Abstract
Summary:
Nexplorer is a web-based program for interactive browsing and manipulation of character data in NEXUS format, well suited for use with alignments and trees representing families of homologous genes or proteins. Users may upload a sequence family data set, or choose from one of several thousand already available. Nexplorer provides a flexible means to develop customized views that combine a tree and a data matrix or alignment, to create subsets of data, and to output data files or publication-quality graphics.
Availability:
Web access is from http://www.molevol.org/nexplorer
Contact:
arlin.stoltzfus@nist.gov
INTRODUCTION
Computational genomics depends increasingly on the comparative analysis of sequences and other data in a phylogenetic context. The task of organizing and analyzing data by way of a phylogenetic tree requires appropriate tools. While various programs exist to plot and manipulate phylogenetic trees, few researchers are interested in viewing and manipulating trees for their own sake. Instead, the usual purpose of manipulating a tree, e.g., re-rooting, selecting a sub-tree, re-ordering branches, etc., is to organize the data that are conceptually linked to the tips of the tree, as an aid to data exploration and analysis. Of interest, then, are software tools to visualize and manipulate trees and data together. An early program that achieved notable success in this regard is MacClade (http://macclade.org), some of the functionality of which is incorporated in the Mesquite Java library (http://mesquiteproject.org). To aid in phylogeny-based analysis of comparative data, we have developed Nexplorer, a web-based program that combines a useful set of features for viewing and manipulating data with the capacity to generate publication-quality graphics of a tree with a sequence alignment or other data.
FEATURES
A Nexplorer session begins by uploading a set of data or choosing an available set. The user then generates a view of the data, optionally manipulates the data, optionally carries out further rounds of visualization and manipulation, and generates output in the form of a data file (to save alterations) or a graphics file.
Input data: Users can upload their own data in the NEXUS file format that has been used successfully by molecular systematists for many years (Maddison, et al., 1997). Alternatively, the Nexplorer server provides pre-assembled data sets, including value added versions of 684 KOGs families (Sverdlov, et al., 2005) and of 7226 families from Pandit release 12.0 (Whelan, et al., 2003). Each family has a protein sequence alignment, a corresponding nucleotide sequence alignment, a matrix of intron data, a phylogeny, and meta-data including taxonomic identifiers. A simple search interface is provided to identify families of interest.
Viewing and manipulating data: Fig. 1 shows a subtree selected from a family of ATP Synthase subunit C proteins (Pfam 00137), along with a matrix of intron data from the corresponding genes. The two most useful options for customizing such a view are to assign a taxonomic coloring scheme, and to choose which set of data to view. The choice of data depends on which CHARACTERS blocks are in the input file: the pre-assembled data sets have protein, nucleotide, and intron blocks. Taxonomic coloring is highly useful for identifying paralogy, horizontal transfer, and misplaced branches due to errors in phylogenetic inference. Other options affect the width and vertical spacing of the tree, whether to display internal node names and branch support values, and whether branch lengths are to be used (cladogram mode). The default output is a clickable image with hyperlinks (e.g., allowing the user to trace data to an external database such as GenBank) and popup menus that provide options for re-rooting, coloring, ordering branches, and selecting or excluding a sub-tree. Some options, such as re-rooting a tree or excluding a sub-tree, change not only the view, but the underlying data object, a NEXUS object represented using the NEXPL library (Liang, et al., 2005). Users interested in automation, or in a greater range of options for manipulating data, should install the command-driven program nextool included in the NEXPL library.
Fig. 1.
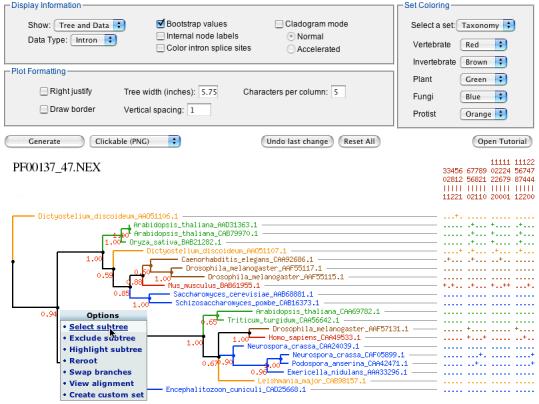
Screenshot showing the Nexplorer interface with intron data for a subset of the ATP Synthase Subunit C family, with control panel (above), data matrix (right), and gene phylogeny (left). The pop-up menu shows node-specific operations. Taxonomic coloring (key, upper right) reveals sub-trees representing paralogous sub-families, each with genes from plants, animals, fungi and protists (custom color schemes can be applied based on user-defined sets in the input file, which will appear automatically in the “Set Coloring” box). The data matrix shows the presence (“+”) or absence (“.”) of introns at the 19 positions listed above the matrix in codon-phase notation (e.g., “30-1” indicates codon 30, phase 1). The red numbers at nodes are branch support values (here, Bayesian posterior probabilities). This screenshot shows interface elements but has limited resolution: for unlimited resolution, the user may select PostScript instead of “Clickable (PNG)”.
Output options: The default output is a PNG image-map. To generate publication-quality graphics, the user may select PostScript or PDF output. If NEXUS output is chosen, a data file is generated that reflects any user modifications such as re-rooting.
Nexplorer also implements specialized features that include the ability to assign colors to arbitrary sets of entities specified in the SETS block of the input file, to display a histogram of weights (for alignment columns) specified in an ASSUMPTIONS block, and to display ancestral character states data from a HISTORY block.
IMPLEMENTATION
Nexplorer has a three-tiered architecture. The user interface is a Perl CGI application (run by the Apache2 server under Linux 2.4.21), utilizing menus and hyperlinks mapped to PNG images, and requiring a JavaScript-2-compliant browser. The middle layer makes extensive use of NEXPL, a NEXUS API in Perl (Liang, et al., 2005). The back-end consists mainly of NEXUS files and a database that links sequence identifiers with a taxonomic hierarchy.
ACKNOWLEDGEMENTS
This work was supported by NIH grant R01-LM007218 to A.S and NIH grant GM060654 to W.G.Q. The identification of specific commercial software products is for the purpose of specifying a protocol, and does not imply a recommendation or endorsement by the National Institute of Standards and Technology.
REFERENCES
- Liang CL, Yang PJ, Hladish T, Stoltzfus A. NEXPL: a NEXUS Applications Programming Interface in Perl. 2005 http://www.molevol.org/camel/software. Available at.
- Maddison DR, Swofford DL, Maddison WP. NEXUS: an extendible file format for systematic information. Systematic Biology. 1997;46:590–621. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- Sverdlov AV, Rogozin IB, Babenko VN, Koonin EV. Conservation versus parallel gains in intron evolution. Nucleic Acids Res. 2005;33:1741–1748. doi: 10.1093/nar/gki316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, de Bakker PI, Goldman N. Pandit: a database of protein and associated nucleotide domains with inferred trees. Bioinformatics. 2003;19:1556–1563. doi: 10.1093/bioinformatics/btg188. [DOI] [PubMed] [Google Scholar]


