfamily can be divided into three major classes: tyrosine-specific and low-molecular weight phosphatases, which strictly dephosphorylate phosphotyrosine residues, and dual-specific phosphatases, which use protein substrates that contain phosphotyrosine, phosphoserine, and phosphothreonine. Tyrosine-specific PTPases can be further divided into two groups: receptor-like PTPases, exemplified by CD45 and PTPα, and intracellular PTPases, exemplified by PTP1B and SHP-2 (Src homology region 2-domain phosphatase). Dual-specific phosphatases include cell-cycle regular Cdc25 phosphatases, MAP kinase phosphatases, and the tumor suppressor PTEN (phosphatase and tensin homologue deleted on chromosome 10).6 The cellular function as well as substrates of many PTPases remain unknown.
The PRL phosphatases (PRL-1, PRL-2, and PRL-3), which have been identified throughout the past few years, are three closely related PTPases. Their sequences contain the PTP active site signature sequence CX5R,7–9 but encode relatively small proteins of ∼22 kd with at least 75% amino acid sequence similarity and contain a C-terminal CAAX sequence for prenylation.10,11 Their catalytic domain is most similar to that of dual-specificity phosphatases and their closest relatives are Cdc14 and PTEN.7 Prl-1 was first identified as an immediate-early gene whose expression is induced in rat regenerating liver.12 Overexpression of PRL-1 and PRL-2 can lead to epithelial cell transformation and the transfected cells can form tumors in nude mice.7,8 PRL-2 was also overexpressed in prostate cancer.13 Saha and colleagues14 demonstrated that the expression of PRL-3 is associated with liver metastasis of colon cancer. They compared the global gene expression profile of metastatic colorectal cancers with that of primary tumors, benign tumors, and normal colorectal epithelium, and found that prl-3 is the only one gene that was highly expressed in all 18 metastases examined. They also demonstrated that PRL-3 mRNA expression was elevated in nearly all metastatic lesions derived from colorectal cancers, regardless of the site metastasis (liver, lung, brain, or ovary).15 Recently, Zeng and colleagues16 found that Chinese hamster ovary cells stably expressing PRL-3 exhibited enhanced motility, invasive activity, and induced metastatic tumor formation in nude mice. These findings may provide a new target for the early diagnosis of metastasis as well as the drug discovery because there are few therapeutic targets for metastatic cancer. However, more direct evidence is required to confirm the role of PRL-3 in promoting tumor metastasis. In this study, therefore, we report that PRL-3 can convert low-metastatic tumor cell line into highly metastatic cells both in vitro and in vivo, supporting the causal role of PRL-3 in tumor metastasis by promoting the migration and cell proliferation.
Materials and Methods
Tumor Samples
Twenty-seven cases of human liver carcinomas were collected from Nanjing Drum Tower Hospital. All patients provided informed consent for use of tissues, and the use of tissues was approved by the institutional review committee of our university, with all procedures following institutional guidelines.
Animals
C57BL/6J mice (6 to 8 weeks old) were obtained from the Shanghai Laboratory Animal Center (Shanghai, China). Throughout the experiments, mice were maintained with free access to pellet food and water in plastic cages at 21 ± 2°C and kept on a 12-hour light-dark cycle. Animal welfare and experimental procedures were performed strictly in accordance with the care and use of laboratory animals (National Research Council, Washington, DC) and the related ethical regulations of our university. All efforts were made to minimize the animals’ suffering and to reduce the number of animals used.
Constructs and Cell Lines
Murine PRL-3 cDNA was cloned from heart total RNA of C57BL/6J mouse by reverse transcriptase-polymerase chain reaction (RT-PCR) with the following primers, 5′-GGTGCTCGAGATGGCCCGCATGAACCGGC-3′ and 5′-CGGGGTACCCTACATGACGCAGCATCTGGTC-3′ with Pfu DNA polymerase (Fermentas, Vilnius, Lithuania). The cloned cDNA sequence indicates three different sites in the nucleotide sequence between our results and those of GenBank. These three differences lead to a nonsense amino acid transition and a (Ala101) → (Leu101) transition. The mouse PRL-3 cDNA was cloned into pIRES2-EGFP (Clontech, Palo Alto, CA) at the XhoI and KpnI sites. To express PRL-3 in Escherichia coli, cDNA was C-terminally (His)6-tagged by cloning into pET-29a (Novagen) at NdeI and XhoI sites. Mouse B16 melanoma cell line and macrophage J774A.1 cell line were obtained from the Chinese Academy of Science. These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies Inc., Grand Island, NY) supplemented with 10% fetal bovine serum (Life Technologies Inc.), 100 U/ml penicillin, and 100 μg/ml streptomycin, and incubated at 37°C in a humidified atmosphere containing 5% CO2 in air.
Purification of PRL-3 from E. coli Cells and DiFMUP PTPase Activity Assay
Prokaryotic expression and purification of recombinant PRL-3 were performed as described.17 Briefly, the E. coli cells containing a C-terminal (His) 6-tag PRL-3 fusion construct were solubilized in protein extraction reagent, sonicated, and centrifuged. The collected supernatant was loaded onto a Ni-chelation affinity column. Then the PRL-3 was eluted with the elution buffer containing imidazole. For the PTPase activity assay, 4 μg of purified PRL-3 were added to a reaction mixture containing 20 mmol/L Tris-HCl, pH 8.0, 10 mmol/L dithiothreitol, 0.01% Triton X-100, and 4 μmol/L DiFMUP (6,8-difluoro-4-methylumbelliferyl phosphate; Molecular Probes, Eugene, OR). Total reaction volume was 200 μl. The fluorescence values were detected using a fluorimeter (Analyzer Fluorat-02 mol/L, Lumex) every 10 minutes, at an excitation/emission wavelength of 300 to 400 nm/475 nm.
Stable Expression of PRL-3 in B16 Melanoma Cells
B16 melanoma cells were cultured to ∼60% confluence in a 35-mm plate and transfected with pIRES2-EGFP using the LipofectAMINE (Life Technologies Inc.). After transfection (48 hours), cells were passaged to a 100-mm dish and geneticin (G418 sulfate; Sigma Chemical Co., St. Louis, MO) was added to final concentration of 1.1 mg/ml. Resistant cells were allowed to grow for 10 days. Individual G418-resistant colonies were picked and screened for monocolony in 96-well plates by limited dilution.
Northern Blot
The total RNA of transfected B16 melanoma cells was extracted using Trizol reagent (Life Technologies Inc.). Fractionated RNA was transferred to Zeta-Probe blotting membrane (Bio-Rad, Richmond, CA). Northern blot hybridization was performed as described.9 Both prehybridization and hybridization were performed using sodium phosphate/sodium dodecyl sulfate buffer [0.5 mol/L sodium phosphate, pH 7.2, 7% sodium dodecyl sulfate, 8% formamide, 1 mmol/L ethylenediaminetetraacetic acid (EDTA)] at 55°C. Hybridization was usually performed for 18 to 24 hours. The membrane was washed with 40 mmol/L sodium phosphate (pH 7.2), 5% sodium dodecyl sulfate, and 1 mmol/L EDTA at 50°C. The membrane was stripped by boiling in 0.1× standard saline citrate buffer and 1% sodium dodecyl sulfate for 15 minutes twice followed by a 10-minute cooling process. Stripped membrane was reprobed as above.
Construction of PRL-3-Inactivating Mutations and Transiently Transfection in Cells
The point mutations D72A and C104S were introduced by PCR (MutaBest kit, Takara) using following primers: for PRL-3 (D72A), 5′-CCCTTTGATGCTGGAGCGCCC-3′ (A to C, italicized) and 5′-CCAGTCCACAACAGTGATGCC-3′; for PRL-3 (C104S), 5′-CTTGTGCACTCTGTGGCGGGC-3′ (G to C, italicized) and 5′-TACGCAGCTTCCCGGGTCATT-3′. The cDNA fragments containing point mutation were cloned into pIRES2-EGFP by XhoI/KpnI double digestion. The constructs were confirmed by sequencing. Cells (50 to 70% confluent) were transiently transfected with pIRES2-EGFP-PRL-3 (D72A) or pIRES2-EGFP-PRL-3 (C104S) using the LipofectAMINE, as per the manufacturer’s instructions, and analyzed 30 hours after transfection.
Semiquantitative RT-PCR and Real-Time PCR
RNA samples were treated by DNase and subjected to semiquantitative RT-PCR. First-strand cDNAs were generated by reverse transcription using oligo (dT). The cDNAs were amplified by PCR for 30 cycles (94°C for 30 seconds, 60°C for 1 minute, and 72°C for 1 minute 30 seconds) using TaqDNA polymerase (Promega Corp., Shanghai, China). The sequences of specific primers were as follows: PRL-3 sense, 5′-CTTCCTCATCACCCACAACC-3′, anti-sense, 5′-TACATGACGCAGCATCTGG-3′; β-actin sense, 5′-GGTACCACCATGTACCAGG-3′, anti-sense, 5′-ACATCTGCTGGAAGGTGGAC-3′. The resultant PCR products were 468 bp (PRL-3) and 161 bp (β-actin). PCR products were electrophoresed on a 2% agarose gel and visualized by ethidium bromide staining. The Gel Imaging and Documentation DigiDoc-It System (version 1.1.23; UVP, Inc., Unpland, CA) was used to scan the gels and the intensity of the bands was assessed using Labworks Imaging and Analysis Software (UVP, Inc.). Quantitative PCR was performed with the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) using SYBR Green I dye (Biotium, Inc. USA), and threshold cycle numbers were obtained using ABI Prism 7000 SDS software version 1.0. The primer sequences used in this study were as follows: PRL-3 forward, 5′-TCAACAGCAAGCAGCTCACC-3′; PRL-3 reverse, 5′-CTTGTGCGTGTGTGGGTCTT-3′; GAPDH forward, 5′-CATGGCCTTCCGTGTTCCTA-3′; GAPDH reverse, 5′-GCGGCACGTCAGATCCA-3′. Conditions for amplification were one cycle of 94°C for 5 minutes followed by 35 cycles of 94°C for 30 seconds, 60°C for 1 minute, and 72°C for 30 seconds.
Anti-Sense Oligodeoxynucleotide (ODN) Treatment of Cells
Phosphorothioate-modified ODNs with the following sequences were commercially synthesized (Sangon, Shanghai, China): PRL-3 anti-sense ODN (5′-GGTAGCTCACCTCCACAG-3′), the scrambled ODN (5′-GGTTGCACAGCTGCAGAG-3′), and a randomized, equal-length ODN (5′-AAACATGGCCTCTGGTGT-3′). The sequence of the ODN (anti-sense) is complementary to PRL-3 bp 43 to 50 from transcription start site. This sequence is not related to PRL-1 and PRL-2 mRNA. At the day before transfection, cells were seeded in 60-mm dishes in growth medium without antibiotics. In the day of transfection, subconfluent cells were washed with fresh DMEM and transfected with 500 nmol/L ODN using Oligofectamine reagent (Life Technologies Inc.) as instructed by the manufacturer. After 4 hours, the growth medium containing three times the normal concentration of serum was added. Thirty hours after initial transfection, the cells were harvested and subjected to migration and adhesion analysis.
Cell Migration Assay and Invasion Assay
Cell migration assay was performed using 8.0-μm pore size Transwell inserts (Costar Corp., Cambridge, MA) as described previously with some modifications.18 In brief, the under surface of the membrane was coated with fibronectin (10 μg/ml) (Sigma Chemical Co.) in phosphate-buffered saline (PBS), pH 7.4, for 2 hours at 37°C. The membrane was washed in PBS to remove excess ligand, and the lower chamber was filled with 0.6 ml of DMEM with 10% fetal bovine serum (FBS). Cells were serum-starved overnight (0.5% FBS), harvested with trypsin/EDTA, and washed twice with serum-free DMEM. Cells were resuspended in migration medium (DMEM with 0.5% FBS) and 1 × 105 cells in 0.1 ml were added to the upper chamber. Sodium orthovanadate (50 μmol/L) (Sigma Chemical Co.) or potassium bisperoxo (bipyridine) oxovanadate V [bpV (bipy)] (30 μmol/L) (Calbiochem, La Jolla, CA) tested in the migration assay were added to both upper and lower chambers. After 12 hours at 37°C, the cells on the upper surface of the membrane were removed using cotton tips. The migrant cells attached to the lower surface were fixed in 10% formalin at room temperature for 30 minutes, and stained for 20 minutes with a solution containing 1% crystal violet and 2% ethanol in 100 mmol/L borate buffer (pH 9.0). The number of migrated cells on the lower surface of the membrane was counted under a microscope in five fields at ×100. For cell invasion assay, the cells (1 × 105 in 0.1 ml of DMEM with 0.5% FBS) were seeded onto Matrigel-coated 8-μm filter invasion chambers (BD Biosciences) according to the manufacturer’s protocol. DMEM supplemented with 10% FBS (0.6 ml) was added to the lower chamber as a chemoattractant. After 24 hours at 5% CO2 and 37°C, the Matrigel was removed with a cotton swab, and the filters were fixed in 10% formalin and stained with crystal violet. Cells that had traversed the Matrigel and passed through the pores of the filter were counted under a microscope in five fields at ×100.
Cell-Spreading Assay
For cell-spreading experiments, 5 × 105 cells were plated into 35-mm-diameter tissue culture dishes precoated with fibronectin (10 μg/ml), polylysine (25 μg/ml) (Sigma Chemical Co.), or concanavalin A (25 μg/ml) (Sigma Chemical Co.). Random fields were photographed after 10, 30, 60, and 120 minutes of incubation in serum-free DMEM using a phase-contrast microscope. Photographs were evaluated for the percentage of spread cells. Unspread cells were described as phase bright, whereas spread cells were flat and phase dark. Five fields were counted for each cell line and, in each field, more than 100 cells were counted. The experiment was repeated three times.
Cell Adhesion Assay
The cell adhesion assay was performed essentially as described with some modifications.19 In brief, 96-well, flat-bottom culture plates were coated with 50 μl of fibronectin (10 μg/ml), laminin (10 μg/ml) (Calbiochem), or collagen type I (50 μg/ml) (Sigma Chemical Co.) in phosphate-buffered saline overnight at 4°C. Plates were then blocked with 0.2% bovine serum albumin for 2 hours at room temperature followed by washing three times with DMEM. The cells were harvested with trypsin/EDTA, washed twice, and resuspended in DMEM medium. Cells (2.5 × 104/well) were added to each well in triplicate and incubated for 30 minutes at 37°C. Plates were then washed three times with DMEM medium to remove unbounded cells. Cells remaining attached to the plates were quantified with an 3-(4 5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay.20 MTT (Sigma Chemical Co.) solution (40 μl per well, 2 mg/ml in PBS) was added and the cells were incubated for 4 hours at 37°C. After discarding the supernatant, 200 μl of dimethyl sulfoxide were added and the absorbance of the color substrate was measured with an enzyme-linked immunosorbent assay reader (Sunrise Remote/Touch Screen; TECAN, Austria) at 540 nm. After subtraction of background cell binding to bovine serum albumin-coated wells, the percentage of adherent cells was calculated by dividing the optical density of the adherent cells divided by that of the initial cell input.
Cell Proliferation Assay
Cells (1 × 104/well) were prepared in 96-well plates in serum-containing medium (day 1). On day 2, 500 nmol/L of phosphorothioate ODNs were introduced into the medium using Oligofectamine reagent. The cells were cultured for 24, 48, and 72 hours, respectively. Cell proliferation was measured by cellular uptake of MTT as described above. Because the cell viability, determined by the trypan blue exclusion test, was more than 95% throughout the experiments, the concentration of ODNs was not toxic.
Tumor Growth Assay
Tumor growth assay was performed essentially as described previously.21 Briefly, subconfluent cultures of the melanoma cells were harvested and washed three times with PBS. Melanoma cells (5 × 105 in 0.1 ml of PBS) were injected subcutaneously into the dorsal flanks of 6- to 8-week-old syngeneic C57BL/6J mice. Tumors were allowed to propagate for the indicated days. Tumor growth was monitored by measuring the dimensions (length and width) of the growing tumors with calipers every 3 days. Tumor size (cm3) was calculated by this formula: 4π/3× (length/2) × (width/2)2.
Tail Vein Metastasis Assay
To produce experimental metastasis, the C57BL/6J mice were injected intravenously with 106 cells in 0.2 ml of PBS via tail vein. After 22 days, the mice were euthanized, and their lungs and livers were resected and photos were taken (Nikon Coolpix 4500) before fixation in Bouin’s solution for further analysis. The numbers of metastatic nodules on the surface of the organs were counted macroscopically.
Melanin Assay
The content of melanin was assayed according to the procedure described by Johnston and colleagues.22 Cells were plated in tissue culture dishes containing 10 ml of DMEM medium. Twenty-four hours after plating, cells were harvested by trypsin, washed twice with PBS, and the number of cells was counted by the trypan blue method. Then the cell pellets were dissolved in 1 ml of 10% dimethyl sulfoxide in 1.0 N sodium hydroxide by incubation at room temperature for 30 minutes. The absorbance values were measured at 470 nm and compared to a standard curve prepared from synthetic melanin (Sigma Chemical Co.) dissolved in the same solution. The melanin content was displayed as quality per 106 cells. For tissues, the 100-mg sample was removed, washed twice with PBS, and dissolved in solution as for cells. The tissue melanin content was displayed as quality per 100-mg sample.
Histology
Tissues were fixed in Bouin’s solution for 24 to 48 hours. After washing in fresh PBS, fixed tissues were dehydrated, cleared, and embedded in paraffin (Paraplast regular, Sigma). Sections (5 μm) were collected on microscope slides, deparaffinized, and stained with hematoxylin and eosin (H&E) as routine procedures.
Immunofluorescence Microscopy
EGFP-PRL-3 fusion protein expression constructs were transfected into COS-7 cells seeded on glass chamber slides (Nalge Nunc, New Zealand) using the LipofectAMINE. Forty-eight hours after transfection, the cells were washed with PBS and fixed with 4% paraformaldehyde at room temperature for 30 minutes followed by washing with PBS twice. The expression of fusion proteins was detected by fluorescence microscope. Nuclei were counterstained with 2 μg/ml 4′,6-diamidino-2-phenylindole (Sigma).
Statistical Analysis
Data were expressed as mean ± SEM. Student’s t-test was used to evaluate the difference between two groups. P < 0.05 was considered to be significant.
Results
Overexpression of PRL-3 in Human Liver Carcinomas and Mouse Melanoma Cell Lines
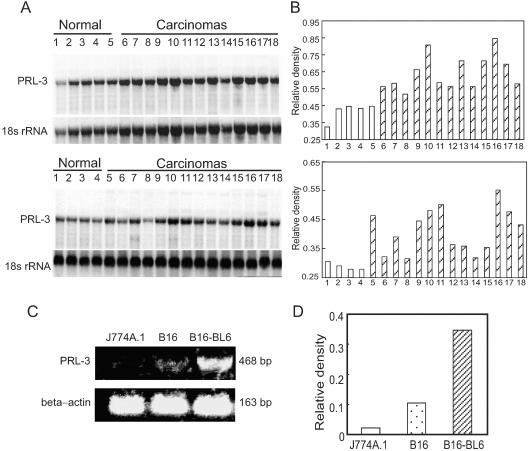
We first analyzed the expression level of PRL-3 in the human liver carcinomas. Northern hybridization analysis showed that PRL-3 was overexpressed in almost all of 27 carcinoma samples examined as compared with the normal liver samples (Figure 1, A and B). Next, the murine lowly metastatic B16 melanoma cells (B16 cells) and its highly metastatic variant subline B16-BL6 cells were analyzed for the expression of PRL-3 by RT-PCR. As shown in Figure 1, C and D, B16 cells only expressed a low level of PRL-3 mRNA. Compared with B16 cells, B16-BL6 cells showed a significantly greater PRL-3 expression. As a negative control, PRL-3 transcript was not detected in the mouse macrophage J774A.1 cell line. We also have examined the mRNA expression pattern in mouse tissues including normal epidermis by using Northern blotting. We detected the strong signal of PRL-3 transcript in the heart (Figure 2A) and skeletal muscles but not in the epidermis (data not shown).
Figure 1.
Expression of PRL-3 in liver carcinomas and tumor cell lines. A: Northern blot confirmed that the expression of the PRL-3 gene was enhanced in human liver carcinoma samples. Total RNA (10 μg/lane) extracted from the samples was used for analysis. The blot was sequentially probed to detect 18s rRNA. Top: Lanes 1 to 5, normal liver tissues; lanes 6 to 18, liver carcinoma samples. Bottom: Lanes 1 to 4, normal liver tissues; lanes 5 to 18, liver carcinoma samples. B: Histogram showing the PRL-3 transcript normalized to 18s rRNA. The values were presented as the percentage of 18s rRNA. C: Representative RT-PCR analysis for PRL-3 on mouse melanoma cell lines. The highly metastatic cell line B16-BL6 showed a higher level of mRNA encoding PRL-3 compared to the lowly metastatic cell line B16, as detected by RT-PCR. J774A.1 cells were PRL-3-negative. β-Actin was used as an internal control. D: The expression of PRL-3 mRNA comparing β-actin mRNA was quantified by densitometry. The values of transcript level for PRL-3 were normalized to β-actin transcript levels and were presented as the percentage of β-actin transcript.
Figure 2.
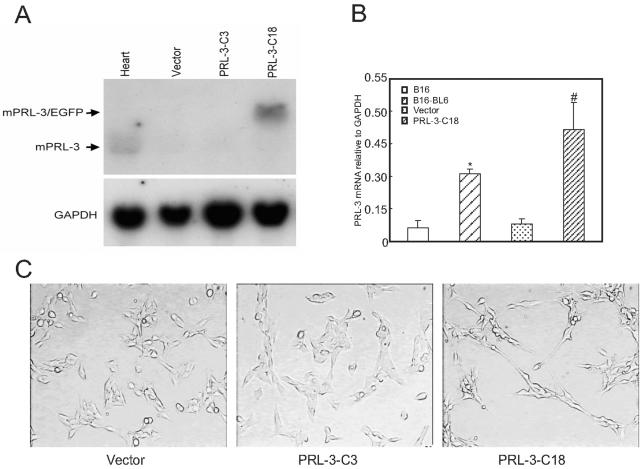
The transfection of PRL-3 into B16 cells and changes in the morphology after stable expression. A: Expression of PRL-3 mRNA by the transfectants was analyzed by Northern blot. Thirty μg of total RNA were subjected to electrophoresis in gels containing formaldehyde, blotted onto nitrocellulose filters, and hybridized with radiolabeled cDNAs corresponding to mouse PRL-3 (top) and mouse glyceraldehydes-3-phosphate dehydrogenase (GAPDH) (bottom). The different lanes were hybridized with RNA from mouse heart (as a positive control), mock vector-transfected cells (vector), or PRL-3-transfected cells (PRL-3-C3 and C18). Because pIRES2-EGFP-PRL-3 has transcript containing both EGFP and PRL-3, the detected band has a little shift in contrast to the endogenous PRL-3 as demonstrated in normal mouse heart (left lane). B: PRL-3 mRNA expression in melanoma cell lines and the transfectants. The expression of PRL-3 in B16 cells, B16-BL6 cells, mock vector-transfected cells, or PRL-3-transfected cells was evaluated using real-time PCR and compared with that of GAPDH. Data are the mean ± SEM of three independent experiments. *, P < 0.05 versus B16; #, P < 0.05 versus mock vector. C: The morphological difference between mock vector- and PRL-3/vector (PRL-3 C18)-transfected cells. The cells were plated in the dishes for 24 hours and then photographed.
Expression of PRL-3 Alters the Morphology of B16 Cells
To address the function of PRL-3 in tumor metastasis, we established stably transfected B16 melanoma cells that overexpress PRL-3. B16 cells were transfected with pIRES2-EGFP mock vector or pIRES2-EGFP vector expressing native PRL-3. As revealed by Northern blotting analysis, a higher level of PRL-3 expression was seen in the vector expressing PRL-3-transfected clone 18 but not in the vector-only transfected clones (Figure 2A). The heart is the organ that expresses the highest endogenous PRL-3. Therefore we used the RNA from mouse heart as a positive control for our Northern blotting assay. To compare the magnitudes of PRL-3 expression in the melanoma cell lines and the transfectant clones, we analyzed PRL-3 mRNA levels in these cells using real-time quantitative RT-PCR and SYBR green fluorogenic detection. As shown in Figure 2B, PRL-3 showed 3.5-fold and 5.0-fold greater expressions in B16-BL6 cells versus B16 cells, and in PRL-3-transfected clone 18 versus vector-only transfected clones, respectively. Figure 2C show the morphological property of the cells in culture. Compared with the mock vector transfectants or the PRL-3 negatively transfected clone 3, the transfection with PRL-3 (PRL-3-C18) made B16 cells display more elongated cell shape. They lose epithelial cell morphology and assume a fibroblast-like appearance, with a bipolar and spindle shape, form extensive filopodia, and do not flatten onto the plates. This fibroblast-like morphology of PRL-3-transfected cells was not found to be changed because of the increase in cell density under the cell culture condition (data not shown). It should also be indicated that B16-BL6 cells display less elongated morphology than PRL-3-C18 cells do.
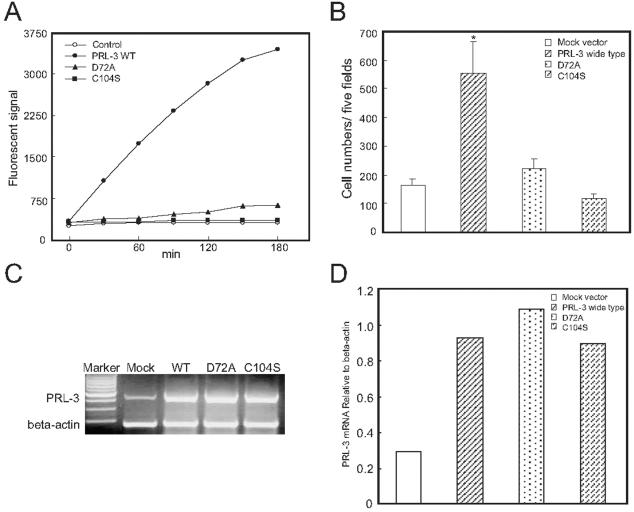
Cloned PRL-3 Is Active in a DiFMUP PTPase Assay and Is Inhibited by Orthovanadate
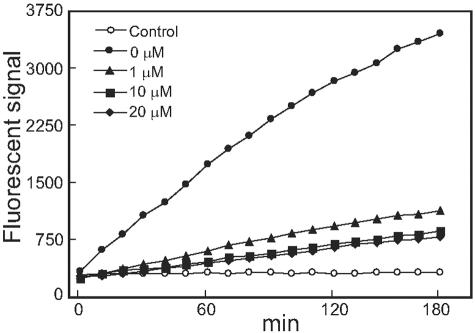
To measure PRL-3 PTPase activity in vitro, a rapid and sensitive activity assay using DiFMUP as substrate was established. Figure 3 demonstrates the time course of PRL-3 activity. Recombinant PRL-3 that we have cloned from E. coli cells showed a gradually increased phosphatase activity within 3 hours. Such phosphatase activity was blocked by the PTPase inhibitor sodium orthovanadate in a concentration-dependent manner.
Figure 3.
Time course of PRL-3 in DiFMUP PTPase activity assay. Four or zero (control) μg of recombinant PRL-3 and 4 μmol/L DiFMUP were incubated for the indicated time and the fluorescence values were detected using a fluorimeter. The inhibitory effect of sodium orthovanadate on PRL-3 was performed by addition of sodium orthovanadate (1 to 20 μmol/L) to the DiFMUP activity as described in Materials and Methods. 0 μmol/L, no sodium orthovanadate was added.
Expression of PRL-3 Enhances in Vitro Migratory and Invasive Ability of B16 Cells
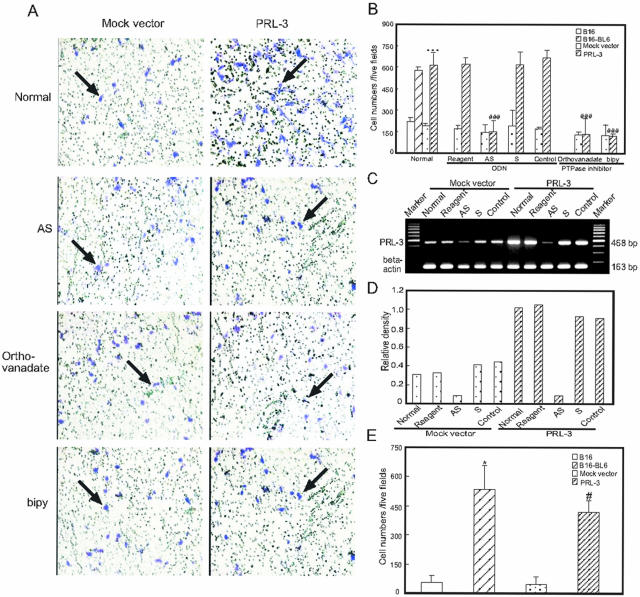
To determine whether PRL-3 can affect an important biological property of metastatic melanoma cells, we used an in vitro migration assay. After 12 hours of culture, cells that migrated onto the other side of the pored filters were checked under a microscope and the cell number for each filter was counted. As shown in Figure 4, A and B, the pooled B16 cells transfected with PRL-3-expressing vector exhibited significant increase of cell number as compared with pooled mock vector transfectants (P < 0.001). Against the migration, the tyrosine phosphatase inhibitor sodium orthovanadate (50 μmol/L) or bpV (30 μmol/L) almost completely blocked the increase. To further test whether the enhanced migratory capability of the cells was mediated by PRL-3, the cells were transfected with an anti-sense phosphorothioate ODN to PRL-3 (500 nmol/L). Evidence of a successful decrease in PRL-3 mRNA level was seen 30 hours after ODN transfection on RT-PCR, compared with both untransfected and scrambled or randomized control ODN-transfected cells (Figure 4, C and D). Treatment with PRL-3 anti-sense ODN resulted in a substantial reduction of the migrated cell number whereas either the scrambled or randomized sequence control ODN had no such effects (Figure 4, A and B). Next, the invasive ability of cells was evaluated in vitro using invasion chambers. PRL-3-transfected cells exhibited a remarkable increase in invasiveness when compared to the respective mock vector transfectants (Figure 4E). A 9.1-fold increase in the number of PRL-3-transfected cells with an invasive ability was observed. Similar results were obtained with B16-BL6 cells compared to B16 cells (9.7-fold increase).
Figure 4.
Effect of PRL-3 expression on cell migration and invasion. B16 cells were transfected with mock vector or PRL-3-expressing vector. A single-cell suspension (100 μl, 1 × 106 cells/ml) of cells were plated into the upper wells of Transwell inserts containing 8-μm pore polycarbonate membranes precoated with fibronectin on the under surface. Cells were allowed to migrate for 12 hours at 37°C, and then they were fixed and stained. A: Representative membranes stained with crystal violet. The left and right panels show untreated (top), anti-sense ODN (top middle), sodium orthovanadate (bottom middle), or bpV treated (bottom) mock vector-transfected cells and PRL-3-expressing cells, respectively. The arrows indicated the migrant cells. B: Quantitative analysis of the number of the cells migrated to the lower side of the membrane. Results represent the average of triplicate samples from three independent experiments. Normal, untreated transfectants; Reagent, treated with oligofectamine only; AS, PRL-3 anti-sense ODN; S, scrambled ODN; control, randomized ODN. Data are mean ± SEM of three independent experiments and each experiment includes triplicate sets. ***, P < 0.001, versus mock vector; ###, P < 0.001, versus reagent. C: Representative suppression of PRL-3 mRNA with anti-sense ODN treatment of transfectants was analyzed by RT-PCR. Cultures were treated with ODNs at 500 nmol/L. β-actin was used as an internal control. D: The level of PRL-3 mRNA comparing β-actin mRNA was quantified by densitometry. E: Histogram of the in vitro invasion assays performed with B16 cells, B16-BL6 cells, PRL-3 transfectants, and mock vector-transfected cells. Results are expressed as number of cells in five random fields per membrane. Data are mean ± SEM of three independent experiments and each experiment includes triplicate sets. *, P < 0.05 versus B16; #, P < 0.05 versus mock vector.
Inactive PRL-3 (D72A or C104S) Reduced the in Vitro Migratory Ability of B16 Cells
To explore whether PRL-3 enhances cell motility and invasion through its catalytic activity as a phosphatase, we created two mutant forms of pIRES2-EGFP-PRL-3, pIRES2-EGFP-PRL-3 (D72A) and pIRES2-EGFP-PRL-3 (C104S), by, respectively, mutating the Asp72 of the conserved aspartic acid residue to Ala and the essential Cys104 of the phosphatase active site signature motif to Ser as described before, because the relevant region (between 64 to 112 residues) of the PRL-1 and PRL-3 are very similar.23 We reasoned that the two mutations would have similar effects on PRL-3’s enzyme activity. Indeed, as shown in Figure 5A, the mutation of D72A resulted in a significant loss of phosphatase activity and C104S mutation abolished the PTPase activity completely. In the in vitro migration assay, a reduced migratory ability was noted in both pIRES2-EGFP-PRL-3 (D72A) and pIRES2-EGFP-PRL-3 (C104S) transiently transfected B16 cells as compared with pIRES2-EGFP-PRL-3 (wide type) transiently transfected cells (Figure 5B). Figure 5, C and D, shows the evidence of successful increase in PRL-3 (wide type, D72A mutation, or C104S mutation) mRNA level 30 hours after transiently transfection in B16 cells on RT-PCR, compared with mock vector-transfected cells.
Figure 5.
Effect of PRL-3 mutations on the phosphatase activity and cell migration. A: Catalytic activity of PRL-3 (wild type, D72A mutation, or C104S mutation) proteins in DiFMUP PTPase activity assay. All reactions were performed in triplicate. The data are representative of three independent experiments. B: Quantitative analysis of the cells migrated to the lower side of the membrane. A single-cell suspension (100 μl, 1 × 106 cells/ml) of cells 30 hours after transiently transfection were added into the upper wells of Transwell inserts containing 8-μm-pore polycarbonate membranes precoated with fibronectin on the under surface. Cells were allowed to migrate for 12 hours at 37°C, and then they were fixed and stained. Results represent the average of triplicate samples from three independent experiments. C: Representative expression of PRL-3 mRNA 30 hours after transiently transfection in B16 cells was analyzed by RT-PCR. D: The level of PRL-3 mRNA comparing β-actin mRNA was quantified by densitometry.
Expression of PRL-3 Alters the Adhesive Ability of B16 Cells to Various Matrix Proteins
Because cell adhesion is a prerequisite to cell migration, we next investigated whether PRL-3 could affect cell adhesion to various extracellular matrix substrates such as fibronectin, laminin, and collagen type I. As shown in Figure 6, A and B, after incubation for 30 minutes, PRL-3 transfectants showed a significantly increased adhesion to fibronectin and laminin compared with the mock vector transfectants, respectively, whereas the adhesion behaviors of the cells to type I collagen showed an opposite result (Figure 6C). The anti-sense ODN to PRL-3 specifically recovered the adhesive ability to normal levels from the increase (Figure 6, A and B) and decrease (Figure 6C) caused by PRL-3 overexpression. Adhesion of the PRL-3-transfected cells treated with anti-sense ODN decreased by 40% for fibronectin and 52% for laminin, and increased by 31% for collagen type I compared with that seen with untreated and scrambled or randomized control ODN-treated cells. On the contrary, the anti-sense ODN had little effect on adhesion of mock vector transfectants.
Figure 6.
Effect of PRL-3 expression on B16 melanoma cell adhesion to various matrix proteins. Mock vector-transfected and vector/PRL-3-transfected cells were added to the wells of 96-well plates coated with fibronectin (10 μg/ml) (A), laminin (10 μg/ml) (B), or collagen I (50 μg/ml) (C). After incubation at 37°C for 30 minutes, nonadherent cells were gently washed away, and the amount of the cell adhesion was determined using an enzyme-linked immunosorbent assay reader after addition of MTT as indicated in Materials and Methods. For anti-sense ODN treatment, the cells were transfected with 500 nmol/L of ODNs before adhesion assay. Normal, untreated cells; reagent, treated with oligofectamine only; AS, PRL-3 anti-sense ODN; S, scrambled ODN; control, randomized ODN. Data are mean ± SEM of three independent experiments and each experiment includes triplicate sets. **, P < 0.01, versus mock vector; #, P < 0.05; ##, P < 0.01, versus reagent.
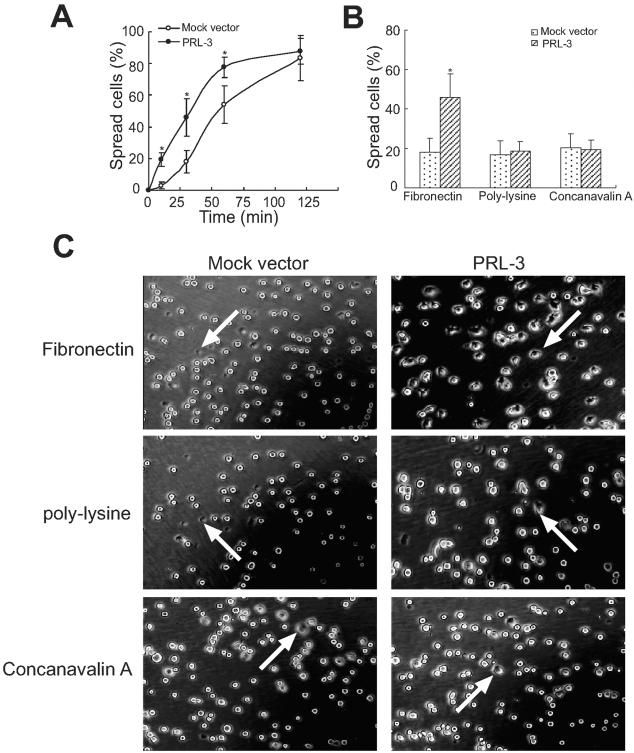
PRL-3 Transfectants Spread Faster on Fibronectin
To further explore the mechanism underlying the effect of PRL-3 on B16 cell migration, we examined cell spreading on fibronectin. After detachment and replating onto fibronectin-coated plates, expression of PRL-3 led to a significant enhancement in cell spreading, as compared with that of mock vector transfectants (Figure 7A). After seeding for 30 minutes on fibronectin, ∼50% of the PRL-3 transfectants were judged as spreading, based on the loss of the rounded phase and bright appearance, whereas only ∼20% of the mock vector-transfected control cells were spread at this time. Further, we compared the effect of PRL-3 overexpression on cell interactions with integrin versus nonintegrin ligands. PRL-3 expression neither improved the spreading efficiency of B16 cells on polylysine nor concanavalin A as nonintegrin substrates (Figure 7B). Figure 7C shows the representative photographs of cell spreading.
Figure 7.
Enhanced cell spreading of PRL-3-transfected B16 cells. Cells were kept in suspension and then replated onto fibronectin-, polylysine-, or concanavalin A-coated cell culture dishes, incubated at 37°C, and then photographed at 10, 30, 60, and 120 minutes. A: Time course of cell spreading on fibronectin. B: Quantitative evaluation of cell spreading efficiency was obtained by calculating the percentage of cells spread on fibronectin, polylysine, or concanavalin A at 30 minutes. Shown are mean percentages of spread cells per field ± SEM of three independent experiments. At least 10 different fields for each sample were averaged. A total of ∼500 cells were counted for each bar. *, P < 0.05, versus mock vector. C: Representative photographs from mock vector- and PRL-3-transfected cells after spread for 30 minutes. The arrows indicated the spreading cells with flat shape and dark phase.
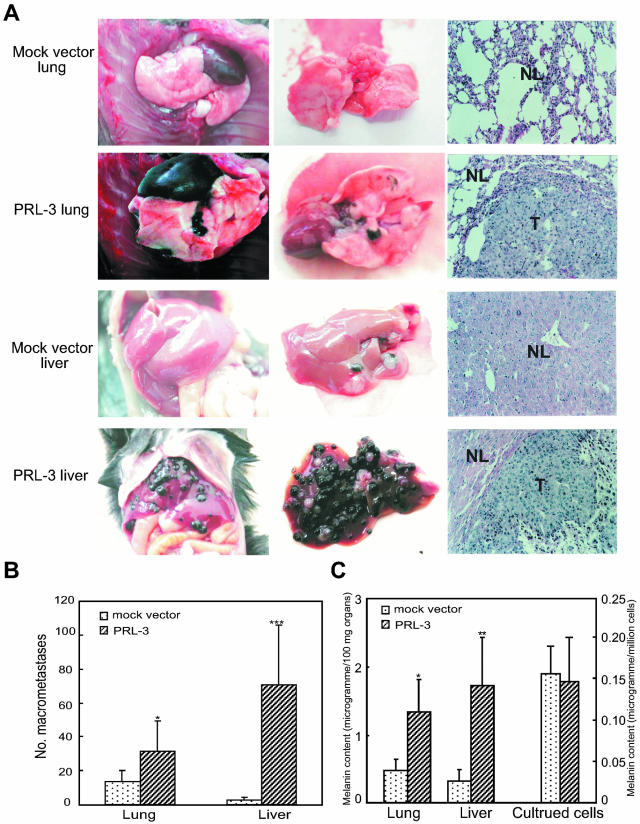
PRL-3 Expression Promotes Metastatic Tumor Formation
Male C57BL/6J mice were injected by the PRL-3-expressing B16 cells or mock vector transfectants into the tail vein. One of the mice carrying PRL-3 transfectants died on day 22 after injection and was found to have had extensive tumor formation in the lung and liver, and then at this time, the organs of all other mice were examined. The left and middle panels in Figure 8A show the gross morphology of the respective lungs and livers. PRL-3 transfectants formed numerous large, well-defined metastatic lung and liver lesions that could also be readily detected by histological analysis. In contrast, the lungs and livers from mice injected with only expression vector-transfected cells showed much fewer or no metastatic lesions. A statistically significant increase in the number of grossly detectable lung and liver metastases was seen in mice injected with PRL-3 transfectants, as compared with that in mice implanted with mock vector transfectants (Figure 8B). In addition, one of six mice carrying PRL-3 transfectants had tumor formation in the kidney and colon, besides in the lung and liver (data not shown). Histological examination of lung and liver tissue sections from the mice carrying PRL-3 transfectants revealed large confluent tumor nodules that invaded most of the lung and liver parenchyma (Figure 8A, right). However, histological examination of the organs from the mice carrying mock vector transfectants revealed either absence of detectable tumor lesions or fewer microscopic tumor nodules. The lungs and livers in mice transplanted with PRL-3 transfectants displayed a high level of melanin production than that in mice with only mock vector transfectants whereas there was no difference in melanin content produced by the two kinds of transfectants in culture (Figure 8C). We also compared the levels of mRNA encoding PRL-3 in the lungs and livers of mice carrying PRL-3 transfectants and mock vector transfectants by Northern blotting. PRL-3 transcript was not detected in the lungs and livers from mice carrying control cells. In contrast, the levels of PRL-3 mRNA were elevated in the lungs and livers of mice carrying PRL-3 transfectants (data not shown).
Figure 8.
PRL-3 expression promotes B16 melanoma cell metastasis in vivo. B16 melanoma cells (1 × 106) transfected with mock expression vector or PRL-3-expressing vector were injected intravenously into C57BL/6J mice via tail vein. After 22 days, lungs and livers from the mice were resected and analyzed for metastasis. A: Representative example of lungs and livers from the mice. The left and middle panels show the stereomicrographs of lungs and livers. The right panels show the histological photomicrographs of lung or liver tissue section stained with H&E (×100). NL, normal lung or liver tissue; T, metastatic tumor lesions. B: Quantitative evaluation of macroscopically detectable lung and liver metastases. After fixation in Bouin’s solution, the number of macroscopically visible metastases on the lung and liver surface was quantified. C: Melanin content was determined as described in Materials and Methods for the lung and liver tissues from the mice and the cells in culture. Data are mean ± SEM of six mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001, versus mock vector.
Expression of PRL-3 Increases Cell Proliferation in Vitro and Tumor Growth in Vivo
Compared with the mock vector transfectants, the PRL-3 transfectants showed a significantly enhanced proliferation as determined by MTT assay (Figure 9A). The cell number of the PRL-3 transfectants was 1.6-fold greater than the mock vector transfectants after 72 hours in culture. This result was confirmed using direct cell counts or BrdU incorporation assay (data not shown). After PRL-3 anti-sense ODN treatment, the proliferation of PRL-3 transfectants was back to the level of PRL-3-nontransfected cells (Figure 9A). In contrast to the effect of the PRL-3 anti-sense ODN, the scrambled or randomized sequence control ODN had no effects on the cell proliferation (data not shown). The time course of the suppression in PRL-3 expression after addition of anti-sense ODN to the PRL-3 transfectants was examined. As shown in Figure 9, B and C, the exposure of the PRL-3 transfectants to 500 nmol/L anti-sense ODN caused a rapid decrease in PRL-3 expression within 24 hours, and suppressive effects continued until at least 72 hours after the initial addition of anti-sense ODN.
Figure 9.
Effect of PRL-3 expression on cell proliferation of B16 cells. A: Mock vector transfectants and PRL-3 transfectants were seeded into 96-well plates (1 × 104 cells/well) in DMEM with 10% FBS. After 24, 48, and 72 hours, the amount of cells in each well was determined by MTT as indicated in Materials and Methods. For anti-sense ODN treatment, cultured cells were treated with oligofectamine reagent alone (reagent), PRL-3 anti-sense ODN (AS), scrambled ODN (not shown), or randomized ODN (not shown) at 500 nmol/L. Data are the mean ± SEM of three independent experiments and each experiment includes triplicate sets. ***, P < 0.001, versus mock vector; ##, P < 0.01, ###, P < 0.001, versus reagent-treated only. B: Time course of PRL-3 mRNA levels after anti-sense ODN treatment. The level of PRL-3 mRNA comparing β-actin mRNA was quantified by densitometry. C: Representative RT-PCR analysis for PRL-3 on the cells after treatment with anti-sense ODN for 24, 48, and 72 hours.
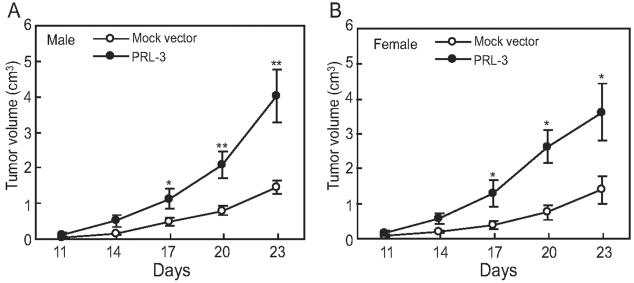
Next, the effect of PRL-3 on tumor growth in syngeneic mice (C57BL/6J) was examined. The melanoma B16 transfectants expressing PRL-3 were injected subcutaneously into the dorsal flanks of male or female C57BL/6J mice and tumor development was compared to that derived from mock vector transfectants. As shown in Figure 9, expression of PRL-3 led to a pronounced increase in B16 cell growth starting from day 17, up to 2.8-fold increase in male mice (Figure 10A) and 2.6-fold increase in female mice (Figure 10B) of tumor volume at day 23 after cell injection.
Figure 10.
Effect of PRL-3 expression on in vivo tumor growth. Mock vector transfectants and PRL-3 transfectants (5 × 105 cells/mouse) were injected subcutaneously into the dorsal flanks of 6- to 8-week-old male (A) or female (B) C57BL/6J mice. The diameters of the formed tumor were measured every 3 days continuously from the 2nd week to 4th week after injection, and the tumor volumes were measured and indicated as mean ± SEM of eight mice in each group. *, Significant difference when compared with control (*, P < 0.05; **, P < 0.01).
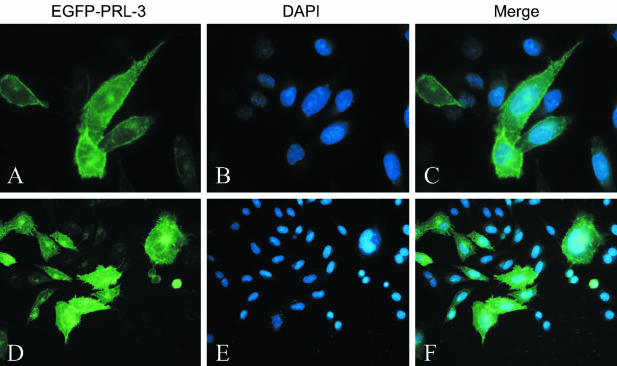
PRL-3 Is Associated with Plasma Membrane and Intracellular Punctate Structures in Transiently Transfected Cells
To address the localization of PRL-3 in cells, we transfected the construct expressing EGFP-tagged PRL-3 in COS-7 cells. As shown in Figure 11, the fusion protein EGFP-PRL-3 was primarily confined to the membrane compartments, including cytoplasmic membrane and nuclear membrane, as well as intracellular punctate structures scattered throughout the entire cytoplasm but concentrated in the perinuclear region, consistent with the previous study.16 More interestingly, we also found that EGFP-PRL-3 was enriched at the metaphase plate in the progression of cells through mitosis (Figure 11, D and E), which implies a possible role for PRL-3 in cell-cycle regulation, although it needs to be carefully confirmed.
Figure 11.
Intracellular localization of PRL-3. The COS-7 cells were transiently transfected with expression vector encoding EGFP-PRL-3 fusion protein. Forty-eight hours after transfection cells were fixed and analyzed by fluorescence microscope. PRL-3 was detected by fusion protein EGFP-PRL-3 (green, A and D). Nuclei were counterstained with 4′, 6-diamidino-2-phenylindole (blue, B and E). Original magnifications, ×100.
Discussion
In this study, we observed the higher level expression of PRL-3 in human liver carcinoma samples as compared with the normal liver samples (Figure 1, A and B). This data implied a possibility of the PRL-3 involvement in tumors other than colorectal cancers as proposed by Saha and colleagues.14 Interestingly, we also found that PRL-3 was highly expressed in metastatic B16-BL6 melanoma cells but not in their lowly metastatic parental B16 cells (Figure 1, C and D). These findings suggested a potential link between PRL-3 expression and tumor metastasis at least in colorectal cancer14 and mouse melanoma cells. For patients with solid tumors, the biggest threat to survival is tumor cell metastasis and metastases associated with resistance to conventional therapies. It has become a major challenge, therefore, to identify the underlying molecular switch of the metastatic state.24,25 Unfortunately, the specific molecular changes that promote the spread of tumor cells from the original site to other parts of the body are still primarily unclear. Does PRL-3 function as one of the key players for the transition from benign to metastatic tumor?
Interestingly, B16 mouse melanoma cells transfected with PRL-3-expressing vector showed a quite different morphology from its parental B16 cells and cells transfected with mock vector. PRL-3 transfectants display a fibroblast-like appearance with a bipolar and spindle shape, form extensive filopodia, and pile up in layer on layer in the culture dish (Figure 2). Such morphological change is quite similar to what is referred as epithelial-mesenchymal transition. During epithelial-mesenchymal transition, epithelial cells acquire fibroblast-like properties and exhibit reduced cell-cell adhesion and increased motility. Because epithelial-mesenchymal transition may endow cancer cells with enhanced motility and invasiveness, it has been suggested to be a crucial process during the invasion and metastasis of carcinoma cells.26 Because the phosphorylation and dephosphorylation of cytoskeletal proteins can regulate the shape of cultured cells,27–29 the changes in cell shape of the PRL-3-overexpressing clone might be related to the cytoskeleton rearrangement induced by enhanced PRL-3 activity. However, whether the effect of PRL-3 transfection on cell cytoskeleton is direct or indirect needs to be investigated in detail, and another possibility for the indirect effect of PRL-3 on cytoskeleton through other signaling molecules could not be excluded. Indeed, the cytoskeleton not only controls cell morphology but also regulates cell growth, migration, differentiation, and gene expression, which are fundamental to embryogenesis, carcinogenesis, and wound healing.30
Because lamellipodia, filopodia, and membrane ruffles are essential for cell motility,31–33 stable expression of PRL-3 might induce enhanced cell motility. Indeed PRL-3-expressing cells display significantly enhanced migratory ability in quantitative transwell-based assays. And PRL-3-specific anti-sense ODN or PTPase inhibitor sodium orthovanadate and bpV could block this effect of PRL-3 almost completely (Figure 4B). This result suggests that the enhanced migratory capacity of the cells was mediated by PRL-3 and further assay using RT-PCR supported the suggestion (Figure 4, C and D). The invasive capability of the cells was enhanced by PRL-3 expression compared with the control by using invasion chambers (Figure 4E). Furthermore, the results that the mutant PRL-3 (D72A or C104S) expression greatly reduced the cell migration suggested that the phosphatase activity of PRL-3 was necessary for the migration of these PRL-3-transfected cells.
Cell migration involves protrusion of actin-rich lamellipodia or filopodia at the leading edge of the cell, formation and stabilization of extracellular matrix attachments at the newly extended cell periphery, detachment of old adhesion sites at the rear of the cell, and contraction of actomyosin-based cytoskeletal filaments in the cell body to move the bulk of the cell forward.34 These events are regulated by multiple signaling cascades, including integrin-mediated signaling, tyrosine kinase and phosphatase signaling, mitogen-activated protein kinase signaling, and cytoskeletal reorganization.35–37 Preliminary results indicated FAK phosphorylation is greatly increased in PRL-3-expressing cells (data not shown).
Cell migration also depends on adhesiveness to the substratum.38–40 By examining the effects of PRL-3 expression on the adhesion specificity of cells to various extracellular matrix proteins, we demonstrated that PRL-3 facilitated the adhesive ability of B16 cells to fibronectin and laminin, while impairing this ability to collagen type I (Figure 6). This adhesive preference was also found between B16-BL6 (highly metastatic) and B16 (poorly metastatic) melanoma cell lines (data not shown), and between highly (HT-29LMM) and poorly (HT-29P) liver-metastatic colon carcinoma cells when they adhered to extracellular matrix proteins in vitro.41 These results suggested that adhesion of the cells is mediated at least in part by different integrins, depending on the ECM components used in the adhesion experiments. Importantly, the changes in its adhesive abilities of the cells were also revised by the treatment of PRL-3-specific anti-sense ODN.
The altered adhesion properties of PRL-3 transfectants may be caused by differences in the integrin expression. One of the earliest phenotypic consequences of integrin signaling is cell spreading. The good attachment of PRL-3 transfectants to fibronectin is also reflected in their ability to spread. Expression of PRL-3 led to a significant elevated rate in cell spreading, compared to that of mock vector transfectants (Figure 7A). Up-modulation of cell spreading by PRL-3 could have resulted from effects on integrin-mediated cell spreading or on cytoskeletal processes required for cell spreading. Our results that PRL-3 did not facilitate the cells to spread on nonintegrin substrates including polylysine and concanavalin A can exclude the latter possibility (Figure 7B).
All above findings suggest that PRL-3 expression plays a key role in the acquisition of metastatic potential of tumor cells. Thus, in the in vivo metastasis study, we confirmed that PRL-3 could facilitate lung metastasis of B16 cells after injection into the lateral tail of the mice. More interestingly, besides enriched metastases in lungs, there were numerous macroscopically visible metastases occurring in livers and a statistically significant increase in the number of grossly detectable liver metastases was seen in mice at 22 days after injection of the PRL-3 transfectants, although the lung was the first organ passed through after intravenous injection (Figure 8). In contrast, no or few tumors were found in lungs and livers of the mice injected with mock vector transfectants. Recently, the work of Zeng and colleagues16 also showed that PRL-3-expressing Chinese hamster ovary cells formed lung and liver metastatic tumors in the same experimental metastasis model in nude mice. This suggests that PRL-3 can act as a key player to initiate and maintain tumor cell growth in the foreign visceral organs by transforming the tumor cell characteristics and subsequently enhancing their metastatic ability. The mechanisms underlying the facilitated metastasis of the tumor cells by PRL-3 need to be investigated. That PRL-3 could accelerate proliferation rate of the tumor cells in vitro and increase the tumor growth in vivo (Figures 9 and 10) may partly explain the strong tumorigenesis of PRL-3, because deregulated cell proliferation and suppressed cell death together provide the underlying platform for neoplastic progression.42
The subcellular localization of a particular protein can sometimes give clues to its physiological function. By using immunofluorescence microcopy, we found that PRL-3 localized to cytoplasmic membrane, which was consistent with the predicted prenylation in its C-terminal CAAX motif (Figure 11). The localization of PRL-3 to the metaphase plate in the progression of cells might be related to the cell mitosis.
Overall, this study gave a direct evidence for the metastasis-enhancing activity of PRL-3, especially in the melanoma cells other than the colorectal tumor cells. The high expression of PRL-3 in human liver carcinomas also revealed a possibility that PRL-3 may be a common factor in the tumorigenesis and metastasis of cancers. Because no protein substrate has yet been identified for PRL-3, and further experiments will be required to determine the biochemical mechanisms through which PRL-3 influences neoplastic growth and to establish its causative role in the metastatic process. Not only could the enzyme provide a good target for chemotherapeutic drugs, but it may also provide a molecular marker to help clinicians assess tumor aggressiveness.43 The data presented here including the changes in cell migration, adhesion, spreading, and growth by PRL-3 may be helpful for the further elucidation of PRL-3’s functions as well as for its usefulness in cancer therapy.
Acknowledgments
We thank Xiaoyan Yang, Yin Lu, Xinxin Gu, Lizhen Chen, Jun Wang, and Jian Huang for technical support and helpful discussion.
Footnotes
Address reprint requests to Qiang Xu, Ph.D., School of Life Sciences, Nanjing University, 22 Han Kou Rd., Nanjing 210093, China. E-mail: molpharm@163.com or to Xiang Gao, Ph.D., Model Animal Genetics Research Center, Nanjing University, 22 Han Kou Rd., Nanjing 210093, China. E-mail: gaoxiang@nju.edu.cn.
Supported by the State Key Project Foundation of “the 10th 5-year plan” of China, the National Natural Science Foundation of China (no.30300425), the Natural Science Foundation of Jiangsu Province (no.BK2003069), and the 973 program of China (no.2002CB513000).
X.W. and H.Z. contributed equally to this work.
References
- Boyd D. Invasion and metastasis. Cancer Metastasis Rev. 1996;15:77–89. doi: 10.1007/BF00049488. [DOI] [PubMed] [Google Scholar]
- Bashyam MD. Understanding cancer metastasis: an urgent need for using differential gene expression analysis. Cancer. 2002;94:1821–1829. doi: 10.1002/cncr.10362. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Streuli M. Protein tyrosine phosphatases in signaling. Curr Opin Cell Biol. 1996;8:182–188. doi: 10.1016/s0955-0674(96)80064-0. [DOI] [PubMed] [Google Scholar]
- Tonks NK, Neel BG. From form to function: signaling by protein tyrosine phosphatases. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- Zhang ZY. Protein tyrosine phosphatases: prospects for therapeutics. Curr Opin Chem Biol. 2001;5:416–423. doi: 10.1016/s1367-5931(00)00223-4. [DOI] [PubMed] [Google Scholar]
- Diamond RH, Cressman DE, Laz TM, Abrams CS, Taub R. PRL-1, a unique nuclear protein tyrosine phosphatase, affects cell growth. Mol Cell Biol. 1994;14:3752–3762. doi: 10.1128/mcb.14.6.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates CA, Michael RL, Stayrook KR, Harvey KA, Burke YD, Randall SK, Crowell PL, Crowell DN. Prenylation of oncogenic human PTP (CAAX) protein tyrosine phosphatases. Cancer Lett. 1996;110:49–55. doi: 10.1016/s0304-3835(96)04459-x. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Hong W, Tan YH. Mouse PRL-2 and PRL-3, two potentially prenylated protein tyrosine phosphatases homologous to PRL-1. Biochem Biophys Res Commun. 1998;244:421–427. doi: 10.1006/bbrc.1998.8291. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Protein prenylation. Mad bet for Rab Nature. 1993;366:14–15. doi: 10.1038/366014a0. [DOI] [PubMed] [Google Scholar]
- Moores SL, Schaber MD, Mosser SD, Rands E, O’Hara MB, Garsky VM, Marshall MS, Pompliano DL, Gibbs JB. Sequence dependence of protein isoprenylation. J Biol Chem. 1991;266:14603–14610. [PubMed] [Google Scholar]
- Mohn KL, Laz TM, Hsu JC, Melby AE, Bravo R, Taub R. The immediate-early growth response in regenerating liver and insulin-stimulated H-35 cells: comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Mol Cell Biol. 1991;11:381–390. doi: 10.1128/mcb.11.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Holmes DI, Powell SM, Lu QL, Waxman J. Analysis of stromal-epithelial interactions in prostate cancer identifies PTPCAAX2 as a potential oncogene. Cancer Lett. 2002;175:63–69. doi: 10.1016/s0304-3835(01)00703-0. [DOI] [PubMed] [Google Scholar]
- Saha S, Bardelli A, Buckhaults P, Velculescu VE, Rago C, St Croix B, Romans KE, Choti MA, Lengauer C, Kinzler KW, Vogelstein B. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- Bardelli A, Saha S, Sager JA, Romans KE, Xin B, Markowitz SD, Lengauer C, Velculescu VE, Kinzler KW, Vogelstein B. PRL-3 expression in metastatic cancers. Clin Cancer Res. 2003;9:5607–5615. [PubMed] [Google Scholar]
- Zeng Q, Dong JM, Guo K, Li J, Tan HX, Koh V, Pallen CJ, Manser E, Hong W. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res. 2003;63:2716–2722. [PubMed] [Google Scholar]
- Matter WF, Estridge T, Zhang C, Belagaje R, Stancato L, Dixon J, Johnson B, Bloem L, Pickard T, Donaghue M, Acton S, Jeyaseelan R, Kadambi V, Vlahos CJ. Role of PRL-3, a human muscle-specific tyrosine phosphatase, in angiotensin-II signaling. Biochem Biophys Res Commun. 2001;283:1061–1068. doi: 10.1006/bbrc.2001.4881. [DOI] [PubMed] [Google Scholar]
- Senger DR, Perruzzi CA, Streit M, Koteliansky VE, de Fougerolles AR, Detmar M. The alpha (1) beta (1) and alpha (2) beta (1) integrins provide critical support for vascular endothelial growth factor signaling, endothelial cell migration, and tumor angiogenesis. Am J Pathol. 2002;160:195–204. doi: 10.1016/s0002-9440(10)64363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai A, Nosaka Y, Kohsaka H, Miyasaka N, Miura O. CrkL activates integrin-mediated hematopoietic cell adhesion through the guanine nucleotide exchange factor C3G. Blood. 1999;93:3713–3722. [PubMed] [Google Scholar]
- Poulaki V, Mitsiades CS, Kotoula V, Tseleni-Balafouta S, Ashkenazi A, Koutras DA, Mitsiades N. Regulation of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in thyroid carcinoma cells. Am J Pathol. 2002;161:643–654. doi: 10.1016/S0002-9440(10)64220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PC, Montgomery AM, Rosenfeld M, Reisfeld RA, Hu T, Klier G, Cheresh DA. Integrin alpha v beta 3 antagonists promote tumor regression by inducing apoptosis of angiogenic blood vessels. Cell. 1994;79:1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Johnston D, Orlow SJ, Levy E, Bystryn JC. Induction of B16 melanoma melanogenesis by a serum-free synthetic medium. Exp Cell Res. 1992;201:91–98. doi: 10.1016/0014-4827(92)90351-8. [DOI] [PubMed] [Google Scholar]
- Wang J, Kirby CE, Herbst R. The tyrosine phosphatase PRL-1 localizes to the endoplasmic reticulum and the mitotic spindle and is required for normal mitosis. J Biol Chem. 2002;277:46659–46668. doi: 10.1074/jbc.M206407200. [DOI] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Ridley A. Molecular switches in metastasis. Nature. 2000;406:466–467. doi: 10.1038/35020170. [DOI] [PubMed] [Google Scholar]
- Boyer B, Valles AM, Edme N. Induction and regulation of epithelial-mesenchymal transitions. Biochem Pharmacol. 2000;60:1091–1099. doi: 10.1016/s0006-2952(00)00427-5. [DOI] [PubMed] [Google Scholar]
- Oliver CJ, Terry-Lorenzo RT, Elliott E, Bloomer WA, Li S, Brautigan DL, Colbran RJ, Shenolikar S. Targeting protein phosphatase 1 (PP1) to the actin cytoskeleton: the neurabin I/PP1 complex regulates cell morphology. Mol Cell Biol. 2002;22:4690–4701. doi: 10.1128/MCB.22.13.4690-4701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini A, Koch A, Wilms R, Tamura T. The SH2-containing inositol 5-phosphatase (SHIP)-1 is implicated in the control of cell-cell junction and induces dissociation and dispersion of MDCK cells. Oncogene. 2002;21:1477–1484. doi: 10.1038/sj.onc.1205224. [DOI] [PubMed] [Google Scholar]
- Jackson J, Meisinger J, Patel S, Lim ZC, Vellody K, Metz R, Young MR. Protein phosphatase-2A associates with the cytoskeleton to maintain cell spreading and reduced motility of nonmetastatic Lewis lung carcinoma cells: the loss of this regulatory control in metastatic cells. Invasion Metastasis. 1997;17:199–209. [PubMed] [Google Scholar]
- Varedi M, Ghahary A, Scott PG, Tredget EE. Cytoskeleton regulates expression of genes for transforming growth factor-beta 1 and extracellular matrix proteins in dermal fibroblasts. J Cell Physiol. 1997;172:192–199. doi: 10.1002/(SICI)1097-4652(199708)172:2<192::AID-JCP6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol. 2002;12:112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- Hsia DA, Mitra SK, Hauck CR, Streblow DN, Nelson JA, Ilic D, Huang S, Li E, Nemerow GR, Leng J, Spencer KS, Cheresh DA, Schlaepfer DD. Differential regulation of cell motility and invasion by FAK. J Cell Biol. 2003;160:753–767. doi: 10.1083/jcb.200212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker KK, Brock AL, Brangwynne C, Mannix RJ, Wang N, Ostuni E, Geisse NA, Adams JC, Whitesides GM, Ingber DE. Directional control of lamellipodia extension by constraining cell shape and orienting cell tractional forces. EMBO J. 2002;16:1195–1204. doi: 10.1096/fj.02-0038com. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Felsenfeld DP, Galbraith CG. Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol. 1998;8:51–54. doi: 10.1016/s0962-8924(98)80005-6. [DOI] [PubMed] [Google Scholar]
- Mareel M, Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol Rev. 2003;83:337–376. doi: 10.1152/physrev.00024.2002. [DOI] [PubMed] [Google Scholar]
- Crean JK, Finlay D, Murphy M, Moss C, Godson C, Martin F, Brady HR. The role of p42/44 MAPK and protein kinase B in connective tissue growth factor induced extracellular matrix protein production, cell migration, and actin cytoskeletal rearrangement in human mesangial cells. J Biol Chem. 2002;277:44187–44194. doi: 10.1074/jbc.M203715200. [DOI] [PubMed] [Google Scholar]
- Welch MD. The world according to Arp: regulation of actin nucleation by the Arp2/3 complex. Trends Cell Biol. 1999;9:423–427. doi: 10.1016/s0962-8924(99)01651-7. [DOI] [PubMed] [Google Scholar]
- Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- Otsuka A, Hirose K, Kilimann MW, Kamata T. Amphiphysin1 inhibits vitronectin-mediated cell adhesion, spreading, and migration in vitro. Biochem Biophys Res Commun. 2003;301:769–775. doi: 10.1016/s0006-291x(03)00040-8. [DOI] [PubMed] [Google Scholar]
- Alblas J, Ulfman L, Hordijk P, Koenderman L. Activation of Rhoa and ROCK are essential for detachment of migrating leukocytes. Mol Biol Cell. 2001;12:2137–2145. doi: 10.1091/mbc.12.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haier J, Nasralla M, Nicolson GL. Different adhesion properties of highly and poorly metastatic HT-29 colon carcinoma cells with extracellular matrix components: role of integrin expression and cytoskeletal components. Br J Cancer. 1999;80:1867–1874. doi: 10.1038/sj.bjc.6690614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- Marx J. New insights into metastasis. Science. 2001;294:281–282. doi: 10.1126/science.294.5541.281a. [DOI] [PubMed] [Google Scholar]