Abstract
In this study, the enzymes involved in polycyclic aromatic hydrocarbon (PAH) degradation were investigated in the pyrene-degrading Mycobacterium sp. strain 6PY1. [14C]pyrene mineralization experiments showed that bacteria grown with either pyrene or phenanthrene produced high levels of pyrene-catabolic activity but that acetate-grown cells had no activity. As a means of identifying specific catabolic enzymes, protein extracts from bacteria grown on pyrene or on other carbon sources were analyzed by two-dimensional gel electrophoresis. Pyrene-induced proteins were tentatively identified by peptide sequence analysis. Half of them resembled enzymes known to be involved in phenanthrene degradation, with closest similarity to the corresponding enzymes from Nocardioides sp. strain KP7. The genes encoding the terminal components of two distinct ring-hydroxylating dioxygenases were cloned. Sequence analysis revealed that the two enzymes, designated Pdo1 and Pdo2, belong to a subfamily of dioxygenases found exclusively in gram-positive bacteria. When overproduced in Escherichia coli, Pdo1 and Pdo2 showed distinctive selectivities towards PAH substrates, with the former enzyme catalyzing the dihydroxylation of both pyrene and phenanthrene and the latter preferentially oxidizing phenanthrene. The catalytic activity of the Pdo2 enzyme was dramatically enhanced when electron carrier proteins of the phenanthrene dioxygenase from strain KP7 were coexpressed in recombinant cells. The Pdo2 enzyme was purified as a brown protein consisting of two types of subunits with Mrs of about 52,000 and 20,000. Immunoblot analysis of cell extracts from strain 6PY1 revealed that Pdo1 was present in cells grown on benzoate, phenanthrene, or pyrene and absent in acetate-grown cells. In contrast, Pdo2 could be detected only in PAH-grown cells. These results indicated that the two enzymes were differentially regulated depending on the carbon source used for growth.
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous contaminants in soils and sediments and are of environmental concern because of their mutagenic and/or carcinogenic effects. While low-molecular-weight PAHs (composed of two or three rings) are readily degraded by bacteria, PAHs consisting of four rings or more are recalcitrant to biodegradation and persist in the environment (6, 41). The biodegradation of low-molecular-weight PAHs, especially naphthalene, has been extensively studied with pseudomonads, leading to a good understanding of the bacterial catabolic pathway (42). On the other hand, relatively little information is available on the metabolism of high-molecular-weight PAHs (20). A number of bacterial isolates capable of pyrene mineralization have been described. Most of them are actinomycetes and belong to the genus Mycobacterium (2, 7, 37), Rhodococcus (4, 40), or Gordonia (21). A few pyrene-degrading strains have been identified as gram-negative species, including Stenotrophomonas maltophilia, Pseudomonas fluorescens (3), Sphingomonas paucimobilis (22), and Burkholderia cepacia (19).
Early studies on the catabolism of pyrene by a Mycobacterium species led to the identification of several ring oxidation products, including pyrene 4,5-dihydrodiol and 4-phenanthroic acid (15). Later studies conducted with other Mycobacterium strains identified phenanthrene 4,5-dicarboxylic acid as another intermediate metabolite (7, 37). Based on these findings, a pathway of pyrene degradation by Mycobacterium species has been proposed which likely involves a dioxygenase for catalysis of the initial attack of the aromatic substrate (6, 7, 20) (Fig. 1). None of the enzymes involved in the catabolism of pyrene has yet been described, except for a polycyclic aromatic ring dioxygenase recently identified in the pyrene-degrading Mycobacterium strain PYR-1 (24). In this study, a Mycobacterium strain selected for its ability to grow with pyrene as the sole carbon and energy source was used to identify proteins involved in pyrene catabolism. For this purpose, proteins from bacteria grown on pyrene and other carbon sources were subjected to metabolic labeling and two-dimensional (2D) electrophoresis. This approach allowed the detection of several pyrene-specific polypeptides, some of which were identified by N-terminal and internal peptide sequencing as putative catabolic enzymes. Two distinct ring-hydroxylating dioxygenases were found to be coexpressed in PAH-grown cells. The genes encoding the two pyrene-induced dioxygenases have been cloned, sequenced, and overexpressed in Escherichia coli.
FIG. 1.
Proposed metabolic pathway for the biodegradation of pyrene and phenanthrene in Mycobacterium species. Steps 1 to 6 are specific to the degradation of pyrene (20), and steps 7 to 15 represent the o-phthalate pathway of phenanthrene degradation, with steps 9 to 15 being common to the degradation pathways of the two PAHs. The genes encoding the enzymes of the phenanthrene degradation pathway, which have been identified in Nocardiodes sp. strain KP7 (16, 17, 34), are shown in italics. The enzyme activities thought to be involved in the catalysis of each step shown are as follows: (1) ring-hydroxylating dioxygenase; (2) dihydrodiol dehydrogenase; (3) intradiol dioxygenase; (4) decarboxylase; (5) ring-hydroxylating dioxygenase; (6) dehydrogenase-decarboxylase; (7) ring-hydroxylating dioxygenase; (8) dihydrodiol dehydrogenase; (9) extradiol dioxygenase; (10) isomerase; (11) hydratase-aldolase; (12) aldehyde dehydrogenase; (13) ring-cleaving dioxygenase; (14) hydratase-aldolase; (15) aldehyde dehydrogenase.
MATERIALS AND METHODS
Reagents.
Pyrene, phenanthrene, antibiotics, and most other chemicals were purchased from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Silicone oil, type 47V20, was from Sodipro (Echirolles, France). [4,5,9,10-14C]pyrene was from Amersham Biosciences (Orsay, France). Oligonucleotides were purchased from Genome Express (Montreuil, France). Restriction enzymes were from Promega France (Charbonnières) or Fermentas (Euromedex, Mundolsheim, France). Isopropyl-β-d-thiogalactopyranoside (IPTG) was purchased from Eurogentec (Seraing, Belgium).
Bacterial strains, plasmids, and culture conditions.
Mycobacterium strain 6PY1 was isolated from PAH-contaminated soil by successive enrichment cultures with pyrene as the sole carbon source, as will be described elsewhere (J. C. Willison, unpublished results). This bacterium was grown on a mineral salts medium (MSM) (40) supplemented with one of the following substrates used as a sole carbon and energy source: acetate (30 mM), benzoate (5 mM), phenanthrene (0.5 g/liter), or pyrene (0.1 g/liter). The latter two substrates were supplied as solutions in silicone oil, so that the ratio of the organic phase to the aqueous phase was 1:5. Growth took place at 25°C in Erlenmeyer flasks incubated in a rotary shaker at 150 rpm. Bacterial density was measured spectrophotometrically as the optical density at 600 nm (OD600).
E. coli strains and plasmids used in this study are listed in Table 1. Strains DH5α and BL21(DE3)(pLysS) or BL21AI were used for general cloning and protein expression, respectively. Culture was carried out on rich broth (Luria-Bertani [LB]) containing appropriate antibiotics.
TABLE 1.
Bacterial strains and plasmids used
| Bacterial strain or plasmid | Relevant genotype and/or properties | Source or reference |
|---|---|---|
| Strains | ||
| Mycobacterium sp. strain 6PY1 | Wild type | This study |
| E. coli DH5α | F−endA1 hsdR17(rK−mK+) supE44 thi-1 λ−recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 φ80d lacZΔM15 | Invitrogen |
| E. coli XL1-Blue MRF' | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| E. coli XL10-Gold Kan | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB laclqZΔM15 Tn10(Tetr) Tn5 (Kanr) Amy] | Stratagene |
| E. coli BL21(DE3)(pLysS) | B F−dcm ompT hsdS(rB− mB−) gal λ(DE3) [pLysS Camr] | Promega |
| E. coli BL21AI | B F−dcm ompT hsdS(rB− mB−) gal araB::T7RNAP-tetA | Invitrogen |
| Plasmids | ||
| pPCR-Script | Apr, cloning vector | Stratagene |
| pCR2.1 | Apr, Kanr, T-cloning vector | Invitrogen |
| pGEM-T Easy | Apr, T-cloning vector | Promega |
| pCR-Blunt II-TOPO | Kanr, cloning vector | Invitrogen |
| pET9a | Kanr, expression vector | Novagen |
| pBBR1MCS-5 | Gmr, expression vector | 26 |
| pSK-B3 | pCR2.1 containing a 372-bp PCR fragment (5′ end of pdoA1) | This study |
| pSKU03 | pPCR-Script containing pdoA1 | This study |
| pSKU04 | pPCR-Script containing pdoB1 | This study |
| pSKU05 | pET9a containing pdoB1 | This study |
| pSKU06 | pET9a containing pdoA1B1 | This study |
| pSKU07 | λ ZAP phagemid containing a 2.3-kb fragment carrying pdoA2B2 | This study |
| pSKU08 | pCR-Blunt II-TOPO containing pdoA2B2 | This study |
| pSKU09 | pET9a containing pdoA2B2 | This study |
| pSKU10 | pGEM-T Easy with a 1.2-kb PCR insert carrying pdoB2 | This study |
| pHA171 | PT7-7 carrying the phdABCD genes from strain KP7 | 34 |
| pDRCD | pDRIVE containing phdCD | This study |
| pBRCD | PBBR1MCS-5 containing phdCD | This study |
[14C]pyrene mineralization experiments.
[14C]pyrene mineralization experiments were carried out in 33-ml Warburg flasks closed with rubber stoppers fitted on the two outlets. A mixture of 0.4 μCi of [4,5,9,10-14C]pyrene and 100 nmol of unlabeled pyrene dissolved in acetone was introduced into empty flasks, and the solvent was allowed to evaporate. Then, 2 ml of strain 6PY1 suspension with an OD600 of around 3.0 was placed in the main chamber of the flask and 0.4 ml of 1N NaOH was added in the central well. Flasks were incubated at room temperature on a reciprocal shaker. The 14CO2 produced by pyrene mineralization and trapped in NaOH was estimated by scintillation counting. For this purpose, the NaOH solution was withdrawn at time intervals from the central well with a syringe and replaced by the same volume of freshly prepared solution. Results from duplicate flasks were averaged and expressed as the number of nanomoles of [14C]pyrene mineralized into CO2 normalized to the protein content of the bacterial samples. Control flasks were incubated with 0.1 mg of chloramphenicol/ml.
Preparation of bacterial protein extracts.
Cultures were harvested at 4°C by centrifugation for 15 min at 15,000 × g. Pellets were washed twice in 10 ml of 0.05 M Tris-HCl buffer (pH 7.5) and resuspended in two volumes of sonication buffer (0.1 M Tris-HCl [pH 7.5] containing 0.01 M EDTA). The bacterial suspensions were transferred to 10-ml conical glass tubes and subjected to sonication for 10 min on ice with a Vibra cell ultrasonifier (10% of maximum amplitude; 5-s pulse intervals). The lysate was centrifuged for 10 min at 15,000 × g to remove cell debris and then supplemented with 10 mM spermine (Sigma-Aldrich) and further centrifuged for 20 min at 290,000 × g with an Optima TL ultracentrifuge (Beckman Instruments). The supernatant fraction was then extensively dialyzed against 2 mM phosphate buffer (pH 7.5) in a dialysis cassette with a 10,000-Mr cutoff (Slide-A-Lyser; Pierce). Samples were stored at −20°C until needed.
2D-PAGE.
Protein samples were analyzed according to the 2D polyacrylamide gel electrophoresis (2D-PAGE) method described by Görg et al. (11). Samples (250 μg) were adjusted to 9 M urea, 2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.32% (vol/vol) Pharmalytes (pH 3 to 10) or 2% (vol/vol) immobilized pH gradient buffer of the appropriate pH range, 0.02% (wt/vol) bromophenol blue, and 0.15% (wt/vol) dithiothreitol in a total volume of 0.45 ml and then applied to 18-cm dry gel strips carrying an immobilized pH gradient (Amersham Biosciences). Strips were allowed to rehydrate with sample solutions for at least 6 h at room temperature. Linear pH gradient gels in the 4 to 7 range were generally employed, but occasionally a narrower pH gradient in the 4.5 to 5.5 range was used.
Isoelectric focusing was carried out in a Multiphor II electrophoresis unit connected to a Multidrive XL power supply (Amersham Biosciences). Voltage was increased stepwise from 150 to 3,500 V over 5 h and held at this value for at least 12 h, resulting in a minimum of 44,000 V · h. Gel strips were subsequently incubated for 30 min in 5 ml of equilibration buffer (6 M urea, 50 mM Tris-HCl [pH 8.8], 30% glycerol, 1% sodium dodecyl sulfate [SDS], 16 mM dithiothreitol, 0.01% bromophenol blue) and applied on top of a 12.5% polyacrylamide gel. Second-dimension electrophoresis was carried out in a Protean II xi cell (Bio-Rad) at 40 mA for 4 to 5 h by using a standard Tris-glycine buffer system. Occasionally, the second dimension was performed in a Tris-Tricine buffer system (36). The proteins were visualized by silver staining or Coomassie blue staining. Gels were scanned wet with an Agfa scanner (model Duoscan T1200) and subsequently dried between cellophane sheets with an air drier (Easy Breeze gel dryer; Amersham Biosciences).
Determination of N-terminal and internal peptide sequences.
For protein sequence analysis of selected spots, 2D gels were loaded with a sample containing between 0.6 and 0.9 mg of protein. After the second dimension, the gel was transferred onto a polyvinylidene difluoride membrane (Problott; Applied Biosystems, Courtaboeuf, France) by semidry electroblotting (30 min at 170 mA) in 10 mM 3-[cyclohexylamino]-1-propanesulfonic buffer (pH 9.5) containing 10% methanol. After protein staining with Coomassie blue, spots of interest were excised from the polyvinylidene difluoride membrane and then subjected to automated Edman degradation on an Applied Biosystems sequencer.
For peptide sequencing by tandem mass spectrometry (MS/MS), protein spots of interest were cut out from Coomassie blue-stained 2D gels and then subjected to in-gel trypsin digestion as previously described (9). Each gel band was then extracted with 5% (vol/vol) formic acid solution and acetonitrile. Extracts were combined and evaporated to dryness. The residues were dissolved in 0.1% (vol/vol) formic acid and desalted with a Zip Tip (Millipore, Bedford, Mass.). Elution of the peptides was performed with 5 μl of a 50:50:0.1 (vol/vol) acetonitrile-H2O-formic acid solution. The peptide solution was introduced into a glass capillary (MDS Protana, Odense, Denmark) for nanoelectrospray ionization. MS/MS experiments were carried out on a quadrupole time of flight hybrid mass spectrometer (Micromass, Manchester, United Kingdom). Collision-induced dissociation of selected precursor ions was performed by using argon as the collision gas and with collision energies of 40 to 60 eV. Interpretation of MS/MS spectra was achieved manually and with the help of the PepSeq program (MassLynx software; Micromass). Data obtained from N-terminal and MS/MS sequence analyses were used for homology searches in the databases with the use of the BLAST facility (http://www.ncbi.nlm.nih.gov/BLAST) or the FASTA program (33).
In vivo 35S-labeling of proteins.
Acetate-grown cultures of Mycobacterium were harvested in mid-log phase and resuspended to an OD600 of 2.0 in MSM. Portions of this suspension (25 ml) were transferred to 100-ml flasks containing one of the following carbon sources: pyrene (5 mg), phenanthrene (25 mg), benzoate (5 mM), acetate (25 mM), or none. PAHs were supplied as 5-ml solutions in silicone oil. After 1 h of incubation at 25°C, bacterial suspensions were supplemented with 0.14 mCi (5.18 MBq) of a 35S-labeled mixture of methionine and cysteine (Easytag Express protein-labeling mix; NEN Life Science Products) combined with a mix of the two nonlabeled amino acids as carriers (12 μM Met plus 6 μM Cys). Bacteria were further incubated for variable time periods depending on the carbon source added: 6 h (benzoate and acetate), 17 h (phenanthrene), or 32 h (pyrene and no carbon source added). Then cells were harvested by centrifugation, washed once in MSM, and resuspended in 0.25 ml of 0.1 M Tris-HCl-10 mM EDTA (pH 8.0). Bacterial extracts were prepared as described above and analyzed by 2D-PAGE (375-μg protein samples were used). Gels were stained with Coomassie blue, dried, and exposed to X-ray films for 4 days (Hyperfilm-βmax; Amersham Biosciences).
General DNA manipulations.
Restriction enzyme digestions, ligations, and agarose gel electrophoresis were performed by using standard procedures (35). DNA fragments were purified from agarose gels by using a Jetquick gel extraction spin kit (Genomed GmbH, Bad Oeynhausen, Germany). Mycobacterium DNA was isolated by using the following method adapted from that of Gonzalez-y-Merchand et al. (10). Benzoate-grown cells (from 2.8 liters of culture) were harvested in mid-log phase, resuspended in TE buffer (10 mM Tris-HCl, pH 8.0, containing 1 mM EDTA), and treated with 10 mg of lysozyme/ml at 37°C for 30 min. The suspension was adjusted to 1% SDS and subjected to three freeze-thaw cycles consisting of a freezing step with liquid nitrogen followed by heating at 65°C for 10 min. Cetyltrimethylammonium bromide (11.3 g/liter) was added to facilitate the removal of polysaccharides and proteins during two chloroform-isoamyl alcohol extraction steps. Two more phenol-chloroform-isoamyl alcohol extractions were then performed before the precipitation of nucleic acids with isopropanol. DNA was washed in 70% ethanol and dissolved in TE buffer.
For Southern hybridization experiments, DNA fragments used as probes were labeled with digoxigenin by using a random prime labeling kit (Roche Diagnostics, Meylan, France). Mycobacterium genomic DNA samples (1 to 2 μg) were digested to completion with restriction endonucleases EcoRI, EcoRV, HindIII, BamHI, PstI, and SmaI and then analyzed by electrophoresis on a 1% agarose gel. The DNA fragments were denatured with NaOH and transferred by Southern blotting onto nylon membrane (Biodyne; Pall Life Sciences, Saint-Germain-en-Laye, France). The membrane was prehybridized for 1 h and then hybridized overnight at 65°C. Posthybridization washings and detection were performed according to the protocol of the supplier (Roche Diagnostics).
Construction of a genomic library.
Genomic DNA from Mycobacterium sp. strain 6PY1 was digested with restriction enzyme BamHI, and the fragments obtained were cloned into predigested ZAP Express vector according to the supplier's instructions (Stratagene Europe, Amsterdam, The Netherlands). The ligated DNA was packaged by using the Gigapack III gold packaging extract (Stratagene) and propagated as recombinant phages in E. coli XL1-Blue MRF'. Clones of E. coli containing corresponding phagemids were transferred to 96-well microtiter plates to facilitate screening of the library.
Cloning of the genes encoding dioxygenase components Pdo1 and Pdo2 from strain 6PY1.
Two degenerate primers (K7-F, 5′-CARACSGARACSACSGA-3′, and ISPα1-R, 5′-CCASCCRTGRTASSWRCA-3′) were designed based on the N-terminal sequence of protein P10 and an internal consensus sequence (CSYHGW) found in most dioxygenase α subunits. With these two primers, a 372-bp DNA fragment was amplified by PCR with Mycobacterium DNA as the template. This fragment was cloned into E. coli by using the pCR2.1 cloning kit (Invitrogen), resulting in plasmid pSK-B3, and its sequence was determined. The genes encoding Pdo1 were amplified by PCR by using strain 6PY1 genomic DNA as the template and the Expand High-Fidelity DNA polymerase (Roche Diagnostics). The following primers were employed: pdoA1-F (5′-GGCATATGCAAACGGAAACGACCGA) and pdoA1-R (5′-GGGATATCTCAAGCACGCCCGCCGAATG) for pdoA1 and pdoB1-F (5′-GGCATATGAACGCCGTTGCCGTGGA) and pdoB1-R (5′-GGGGATCCTACAGGACTACCGACAG) for pdoB1 (underlined sequences denote restriction sites for NdeI, EcoRV, and BamHI). Primer pdoA1-F was designed after the 5′ end of the 372-bp DNA fragment described above. The other primers were designed after the nucleotide sequences of nidA (accession number AF249301) and nidB (accession number AF249302) from Mycobacterium sp. strain PYR-1. PCR products were ligated into pPCR-Script, giving pSKU03 (pdoA1) and pSKU04 (pdoB1). The DNA sequences of the inserts on both strands were determined. The genes encoding Pdo2 were cloned as follows. A PCR amplification with 6PY1 genomic DNA and two degenerate primers, P3-F (5′-ATGGTNGCNACNGTNGARCA) and P3-R (5′-TCYTCNGCCCANGCRTAYTC), generated a 270-bp fragment which was cloned and sequenced. The same primers were then used to screen the genomic library in ZAP Express by PCR amplification of clones. For this purpose, serial rounds of PCR were carried out on groups of 96, and then of 12, clones, each round consisting of cell lysis by boiling of the colonies in water for 5 min at 95°C, centrifugation, and PCR with 2 μl of supernatant and 10 μl of a mix containing deoxynucleoside triphosphate, primers, and Taq polymerase (Promega). One positive clone was selected, which contained a phagemid with a 2.3-kb insert (pSKU07). The sequence of the insert was determined. A 1.2-kb DNA fragment containing the 3′ end of pdoB2 was generated by PCR by using genomic DNA as a template and primers P3-F and Redcons1-R (5′-TTGGAYAGWGGCGGWCGBTCRTAWGG). The first primer corresponds to the 5′ end of pdoB2, whereas the second primer was designed after a consensus sequence (PYERPPLSK) found upon alignment of reductase components of representative ring-hydroxylating dioxygenases. The 1.2-kb fragment was cloned into pGEM-T Easy to give pSKU10 and sequenced. DNA sequence analyses as well as protein sequence comparison and alignment were performed by using programs available on the Infobiogen server (http://www.infobiogen.fr/services/deambulum/fr/index.html).
Construction of plasmids for protein overexpression.
pdoB1 was isolated from pSKU04 by digestion with NdeI and BamHI and then ligated into pET9a to give pSKU05. This plasmid was digested with XbaI, treated with the Klenow enzyme to generate a blunt-ended product, and further digested with BamHI to generate a 0.6-kb DNA fragment carrying the pdoB1 coding sequence preceded by an efficient ribosome-binding site. This fragment, as well as an NdeI-EcoRV fragment from pSKU03 carrying pdoA1, were ligated between the NdeI and BamHI sites of pET9a to give pSKU06. The resulting plasmid contained the pdoA1 and pdoB1 genes (in that order) located downstream of a T7 promoter, with each gene preceded by a ribosome-binding site sequence optimal for expression in E. coli.
A fragment encompassing the pdoA2B2 gene sequence was amplified as described above by using primers pdoA2-F (5′-GGCATATGTCTACTGTCGGTAAGAA) and pdoB2-R (5′-GGAGATCTTAGAAGAAGTTAGCCAG). The underlined sequences denote NdeI and BglII sites introduced to facilitate subcloning. The PCR product was cloned into pCR-Blunt II-TOPO (Invitrogen) to give pSKU08 and subjected to DNA sequencing to ensure that no error occurred during PCR amplification. The insert of plasmid pSKU08 was isolated by digestion with NdeI and BglII and ligated into pET9a to give pSKU09.
A 1.5-kb DNA fragment carrying the phdC and phdD coding sequences was amplified by PCR by using the Expand High-Fidelity DNA polymerase, plasmid pHA171 as a template, and primers phdCeco (5′-GGAATTCAGGAGATATACATATGCGTGTG) and phdDxba (5′-CTCTAGACTCATGCCGTCGGTAC). The resulting product was treated with an A-addition kit (Qiagen) and cloned into pDRIVE to give plasmid pDRCD. The insert was verified by DNA sequencing and then isolated as an EcoRI-XbaI fragment and cloned into the corresponding sites of pBBR1MCS-5, which gave plasmid pBRCD.
Plasmids pSKU06, pSKU09, and pBRCD were introduced into E. coli strains BL21(DE3)(pLysS) and BL21AI by transformation.
Overexpression and purification of the recombinant Pdo2 dioxygenase component.
E. coli BL21(DE3)(pLysS)(pSKU09) was grown in LB medium containing kanamycin (20 μg/ml) and chloramphenicol (33 μg/ml) to an OD600 of ∼0.6 and then subjected to induction with 0.1 mM IPTG. After a 3-h incubation at 25°C, the cells were harvested by centrifugation and stored at −20°C until needed. The cells were disrupted by sonication (10% of maximum amplitude; 5-s pulse intervals) for 5 min at 4°C in sonication buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 5% [vol/vol] ethanol). After centrifugation of the cell extract for 10 min at 10,000 × g, the supernatant fraction was diluted with the same volume of buffer A (50 mM Tris-HCl [pH 7.5], 5% [vol/vol] ethanol, 5% [wt/vol] glycerol) before being applied to a 30-ml DEAE-cellulose column (DE52; Whatman) equilibrated with buffer A. After the column was washed with the same buffer, a brown fraction was eluted with buffer A containing 250 mM NaCl. The brown fraction was adjusted to 1 M ammonium sulfate and applied to a 20-ml phenyl-Sepharose column (Amersham Biosciences). The column was developed with a linear gradient from 1 to 0 M ammonium sulfate in buffer A by using a fast protein liquid chromatography system (Bio-Tek Kontron Instruments, St. Quentin en Yvelines, France). The brown fraction was dialyzed against buffer A and then loaded onto a 1.7-ml column (Optima 5/10 Q-HyperD 10; Biosepra SA, Villeneuve-la-Garenne, France). The column was developed with a linear gradient from 0 to 250 mM NaCl in buffer A. The purified protein was stored frozen in liquid nitrogen.
Overexpression of the Pdo1 component and purification of the α subunit.
Expression of Pdo1 in E. coli BL21(DE3)(pLysS)(pSKU06) was carried out under conditions similar to those described above, except that recombinant cells were induced with 50 μM IPTG at 15°C for 16 h. The cells were disrupted by sonication (same conditions as described above), and the crude extract was centrifuged for 10 min at 10,000 × g to recover the insoluble fraction. This fraction was solubilized in 40 mM Tris-HCl (pH 8.0) containing 8 M urea and then centrifuged for 10 min at 10,000 × g. This sample was diluted by half in H2O and applied to a 20-ml column of DEAE-Trisacryl LS (Biosepra SA) equilibrated with 20 mM Tris-HCl (pH 8.0) containing 4 M urea (buffer B). The column was developed with a linear gradient from 0 to 0.5 M NaCl in buffer B. Fractions containing a prominent 52-kDa band were pooled and concentrated. The 52-kDa protein was further purified by preparative SDS-PAGE and then eluted from gel slices by using an electroeluter (model 422; Bio-Rad SA). The purified preparation was used to raise rabbit polyclonal antibodies against the Pdo1 α subunit, according to the standard protocol of the supplier (Eurogentec). Antibodies against the Pdo2 α subunit were obtained under similar conditions.
In vivo assays of recombinant Pdo1 and Pdo2 activity.
The PAH dioxygenase activity of recombinant E. coli cells expressing Pdo1 or Pdo2 was assayed as follows. Cells were grown in LB medium to an OD600 of 1.0 and then incubated overnight at 25°C with 0.5 mM IPTG and/or 0.2% dl-arabinose, depending on the strain. Cells were harvested, resuspended in the same volume of M9 minimal medium, and incubated with one-fourth (by volume) of the silicone oil containing 0.1 g of either pyrene or phenanthrene/liter. At various time intervals, samples (0.5 to 1.0 ml) of the upper layer emulsion were withdrawn and briefly centrifuged. The oil phase was extracted with acetonitrile, and the absorbance of the extract was recorded in the 200-to-350 nm range. Amounts of residual pyrene and phenanthrene were calculated from absorption readings at 240 and 252 nm, respectively. Hydrosoluble products were analyzed in 5-ml samples of the aqueous phase, which were centrifuged and then passed through solid-phase extraction columns (Upti-clean C18U, 0.5 g; Interchim, Montluçon, France). Columns were washed with 10 ml of water and then eluted with 1 ml of methanol. Extracts were evaporated under vacuum conditions and stored dry until ready to be analyzed by gas chromatography (GC)-MS. Prior to analysis, extracts were derivatized either with bis(trimethysilyl)trifluoroacetamide:trimethylchlorosilane (99:1) or with n-butylboronate (NBB), both reagents from Supelco (Sigma-Aldrich), according to the manufacturer's instructions. GC-MS analysis was carried out on a HP6890/HP5973 apparatus (Agilent Technologies, Les Ulis, France) equipped with an MDN-12 fused silica capillary column (30 m, 0.25-mm internal diameter, 0.25-μm film; Supelco). The oven temperature was held at 75°C for 3 min and then increased to 250°C at 12°C/min, held at this temperature for 1 min, and then increased to 300°C at 10°C/min, with a final hold time of 10 min. The injector was used in the split mode, with a split ratio of 50:1 and an injection volume of 2.5 μl. Mass detection was carried out in the selected ion monitoring mode for quantification and in the total ion current mode for determination of the mass spectra of diols and their derivatives. The solvent delay prior to data acquisition was 5.5 min for trimethylsilyl (TMS) derivatives and 11 min for NBB derivatives.
The TMS derivatives of phenanthrene and pyrene dihydrodiols were quantified by using purified phenanthrene cis-9,10-dihydrodiol and pyrene cis-4,5-dihydrodiol as standards (Willison, unpublished). GC-MS peak areas were proportional to sample concentration when the selected ion monitoring mode of detection was used, with the selected ions having m/z values of 290 for the TMS derivative of pyrene dihydrodiol and 178, 266, and 356 for the TMS derivative of phenanthrene dihydrodiol. Phenanthrene cis-3,4-dihydrodiol was provisionally identified on the basis of the mass spectra of the underivatized and derivatized compounds and its longer GC retention time with respect to the 9,10-isomer (32).
Other analytical methods.
Whole-cell protein content was determined according to the method of Lowry et al. (29) after bacterial samples were subjected to 0.2 M NaOH for 1 h at 90°C. Routine protein assays were done with the bicinchoninic acid reagent kit (Pierce) by using bovine serum albumin as a standard. SDS-PAGE on mini slab gels and Western blot analysis were performed as previously described (18). Rabbit serum antibodies and secondary goat anti-rabbit immunoglobulin G coupled to peroxidase (Sigma-Aldrich) were used at a 1:5,000 dilution.
Nucleotide sequence accession numbers.
The nucleotide sequences described in this report have been deposited in the DDBJ, EMBL, and GenBank databases under accession numbers AJ494743, AJ494744, and AJ494745.
RESULTS
Induction of pyrene mineralization in Mycobacterium strain 6PY1.
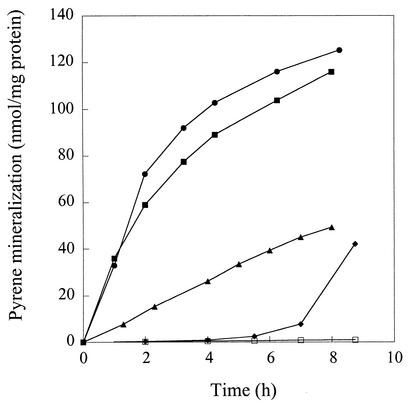
In addition to pyrene, strain 6PY1 can grow with acetate, benzoate, and phenanthrene as alternative carbon sources. The abilities of bacterial cells grown on each of these substrates to mineralize [14C]pyrene were compared (Fig. 2). Pyrene- and phenanthrene-grown cells showed similar activities, with initial mineralization rates around 36 nmol/h/mg of protein. Benzoate-grown cells displayed about fivefold less activity than pyrene- and phenanthrene-grown cells, whereas acetate-grown cells had essentially no activity. After a 6-h incubation with pyrene, mineralization became detectable in acetate-grown cells. This activity was dependent on protein synthesis since it did not occur in cells treated with chloramphenicol. These results suggested that the enzymes responsible for pyrene degradation were not expressed in acetate-grown cells but that intermediate levels of these enzymes might be expected in cells grown on benzoate. As a means of identifying enzymes involved in pyrene degradation, soluble protein extracts from cells grown on pyrene, phenanthrene, acetate, or benzoate were analyzed by 2D-PAGE.
FIG. 2.
Time course of [14C]pyrene mineralization by Mycobacterium strain 6PY1 grown on different C sources. Bacteria were incubated with [14C]pyrene in closed flasks as described in Materials and Methods. The 14CO2 that evolved was continuously trapped in sodium hydroxide and quantified at the times indicated. Results shown are the means of results from duplicate experiments and are expressed as the amount of cumulated pyrene mineralization normalized to the protein concentration of the bacterial samples. Bacteria were grown on either pyrene (▪), phenanthrene (•), benzoate (▴), or acetate (⧫). Acetate-grown cells were also incubated with chloramphenicol (□). The extent of pyrene mineralization at the end of the experiment was 38.9, 44.5, 17.8, and 17.2% for cells grown on pyrene, phenanthrene, benzoate, and acetate, respectively.
Protein profiles of pyrene-grown cells compared to those of cells grown on three other C sources.
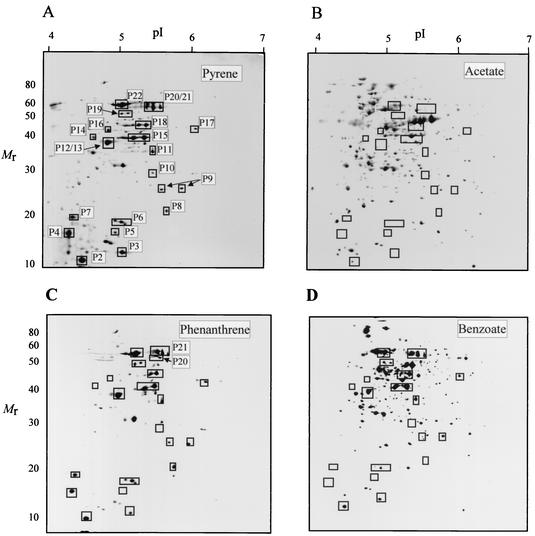
Preliminary 2D-PAGE analyses of cell-free extracts of strain 6PY1 involving a first-step separation in a pH gradient from 3 to 10 indicated that most soluble polypeptides had isoelectric points (pIs) ranging between 4 and 7. For the comparative analysis of proteins from cells grown on four C sources, we therefore performed isoelectrofocusing of polypeptides in the pH 4 to 7 range and further separated those in the 10- to 100-kDa range on 12.5% polyacrylamide gels. Visual inspection of silver-stained protein patterns revealed that 21 protein spots found in pyrene-grown cells were absent in acetate-grown cells (Fig. 3). At least 12 of these polypeptides appeared as major components of the cell extract, as judged from their polypeptide spot size. As revealed by subsequent peptide sequence analysis (see below), many of the peptides of interest (P6, P9, P15, P18, and P21) gave rise to a doublet or triplet of spots with similar Mrs and slightly different pIs. Some other polypeptides were poorly resolved due to very close Mrs and pIs, as was the case for polypeptides P12 and P13. Further analysis revealed the occurrence of two additional pyrene-specific polypeptides, named P1 and P20, which passed undetected on regular 2D gels, as shown in Fig. 3A. Because of its small size, P1 (Mr = 10,900; pI 4.6) was best detected on 2D gels run in a Tris-Tricine buffer system in the second dimension as described in Materials and Methods (data not shown). Peptide P20, which was later identified as a dioxygenase α subunit (Pdo2α; see below), appeared as a minor peptide found very close to the triplet of spots generated by P21 (Fig. 3B). P20 was unambiguously identified as the α subunit of Pdo2 upon 2D-PAGE analysis of a 6PY1 extract from pyrene-grown cells supplemented with purified Pdo2 (Fig. 3C). At this stage of our comparative 2D-PAGE analysis, a total of 16 distinct polypeptides appeared to be specifically synthesized in pyrene-grown cells of strain 6PY1 (Fig. 3). Most of the selected polypeptides were also found in phenanthrene-grown cells, but five proteins were missing in benzoate-grown cells (designated P4, P5, P8, P14, and P20).
FIG. 3.
2D-PAGE analysis of the soluble extract from strain 6PY1 grown on pyrene. (A) Proteins were separated in a pI gradient from 4 to 7 and then on a 12.5% polyacrylamide gel, after which they were silver stained. The pI and molecular mass (in kilodaltons) ranges are indicated. (B) Enlargement of the boxed area indicated in panel A. (C) Same area of a gel loaded with a similar extract supplemented with purified Pdo2. Polypeptides that were absent in acetate-grown cells are indicated by arrows and numbered as a function of increasing Mrs. Polypeptides P1, P3, P7, P10, P16, P17, and P19 do not sppear in this figure.
As an alternate method to detect pyrene-specific polypeptides, we performed metabolic labeling of whole-cell proteins, taking advantage of the fact that pyrene-degrading enzymes were apparently inducible in strain 6PY1 upon incubation with PAHs (Fig. 2). For this purpose, bacteria grown on acetate were washed and resuspended in MSM containing a mixture of [35S]methionine and cysteine and either pyrene, phenanthrene, benzoate, or acetate as carbon sources. Proteins which incorporated radioactivity were subsequently analyzed by 2D-PAGE and autoradiography (Fig. 4). Analysis of cells exposed to pyrene revealed a relatively small number of labeled polypeptide spots (around 40), 31 of which were undetectable in control cells incubated without any carbon source (data not shown) or in cells exposed to acetate (Fig. 4B). The selected set of polypeptides included all those previously found to be missing in acetate-grown cells relative to pyrene-grown cells when steady-state levels of the proteins were compared (Fig. 3). The additional peptides detected by protein labeling, designated P3, P7, P10, P16, P17, and P19, were relatively minor components and either hardly detectable by silver staining or masked by neighboring spots in crowded regions of the 2D gel. Among those polypeptides, three were undetectable in benzoate-induced cells (P7, P10, and P16), two of which (P10 and P16) were also absent in phenanthrene-induced cells (Fig. 4C and D).
FIG. 4.
2D-PAGE analysis of 35S-labeled proteins induced upon incubation of strain 6PY1 with pyrene or other carbon sources. Acetate-grown cells were incubated with either pyrene, phenanthrene, benzoate, or acetate in the presence of 35S-labeled amino acids as described in Materials and Methods. Labeled proteins were then analyzed by 2D-PAGE and autoradiography. Boxed protein spots were not detected in control cells incubated with no carbon source or with acetate. Proteins P21 and P6 represent the α and β subunits of Pdo1, and P20 and P8 represent the α and β subunits of Pdo2.
Identification of selected pyrene-induced proteins by peptide sequencing.
Eleven polypeptides chosen among the 22 pyrene-induced polypeptides detected in strain 6PY1 were subjected to amino acid sequencing, either by N-terminal or internal peptide analysis (Table 2). Three polypeptides (P9, P15, and P18) gave pairs of spots with identical N termini, the same Mrs, and different pIs, whereas two polypeptides yielded triplets (P6 and P21). In such cases, each pair or triplet was assumed to result from some minor posttranslational modification of a single polypeptide. A database search for proteins showing similarities with the available N-teminal sequences yielded a significant match in five cases. Polypeptides P1, P15, and P22 showed high similarity with the ORF131, PhdG, and PhdH products found in the phenanthrene-degrading marine bacterium Nocardioides sp. strain KP7 (34). PhdG and PhdH have been identified as a hydratase-aldolase and an aldehyde dehydrogenase, respectively. The N terminus of protein P6 closely matched the corresponding region of the nidB gene product from Mycobacterium strain PYR-1 which has been recently described as the β subunit of a ring-hydroxylating dioxygenase (24). Protein P21 gave a triplet of spots, none of which yielded more than six residues by N-terminal sequencing. Nevertheless, a significant match was found with the α subunit of benzene dioxygenase and the nidA gene product from strain PYR-1. Gene sequencing later revealed that protein P21 was associated with protein P6 to form a ring-hydroxlating dioxygenase (see below).
TABLE 2.
Sequence analysis of pyrene-induced polypeptides from 6PY1 and possible functions based on sequence similarities
| Polypeptide (Mr) | N terminus or internal peptide sequence(s)a | Best database matchb | % Amino acid identityc | Possible function |
|---|---|---|---|---|
| P1 (10.9) | AIRFLTDDWATAVTDAANADERF(R)IAAKGH | ORF131 (AB031319) | 70 | Unknown |
| P6 (19.0) | N[AM][NV]AVDRD(R)(R)E | NidB (AF249302) | 99 | β Subunit of arene dioxygenase |
| P8 (21.0) | VATVEQQLLLRCE(M)E | PhdB (AB031319) | 62 | β Subunit of arene dioxygenase |
| XEYAWAED | Residues 85-91 | |||
| P9 (25.4) | TVDDQRTGRPMA | None | Unknown | |
| P12 (37.0) | [L/I]YADAG[L/I]DXXKET[L/I]AY | ThtR (O05793) | 89 | Thiosulfate sulfur transferase |
| CR | Residues 217-234 | |||
| P13 (37.2) | ARDDKLTSDDITGV | PhdJ (D89988) | 58 | trans-2′-Carboxylbenzalpyruvate hydratase-aldolase |
| [I/L]GELGADGLFVGR | Residues 111-120 | |||
| AA[L/I][K/Q][K/Q]R | Residues 194-199 | |||
| H[L/I]G[L/I][L/I]SGSAES. . .[L/I]R | Residues 179-192 | |||
| XX[L/I]Q[L/I]DNA[Q/K]F[Q/K]AA | Residues 278-288 | |||
| P15 (39.0) | TTRSSELVPSDMKGLTGFVPA(A)STP | PhdG (AB031319) | 72 | Hydratase-aldolase |
| P16 (42.7) | MRAALAMKVGGKFEI | None | Unknown | |
| P18 (45.4) | STAEISGLTEF | PhdI (AB000735) | 40 | 1-Hydroxy-2-naphthoate dioxygenase |
| MAG[L/I]AGSPYDVVG[L/I]R | Residues 239-253 | |||
| NALLFSVTLQPTLK | Residues 333-346 | |||
| P21 (54.0) | QTETTEX(A) | NidA (AF249301) | 98.5 | α Subunit of arene dioxygenase |
| P22 (58.0) | MINANLIIDGREETSDRTIDV | PhdH (AB031319) | 90 | Aldehyde dehydrogenase |
Bold letters indicate residues that are identical between relevant peptides and best database matches. Italicized letters indicate N-terminal sequences. Letters in parentheses represent uncertain residues; those in brackets represent residues that are equally possible at that position. X, undetermined residue.
Accession numbers are indicated in parentheses. Other numbers indicate positions in the sequences of the protein homologues which best match relevant peptide sequences.
Calculated from available peptide sequences. Proteins P6 and P21, the sequences of which were deduced from the pdoA1 and pdoB1 gene sequences, were compared to the nidB and nidA gene products, respectively, on the basis of the entire sequences.
Proteins P8, P13, and P18, for which N-terminal sequence analysis was inconclusive, were subjected to MS/MS to determine the sequences of internal tryptic peptides. Protein P8 yielded a heptapeptide, EYAWAED, corresponding to a partially conserved region found in β subunits of ring-hydroxylating dioxygenases. This finding suggested the presence of a second arene dioxygenase in strain 6PY1, which we later confirmed by identifying the corresponding genes and isolating the protein (see below). Sequences of four peptides from protein P13 were determined, and all matched internal regions of a single protein characterized as a trans-2′-carboxybenzalpyruvate hydratase-aldolase encoded by phdJ in Nocardioides strain KP7 (34). In addition, protein P13 (Mr, 37,200) was similar in size to the enzyme from strain KP7 (38 kDa). A database search for proteins with similarities to protein P18 with the use of available peptide sequences did not give significant results. However, we observed that two internal peptides from this protein (Table 2) exhibited some sequence similarity to internal regions of the 1-hydroxy-2-naphthoate dioxygenase (PhdI) from strain KP7 (17). Moreover, the molecular properties of protein P18 (Mr, 45,000; pI 5.15) were very close to those deduced from the PhdI protein sequence (molecular mass, 43,076 Da; pI 5.01 to 5.23, depending on software). On the basis of available information, it seems reasonable to propose that protein P18 is a 1-hydroxy-2-naphthoate dioxygenase analogous to the PhdI protein from strain KP7. Protein P12 was hardly resolved from peptide P13 by 2D-PAGE, even when the proteins were isofocused in a 1-U pH range. Nevertheless, we could separate the two protein spots well enough for sequence analysis upon prolonged electrophoresis in the second dimension. An 18-residue-long tryptic peptide from protein P12 was sequenced by MS/MS and found to perfectly match an internal region of the ThtR protein from Mycobacterium tuberculosis (accession number O05793). ThtR was described as a putative thiosulfate sulfur transferase based on sequence similarities to rhodanese, a ubiquitous enzyme that might be involved in Fe-S cluster assembly (5). Should protein P12 also be a sulfur transferase, its possible role in the catabolism of aromatic hydrocarbons in strain 6PY1 remains elusive.
Further comparison of 2D gel patterns revealed that four polypeptides found in extracts from phenanthrene-grown cells were either absent or much less abundant under other culture conditions (data not shown). The N-terminal sequences of those peptides were determined. Searches in the databases yielded a significant match for only one polypeptide, called P23, whose N terminus displayed 60% identity with that of a dihydrodiol dehydrogenase (BphB; accession number D88021) from the biphenyl-degrading strain Rhodococcus erythropolis TA421 (25). A 50% sequence identity was also found between the P23 N terminus and that of the phdE gene product, another dihydrodiol dehydrogenase thought to be involved in phenanthrene degradation in Nocardioides strain KP7 (34). The apparent molecular characteristics of protein P23 (Mr, 27,000; pI 5.15) are also consistent with those of many dihydrodiol dehydrogenases, including PhdE (mass, 28,150 Da; pI 5.1).
Cloning and nucleotide sequence analysis of the genes encoding two distinct arene dioxygenases.
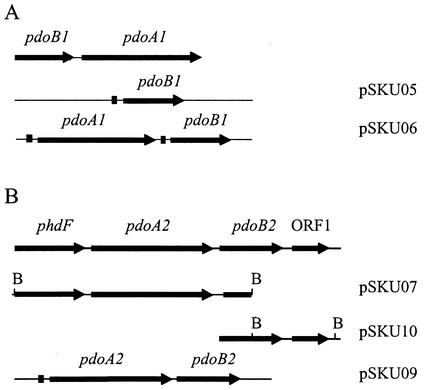
In order to clone the gene encoding protein P21, a DNA fragment was amplified by PCR from the genomic DNA of Mycobacterium strain 6PY1 by using two primers designed from the N terminus of P21 and a consensus sequence (C[P/S]YHGW) found at the iron-sulfur binding site in dioxygenase α subunits. The resulting 372-bp fragment was sequenced and found to be 98% identical to the 5′ end of the nidA gene from Mycobacterium sp. strain PYR-1 (24). Since protein P6 had an N-terminal sequence closely similar to that of the nidB gene product from strain PYR-1, it was likely that proteins P21 and P6 were encoded by genes homologous to the nid genes from strain PYR-1. We then used two sets of primers designed after the 5′ and 3′ ends of nidA and nidB, respectively, and performed PCR amplification with strain 6PY1 DNA as a template. Two fragments were obtained, cloned, and sequenced. One fragment consisted of a 1,377-bp open reading frame (ORF), potentially coding for a 50,336-Da polypeptide. The second fragment comprised 512 bp and might encode a polypeptide with a predicted molecular mass of 19,502 Da. The N termini of the deduced polypeptides exactly matched those found for proteins P6 and P21. These genes displayed high similarity with the nidB and nidA genes from strain PYR-1, and further PCR analysis suggested that they were arranged in the same atypical order, i.e., nidB preceded nidA (Fig. 5). Unlike the nidBA genes from strain PYR-1, the corresponding genes from strain 6PY1 are not inducible by naphthalene, since this strain cannot use this PAH as a growth substrate. They were therefore designated pdoB1(β subunit) and pdoA1(α subunit).
FIG. 5.
Maps representing the two loci containing the dioxygenase-encoding genes from Mycobacterium sp. strain 6PY1. Plasmids used for overexpression of pdoA1B1 (A) and pdoA2B2 (B) in E. coli are also represented. Black squares indicate ribosome-binding sequences optimal for E. coli. B, BamHI.
The occurrence of a second ring-hydroxylating dioxygenase in strain 6PY1 was anticipated after the identification of protein P8 as a potential β subunit distinct from P6. Degenerate primers designed after the N terminus and the available internal sequence of protein P8 allowed for the amplification of a 270-bp DNA fragment. The deduced partial sequence exhibited significant similarity to known β subunits of dioxygenases. The 270-bp fragment was then used to screen a λ library constructed with strain 6PY1 genomic DNA. A positive clone that carried a 2.3-kb insert was isolated. Sequence analysis revealed that the insert contained two partial ORFs and one complete ORF (Fig. 5). The first ORF (partial sequence) translated into a polypeptide sequence of 189 residues which exhibited 88% identity with an extradiol dioxygenase from Nocardioides strain KP7 (phdF gene product) (34). The second ORF comprised 1,401 bp and translated into a 466-residue polypeptide which showed strong similarity with the α subunit of the phenanthrene dioxygenase from strain KP7 (phdA gene product; 73% identity) (34). The sequence of the third ORF, which encodes protein P8, was interrupted at the 3′ end of the insert at a BamHI site. A DNA fragment starting at the 5′ end of this ORF and extending 1.2 kb downstream was generated by PCR and subjected to sequencing. Thereby, the complete sequence of the gene encoding protein P8 was determined. The deduced polypeptide sequence showed 63% identity with the product of phdB which codes for the β subunit of phenanthrene dioxygenase in strain KP7. Hence, a second dioxygenase system was identified in strain 6PY1, encoded by genes designated pdoA2 and pdoB2. Downstream of pdoB2, we found an ORF similar in sequence and location to ORF72 in strain KP7. Further downstream, no sequence analogous to a gene present in the phd operon of strain KP7 could be identified. In this respect, we were unable to detect a reductase-encoding gene analogous to phdD (found 3 kb downstream from phdB in strain KP7) by employing several PCR-based strategies in attempts to amplify such a gene downstream of pdoB2 (data not shown). For instance, a PCR product was obtained by using primers designed to hybridize to pdoB2 and to a consensus sequence found in several reductases, including PhdD (see Materials and Methods), but surprisingly, no sequence relevant to a reductase was found in this fragment (Fig. 5). Also, attempts were made to detect a potential ferredoxin or reductase gene by subjecting total DNA to Southern hybridization with the phdC or phdD gene from KP7 as probes, but no specific signal was obtained. Hence, there was no evidence for a gene coding for a ferredoxin or a reductase in the immediate vicinity of pdoA2B2.
Digestions of total genomic DNA from strain 6PY1 with six restriction enzymes were analyzed by Southern hybridization by using probes specific for pdoA1 or pdoA2. Results showed a single-band pattern in every case (data not shown), indicating that each gene was present as a single copy in the genome and that the probes did not detect an additional homologous gene under the conditions used.
Overexpression and purification of the terminal component of two dioxygenases from strain 6PY1.
To further characterize the two dioxygenase systems found in strain 6PY1, the pdoA1B1 and pdoA2B2 pairs of genes were separately cloned in expression vector pET9a and overexpressed in E. coli BL21(DE3)(pLysS), as described in Materials and Methods. Analysis of crude extracts of IPTG-induced recombinant cells by SDS-PAGE revealed that two polypeptides with Mrs of about 52,000 and 20,000 were overproduced in both cases (data not shown). Parts of the recombinant polypeptides were pelleted with cell debris upon centrifugation of broken cells, indicating that they formed insoluble inclusion bodies in E. coli. When strain BL21AI was used as a host, a greater proportion of the recombinant Pdo1 and Pdo2 proteins was recovered in the soluble fraction, which correlated with the detection of much higher levels of dioxygenase activity in those recombinant cells (see below). The Pdo2 protein could be purified to near homogeneity by using a three-step procedure (see Materials and Methods). The Pdo1 protein appeared to be unstable and lost its brown color at the second step of the purification. Nevertheless, the α subunit of this protein could be purified under denaturing conditions and was used to raise antibodies for immunoblot studies (see below).
The purified Pdo2 enzyme, encoded by pdoA2B2, consisted of two types of subunits, with Mrs of 52,000 (α subunit) and 20,000 (β subunit) in accordance with the expected sizes of the pdoA2 (51.7 kDa) and pdoB2 (19.8 kDa) gene products, respectively (Fig. 6). It appeared as a red-brown protein fraction which displayed a broad absorption band in the visible range, with a maximum around 456 nm. The absorption coefficient at 456 nm was calculated to be 10.6 mM−1 · cm−1, assuming a molecular mass of 215 kDa for an expected α3β3 hexamer. This value is similar to the values previously reported for the terminal component of other dioxygenases (8, 13). These data are consistent with the presence of one Rieske-type (2Fe-2S) cluster per pair of αβ subunits in the recombinant protein.
FIG. 6.
SDS-PAGE analysis of recombinant Pdo2 during purification from E. coli. Lane 1, crude extract; lane 2, soluble fraction; lane 3, DEAE column eluate; lane 4, phenyl-Sepharose column eluate; lane 5, Q-HyperD column eluate; lane M, protein markers. Arrows show the positions of the Pdo2 α and β subunits.
Transformation of pyrene and phenanthrene by recombinant Pdo1 and Pdo2 proteins.
Preliminary attempts to detect pyrene or phenanthrene dioxygenase in strain BL21(DE3)(pLysS) carrying pSKU06 or pSKU09 yielded very low activity levels. In contrast, when strain BL21AI was used as a host, significant conversion of the PAH substrates by arabinose-induced cells was observed. Strain BL21AI(pSKU06) transformed pyrene into a hydrosoluble product which was identified as cis-4,5-dihydroxy-4,5-dihydropyrene by GC-MS. A quantitative estimation of the diol product indicated that approximately 5% of the substrate was converted within 24 h. Strain BL21AI(pSKU09) also converted pyrene to the same dihydrodiol but at a much slower rate (0.18% of pyrene conversion within 24 h). When phenanthrene was provided as the substrate, two oxidation products were detected in the aqueous supernatant from cultures expressing either Pdo1 or Pdo2. GC-MS analysis of the TMS and NBB derivatives identified those products as 3,4-dihydroxy-3,4-dihydrophenanthrene and 9,10-dihydroxy-9,10-dihydrophenanthrene (Fig. 7A). In strain BL21AI(pSKU06) expressing Pdo1, the 9,10-dihydrodiol was the major product formed during the first 6 h of incubation (Table 3) but the 3,4-dihydrodiol became predominant after 24 h (data not shown). On the other hand, strain BL21AI(pSKU09) expressing Pdo2 produced almost exclusively the 3,4-dihydrodiol and only trace amounts of the 9,10-dihydrodiol.
FIG. 7.
Phenanthrene transformation by recombinant strains of E. coli overproducing Pdo2. (A) Mass spectrum and proposed structure of the major oxidation product, phenanthrene cis-3,4-dihydrodiol, recovered from a culture of strain BL21AI(pSKU09) incubated with phenanthrene. The diol product was derivatized with NBB. (B) Time course of phenanthrene transformation by strain BL21AI(pSKU09)(pBRCD) coexpressing pdoA2B2 and phdCD.
TABLE 3.
Production of pyrene and phenanthrene dihydrodiols by E. coli strains overproducing the two dioxygenases from Mycobacterium sp. strain 6PY1a
| Strain | Gene product(s) | Concn of phenanthrene cis-3,4-dihydrodiol (ppm)b | Concn of phenanthrene cis-9,10-dihydrodiol (ppm)b | Concn of pyrene cis-4,5-dihydrodiol (ppm)b |
|---|---|---|---|---|
| BL21AI(pSKU06) | Pdo1 | 0.135 | 0.317 | 0.415 |
| BL21AI(pSKU09) | Pdo2 | 0.913 | 0.00074 | 0.00893 |
| BL21AI(pSKU06)(pBRCD) | Pdo1 and PhdCD | 0.0875 | 0.165 | 0.288 |
| BL21AI(pSKU09)(pBRCD) | Pdo2 and PhdCD | 5.80 | 0.00164 | 0.0187 |
The amount of PAH added to cultures was equivalent to 25 mg/liter of aqueous medium.
The concentrations of dihydrodiols in the growth medium were determined after 6 h of incubation in the presence of the respective PAHs, as described in Materials and Methods. The GC retention times of the TMS derivatives were 18.4 min for phenanthrene cis-3,4-dihydrodiol, 17.5 min for phenanthrene cis-9,10-dihydrodiol, and 20.5 min for pyrene cis-4,5-dihydrodiol.
In these experiments, the activities of Pdo1 and Pdo2 measured in recombinant strains were dependent on a source of reductant delivered by the host. As the E. coli electron carriers might not be as efficient as those from strain 6PY1 in providing the two dioxygenase terminal components with reductant, the observed activities might have been underestimated. This hypothesis was tested by using strains which coexpressed Pdo1 or Pdo2 together with the reductase and ferredoxin components of the phenanthrene dioxygenase from Nocardioides strain KP7 (Table 3). Expression of phdC and pdhD from strain KP7 in strain BL21AI(pSKU09)(pBRCD) resulted in a sixfold increase in the rate of phenanthrene dihydrodiol formation and a twofold increase in the rate of pyrene dihydrodiol formation (Table 3). Using this strain, the rate of phenanthrene removal from the medium was shown to follow first-order kinetics (Fig. 7B), and this finding was correlated with a corresponding accumulation of 3,4-dihydrodiol (data not shown). On the other hand, expression of phdC and pdhD in strain BL21AI(pSKU06)(pBRCD) did not result in a significant increase in Pdo1 activity (Table 3).
Differential expression of the two dioxygenases in strain 6PY1.
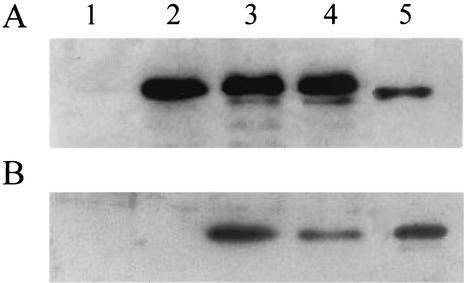
Polyclonal antibodies were raised against the purified large subunits of Pdo1 and Pdo2 and used to detect the presence of each dioxygenase in cell extracts from strain 6PY1. Control immunoblot analysis revealed that the Pdo2 large subunit cross-reacted with the anti-Pdo1α antibodies but that the Pdo1 subunit did not cross-react with anti-Pdo2α. Western blot analysis of strain 6PY1 cell protein extracts prepared from cultures grown with four different C sources indicated that Pdo1 was produced provided that bacteria were supplied with an aromatic compound but that it was undetectable in acetate-grown cells (Fig. 8A). In addition, a faint band that had the same mobility as Pdo2α was detected in the pyrene and phenanthrene extracts. Analysis of the same protein extracts with anti-Pdo2α antibodies revealed that the Pdo2 enzyme was detectable only in PAH-grown cells (Fig. 8B). These findings indicated that Pdo1 synthesis was induced by a single-ring aromatic compound or a PAH but that the Pdo2 biosynthesis required the presence of a PAH. This result is consistent with the results of 2D gel analyses, showing that Pdo1 was detectable in cells induced with each of the three aromatic substrates but that Pdo2 was detectable only in PAH-induced cells (Fig. 4).
FIG. 8.
Immunoblot detection of Pdo1 and Pdo2 in cell extracts of strain 6PY1. Lanes were loaded with the same amounts of cell proteins prepared from bacteria grown on acetate (lane 1), benzoate (lane 2), phenanthrene (lane 3), and pyrene (lane 4). After separation by SDS-PAGE and electrotransfer onto nitrocellulose, proteins were immunodetected by using either anti-Pdo1α (A) or anti-Pdo2α (B) polyclonal antibodies. Lane 5 was loaded with 0.4 μg of Pdo2.
DISCUSSION
In this study, proteins involved in pyrene degradation were thoroughly investigated by using a combination of in vivo protein-labeling experiments and 2D-PAGE analyses. This approach led to the identification of a relatively limited number of pyrene-specific polypeptides, most of which were also detected upon induction with phenanthrene. This finding suggests that the same enzymes are involved in the catabolism of the two PAHs. Accordingly, phenanthrene-grown cells were found to be as active as pyrene-grown cells in mineralizing pyrene (Fig. 2). Likewise, Molina et al. observed that when Mycobacterium strains were exposed to phenanthrene, de novo protein synthesis was not required for rapid mineralization of pyrene (31). In addition, a majority of the pyrene-specific proteins found in strain 6PY1 were similar in sequence to enzymes responsible for phenanthrene degradation in Nocardioides strain KP7 (Table 2). Furthermore, it is remarkable that most bacterial strains able to grow on pyrene that have been described so far were also found to metabolize phenanthrene (2, 4, 7, 19, 21, 37, 40). The fact that the metabolisms of phenanthrene and pyrene share common enzymes is also consistent with the current knowledge of the catabolic pathways of these PAHs (Fig. 1). From the identification of the metabolites produced by various pyrene degraders, essentially Mycobacterium species, it was established that pyrene was initially hydroxylated on the C4 and C5 positions, followed by dehydrogenation and ortho cleavage to give phenanthrene 4,5-dicarboxylic acid (7, 15, 37, 39). This intermediate is further transformed to 3,4-dihydroxyphenanthrene, which is subsequently decomposed to phthalic acid through the action of enzymes known to be involved in phenanthrene degradation (6, 7). It is likely that strain 6PY1 decomposes pyrene through the same succession of reaction steps because the key intermediate metabolites have been identified in pyrene-grown cultures (Willison, unpublished). Moreover, we have identified, on the basis of sequence similarities, four of the seven enzymes responsible for the conversion of 3,4-dihydroxyphenanthrene to phthalic acid, i.e., proteins P13, P15, P18, and P22 (Fig. 1 and Table 2). The question arises of whether the set of enzymes responsible for phenanthrene degradation is sufficient to metabolize pyrene or whether additional enzymes are required, especially for the initial steps of pyrene degradation. We have identified two ring-hydroxylating dioxygenases, one of which likely initiates the oxidative degradation of pyrene. We have demonstrated that either enzyme could convert this PAH into the corresponding 4,5-dihydrodiol. However, given the fact that the recombinant Pdo1 enzyme showed considerably greater activity than Pdo2 in this reaction, the former enzyme probably plays a major role in the oxidative attack of pyrene.
The second step would be catalyzed by a dihydrodiol dehydrogenase. An enzyme of such type was detected in phenanthrene-grown cells, but we do not know whether this enzyme is also present in pyrene-grown cells and whether it could use pyrene 4,5-dihydrodiol as a substrate. The next three steps, involving ortho cleavage of 4,5-dihydroxypyrene, decarboxylation of 4,5-phenanthrene dicarboxylic acid, and dihydroxylation of 4-phenanthroic acid, are catalyzed by unknown enzymes. Perhaps components of such enzymes might be found among the pyrene-specific polypeptides which remain to be identified (Fig. 3 and 4; Table 2). Alternatively, we might have failed to detect some of these enzymes due to limitations of the approach employed based on 2D-PAGE analysis. Especially, proteins of interest could have passed undetected if they were membrane bound or synthesized at very low levels. This is probably the case for the reductase and the ferredoxin, which are expected to associate with the Pdo1 and Pdo2 terminal components to form active dioxygenase complexes.
The present study demonstrates that the biosynthesis of the catabolic enzymes responsible for PAH degradation in strain 6PY1 is under strict metabolic control. In acetate-grown cells, no pyrene mineralization activity was detected and many proteins identified as PAH catabolic enzymes were absent. Likewise, pyrene degradation enzymes were found to be inducible in Mycobacterium strain PYR-1 (14). Several enzymes involved in PAH degradation, including Pdo1 and proteins which appear to be specific to the catabolism of phenanthrene (proteins P13, P15, P18, and P22), were biosynthesized at high levels in benzoate-grown cells (Fig. 4D). This result suggested that the synthesis of these enzymes is induced by a metabolite formed upon degradation of aromatic hydrocarbons rather than by PAHs. A few polypeptides, including the two subunits of Pdo2, were synthesized only in the presence of a PAH, indicating that they are subjected to a regulation mechanism different from that controlling the enzymes induced by benzoate. Finally, we observed that some polypeptides were detectable only in pyrene-induced cells and that other proteins, including a dihydrodiol dehydrogenase, were found exclusively or predominantly in phenanthrene-grown cells. Hence, the synthesis of PAH catabolic enzymes appears to involve several regulatory mechanisms which remain to be elucidated in future studies.
The two dioxygenase terminal components described herein were found to be related to enzymes found in four actinomycete species, including two Rhodococcus strains (27, 38), Nocardioides strain KP7 (34), and Mycobacterium strain PYR-1 (24). In previous reports, phylogenetic analysis indicated that these dioxygenases formed a cluster of enzymes distinct from other dioxygenases (24, 34). Hence, the Pdo1 and Pdo2 enzymes from strain 6PY1 are two new members of the same phylogenetic group. As judged from in vivo activity assays, the two dioxygenases showed markedly different selectivities towards pyrene and phenanthrene. Pdo1 converted pyrene to pyrene cis-4,5-dihydrodiol much more efficiently than did Pdo2, whereas the latter enzyme displayed higher activity in the oxidation of phenanthrene. It might be inferred that in strain 6PY1 the initial attack of each PAH substrate involved a specialized enzyme, Pdo1 for pyrene and Pdo2 for phenanthrene. Interestingly, the dioxygenase most similar to Pdo1 was shown to oxidize pyrene (24), whereas the enzyme with closest similarity to Pdo2 was described as a phenanthrene dioxygenase (34). The Pdo2-catalyzed oxidation of phenanthrene yielded almost exclusively the 3,4-dihydrodiol, which is the natural substrate of the cis-dihydrodiol dehydrogenase, consistent with the current pathway of phenanthrene degradation (Fig. 1). Pdo1 also generated 9,10-dihydroxyphenanthrene, which is probably not metabolized further. This metabolite, but not the 3,4-dihydrodiol, was detected in the culture medium of strain 6PY1 growing on phenanthrene (unpublished results), indicating that it is probably formed as a dead-end product, mainly as a result of Pdo1 activity.
As for other ring-hydroxylating dioxygenases, Pdo1 and Pdo2 certainly require electron carrier proteins for catalysis, namely an NAD(P)H reductase and a ferredoxin. Since those recombinant enzymes showed detectable activity in vivo, electron carriers from E. coli certainly replace at least partially the dioxygenase-specific electron carriers. Likewise, the terminal dioxygenase components from Burkholderia sp. strain RP007 (28), Pseudomonas abietaniphila BKME-9 (30), Rhodococcus sp. strain 19070 (12), and Terrabacter sp. strain DBF63 (23) were found to be active in E. coli with the complementation of nonspecific ferredoxin and reductase components from the host. On the other hand, the dioxin dioxygenase from Sphingomonas sp. strain RW1 was active in E. coli only when the appropriate electron carrier proteins were simultaneously expressed (1). As for the two dioxygenase components described in this study, the source of reductant provided by E. coli was probably suboptimal, as judged from the stimulation of Pdo2 catalytic activity observed in the recombinant strain coexpressing the PhdC and PhdD electron carriers from strain KP7. Further characterization of the Pdo1 and Pdo2 enzymes from strain 6PY1 will require identification of their cognate electron carrier proteins.
Acknowledgments
We thank S. Harayama for providing plasmid pHA171.
This work was supported in part by a grant from the Agence de l'Environnement et de la Maîtrise de l'Energie and by grants from the Centre National de la Recherche Scientifique and the Commissariat à l'Energie Atomique.
REFERENCES
- 1.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldrin, B., A. Tiehm, and C. Fritzsche. 1993. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl. Environ. Microbiol. 59:1927-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boonchan, S., M. L. Britz, and G. A. Stanley. 1998. Surfactant-enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia. Biotechnol. Bioeng. 59:482-494. [DOI] [PubMed] [Google Scholar]
- 4.Bouchez, M., D. Blanchet, and J. P. Vandecasteele. 1995. Degradation of polycyclic aromatic hydrocarbons by pure strains and by defined strain associations: inhibition phenomena and cometabolism. Appl. Microbiol. Biotechnol. 43:156-164. [DOI] [PubMed] [Google Scholar]
- 5.Cerletti, P. 1986. Seeking a better job for an under-employed enzyme: rhodanese. Trends Biochem. Sci. 11:369-372. [Google Scholar]
- 6.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 7.Dean-Ross, D., and C. E. Cerniglia. 1996. Degradation of pyrene by Mycobacterium flavescens. Appl. Microbiol. Biotechnol. 46:307-312. [DOI] [PubMed] [Google Scholar]
- 8.Ensley, B. D., and D. T. Gibson. 1983. Naphthalene dioxygenase: purification and properties of a terminal oxygenase component. J. Bacteriol. 155:505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferro, M., D. Seigneurin-Berny, N. Rolland, A. Chapel, D. Salvi, J. Garin, and J. Joyard. 2000. Organic solvent extraction as a versatile procedure to identify hydrophobic chloroplast membrane proteins. Electrophoresis 21:3517-3526. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-y-Merchand, J. A., I. Estrada-Garcia, M. J. Colston, and R. A. Cox. 1996. A novel method for the isolation of mycobacterial DNA. FEMS Microbiol. Lett. 135:71-77. [DOI] [PubMed] [Google Scholar]
- 11.Görg, A., W. Postel, and S. Gunther. 1988. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 9:531-546. [DOI] [PubMed] [Google Scholar]
- 12.Haddad, S., D. M. Eby, and E. L. Neidle. 2001. Cloning and expression of the benzoate dioxygenase genes from Rhodococcus sp. strain 19070. Appl. Environ. Microbiol. 67:2507-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haddock, J. D., and D. T. Gibson. 1995. Purification and characterization of the oxygenase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heitkamp, M. A., W. Franklin, and C. E. Cerniglia. 1988. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl. Environ. Microbiol. 54:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heitkamp, M. A., J. P. Freeman, D. W. Miller, and C. E. Cerniglia. 1988. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl. Environ. Microbiol. 54:2556-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwabuchi, T., and S. Harayama. 1998. Biochemical and genetic characterization of trans-2′-carboxybenzalpyruvate hydratase-aldolase from a phenanthrene-degrading Nocardioides strain. J. Bacteriol. 180:945-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwabuchi, T., and S. Harayama. 1998. Biochemical and molecular characterization of 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. KP7. J. Biol. Chem. 273:8332-8336. [DOI] [PubMed] [Google Scholar]
- 18.Jouanneau, Y., C. Meyer, I. Naud, and W. Klipp. 1995. Characterization of an fdxN mutant of Rhodobacter capsulatus indicates that ferredoxin I serves as electron donor to nitrogenase. Biochim. Biophys. Acta 1232:33-42. [DOI] [PubMed] [Google Scholar]
- 19.Juhasz, A. L., M. L. Britz, and G. A. Stanley. 1997. Degradation of fluoranthene, pyrene, benz[a]anthracene and dibenz[a,h]anthracene by Burkholderia cepacia. J. Appl. Microbiol. 83:189-198. [Google Scholar]
- 20.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kastner, M., M. Breuer-Jammali, and B. Mahro. 1994. Enumeration and characterization of the soil microflora from hydrocarbon-contaminated soil sites able to mineralize polycyclic aromatic hydrocarbons (PAH). Appl. Microbiol. Biotechnol. 41:267-273. [Google Scholar]
- 22.Kastner, M., M. Breuer-Jammali, and B. Mahro. 1998. Impact of inoculation protocols, salinity, and pH on the degradation of polycyclic aromatic hydrocarbons (PAHs) and survival of PAH-degrading bacteria introduced into soil. Appl. Environ. Microbiol. 64:359-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasuga, K., H. Habe, J. S. Chung, T. Yoshida, H. Nojiri, H. Yamane, and T. Omori. 2001. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the Gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophys. Res. Commun. 283:195-204. [DOI] [PubMed] [Google Scholar]
- 24.Khan, A. A., R. F. Wang, W. W. Cao, D. R. Doerge, D. Wennerstrom, and C. E. Cerniglia. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:3577-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosono, S., M. Maeda, F. Fuji, H. Arai, and T. Kudo. 1997. Three of the seven bphC genes of Rhodococcus erythropolis TA421, isolated from a termite ecosystem, are located on an indigenous plasmid associated with biphenyl degradation. Appl. Environ. Microbiol. 63:3282-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 27.Larkin, M. J., C. C. Allen, L. A. Kulakov, and D. A. Lipscomb. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 30.Martin, V. J. J., and W. W. Mohn. 1999. A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J. Bacteriol. 181:2675-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina, M., R. Araujo, and R. E. Hodson. 1999. Cross-induction of pyrene and phenanthrene in a Mycobacterium sp. isolated from polycyclic aromatic hydrocarbon contaminated river sediments. Can. J. Microbiol. 45:520-529. [PubMed] [Google Scholar]
- 32.Parales, R. E., K. Lee, S. M. Resnick, H. Y. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito, A., T. Iwabuchi, and S. Harayama. 2000. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: expression in Escherichia coli. J. Bacteriol. 182:2134-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, J., R. Grosser, K. Jayasimhulu, W. Xue, and D. Warshawsky. 1996. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl. Environ. Microbiol. 62:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treadway, S. L., K. S. Yanagimachi, E. Lankenau, P. A. Lessard, G. Stephanopoulos, and A. J. Sinskey. 1999. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl. Microbiol. Biotechnol. 51:786-793. [DOI] [PubMed] [Google Scholar]
- 39.Vila, J., Z. Lopez, J. Sabate, C. Minguillon, A. M. Solanas, and M. Grifoll. 2001. Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp. strain AP1: actions of the isolate on two- and three-ring polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 67:5497-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter, U., M. Beyer, J. Klein, and H.-J. Rehm. 1991. Degradation of pyrene by Rhodococcus sp. UW1. Appl. Microbiol. Biotechnol. 34:671-676. [Google Scholar]
- 41.Wilson, S. C., and K. C. Jones. 1993. Bioremediation of soil contaminated with polynuclear aromatic hydrocarbons (PAHs): a review. Environ. Pollut. 81:229-249. [DOI] [PubMed] [Google Scholar]
- 42.Yen, K. M., and C. M. Serdar. 1988. Genetics of naphthalene catabolism in pseudomonads. CRC Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]