Abstract
LacS− mutants of Sulfolobus solfataricus defective in β-glycosidase activity were isolated in order to explore genomic instability and exploit novel strategies for transformation and complementation. One of the mutants showed a stable phenotype with no reversion; analysis of its chromosome revealed the total absence of the β-glycosidase gene (lacS). Fine mapping performed in comparison to the genomic sequence of S. solfataricus P2 indicated an extended deletion of ∼13 kb. The sequence analysis also revealed that this chromosomal rearrangement was a nonconservative transposition event driven by the mobile insertion sequence element ISC1058. In order to complement the LacS− phenotype, an expression vector was constructed by inserting the lacS coding sequence with its 5′ and 3′ flanking regions into the pEXSs plasmid. Since no transformant could be recovered by selection on lactose as the sole nutrient, another plasmid construct containing a larger genomic fragment was tested for complementation; this region also comprised the lacTr (lactose transporter) gene encoding a putative membrane protein homologous to the major facilitator superfamily. Cells transformed with both genes were able to form colonies on lactose plates and to be stained with the β-glycosidase chromogenic substrate X-Gal (5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside).
The recent availability of the Sulfolobus solfataricus P2 genome (36), as well as other archaeal genome sequences (8, 11, 14, 19, 20, 37), has provided a basis for detailed analyses of the frequencies, locations, and phylogenies (18, 23) of several mobile elements. Autonomous insertion sequence (IS) elements (7) and the nonautonomous miniature inverted repeat-like elements (28) were the main types identified, 8% of them belonging to still unclassified families and hence supposed to be archaeon specific. These elements, which constitute ∼10% of the entire genome, were found to be spread out but also mainly clustered in two broad areas (replicore 1 and replicore 2) separated by the putative oriC and terC (replication origin and termination) regions (7); these sequences, fundamental for replication, should act as barriers for the mobility of both classes of transposable elements.
The transposable elements ISC1058, ISC1217, ISC1359, and ISC1439 have been shown to be active, able to spread and disrupt functional genes (24) by spontaneous excision-insertion or copying-insertion events; it has been proposed that they are probably mobilized by integrase and excisionase activities, often encoded by the same transposase open reading frame (ORF) (7, 28, 30) located on the IS elements themselves. Moreover, putative transposases and at least one insertion element have also been found in the sequence of the promiscuous conjugative plasmid pNOB8 of a Japanese Sulfolobus sp., a close relative of S. solfataricus (34, 35), and in strain MT-4 (2). Together, these data suggested that several other IS elements might be active and responsible for insertion-mediated adaptive mutagenesis in Sulfolobus.
Spontaneous β-galactosidase mutants (17, 33) have been shown to arise with relatively high frequencies of ∼10−4 per plated cell (33). These mutants contained a transposable element in the lacS gene with features typical of bacterial and archaeal ISs, including terminal inverted repeats, a putative transposase gene, and short direct flanking repeats. Like Escherichia coli, S. solfataricus forms blue colonies upon exposure to X-Gal (5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside). Disruption of lacS by insertion element transposition results in the formation of colorless colonies on X-Gal (33). In fact, the S. solfataricus lacS gene (12) encodes a β-glycosidase (Ssβ-gly) with broad substrate specificity, including activity against β-galactosides (26) and their chemical analogs, such as the chromogenic indicator X-Gal. Moreover, S. solfataricus is able to grow on lactose and cellobiose (16), which are potential substrates for Ssβ-gly, but lacS gene function and regulation in vivo have not been investigated in great detail.
We were interested in exploiting the genomic instability of S. solfataricus by selecting spontaneous stable lacS gene mutants that can serve as complementable recipients in selectable genetic systems. Recently, some progress has been achieved in developing gene transfer strategies, appropriate vector-host transformation systems, mutant production, and screening methods. In this respect, S. solfataricus has been demonstrated to be a versatile model for the manipulative genetics of crenarchaea, based on the isolation of mobile introns (1), as well as several plasmids and viruses (42), and on the demonstration of genetic transformation of the organism (1, 3, 4, 9, 10, 13, 38).
In this study, we show that a chromosomal rearrangement, mediated by transposition of an IS element, was responsible for an extended deletion in the lacS genetic locus of a spontaneous mutant of S. solfataricus. Moreover, the cell requirement for a putative sugar transporter (27), in addition to the lacS-encoded β-glycosidase for lactose metabolism, was demonstrated by complementation of the lacking function, using a suitable Sulfolobus vector and the lacS gene as an effective genetic marker for the transformation tests.
MATERIALS AND METHODS
Growth conditions of S. solfataricus Gθ and isolation of GθW, Gθwb1, and Gθwb2 mutants.
S. solfataricus Gθ (9) was grown at 75°C and pH 3.8 in mineral medium supplemented with 0.1% yeast extract (Difco), 0.1% Casamino Acids, and 0.1% d-glucose (medium A) or 0.2% tryptone (Oxoid) and 0.2% lactose (medium B). Gelrite (0.8%; Kelco, San Diego, Calif.) was added to solidify the media for plating the cells embedded in soft gelrite layers as described previously (16). For specific selection, cells were also grown in minimal medium, which contained only 0.2% lactose as a nutrient source.
The Gθ strain was grown in 50-ml liquid culture to an A600 of 0.5 to 0.7 (mid-exponential phase), and ∼5 × 105 cells were plated onto solid medium B. The plates were incubated for 5 to 7 days, and the colonies were stained by overlaying them with X-Gal solutions (2 ml per 10-cm-diameter plate of 2-mg/ml X-Gal in medium B) and incubating them at 75°C for 1 h. Spontaneous mutants were isolated, propagated in the same medium, and reisolated as single colonies after being streaked on fresh plates and stained with X-Gal (33).
Isolation of chromosomal DNA and PCR amplification analyses.
Total genomic DNA was extracted from 50-ml cultures of S. solfataricus cells, essentially as described by Martusewitsch et al. (24), with suitable modification for larger preparations. For mutation analysis, 50-ng amounts of such preparations were used as templates in PCR amplifications of the lacS gene with primers IVS1 (5′-GTTTTCCCCCAGGATACCTAAGCTTTG-3′, at positions 662 to 688) and IVS2 (5′-CTCGTTTCCTCTGGTGATCTCACCTC-3′, at positions 884 to 909). Alternatively, another pair of primers corresponding to the 5′ (lacFw; 5′-TACTCATTTCCAAATAGCTTTAGG-3′) and 3′ (lacRe; 5′-TAGTGAGACTTGAGAAAGTTTAGT-3′) ends of the entire coding sequence were used. The reaction was carried out for 35 cycles at a 50°C annealing temperature and using Taq DNA polymerase and chromosomal DNAs from the wild-type and mutant LacS− strains as templates, following a general protocol (31). The amplicons were analyzed by 0.8% agarose gel electrophoresis in Tris-borate-EDTA buffer.
16S ribosomal DNAs (rDNAs) were amplified from genomic DNAs of both the parental Gθ and the deletion mutant (named GθW), using the same protocol; the primers were designed against the positions 119 to 141 and 1370 to 1392 of the corresponding gene sequence of strain P2, identified as sso16SRNA on the whole sequenced genome (http://www-archbac.u-psud.fr/projects/sulfolobus/). The amplified fragments were sequenced by MWG-BIOTECH AG (Ebersberg, Germany) and compared to other 16S rDNA sequences from other Sulfolobus species using the Ribosomal Database Project Phylip interface program, available at the Ribosomal Database Project II web site (http://rdp.cme.msu.edu/cgis/phylip.cgi).
Southern and boundary sequence analyses of the deletion in the mutant Gθ chromosome.
Approximately 5 to 10 μg of chromosomal DNAs were cut with EcoRI and XbaI and then electrophoresed in a 0.8% agarose gel; the DNA digests were blotted and hybridized according to standard procedures (31). The probes were prepared from a recombinant pGEM3 vector containing a 15-kb genomic region, encompassing the lacS gene and extended 5′ and 3′ flanking sequences (see Fig. 3 and 4) from the strain Gθ. These DNA restriction fragments were randomly labeled using the Random Prime DNA labeling kit (Boehringer Mannheim).
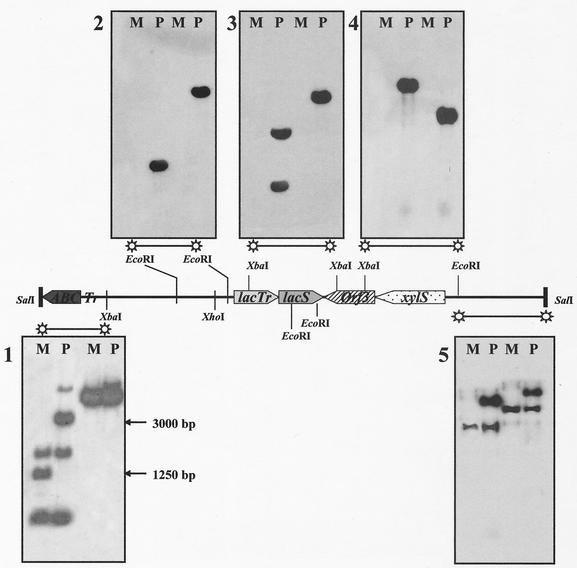
FIG. 3.
Comparative Southern analysis of the wild-type and mutant lacS loci. Chromosomal DNAs digested with EcoRI (two left lanes of each gel) and XbaI (two right lanes of each gel), identifying the lacS locus in S. solfataricus Gθ (P) and in the LacS− mutant GθW (M), were hybridized with DNA fragments progressively covering the upstream and downstream regions of the lacS coding sequence in independent experiments. The extended 15-kbp region used as a source of these probes is represented in the middle as a solid bar containing already-identified ORFs (xylS, lacS, and lacTr) or putative protein-encoding sequences (ABC, ABC transporter) in their relative orientations. On the left, two arrows indicate the sizes corresponding to the differential cross-hybridization signals on the EcoRI digests hybridized with the extreme 5′ DNA fragment used in the analysis.
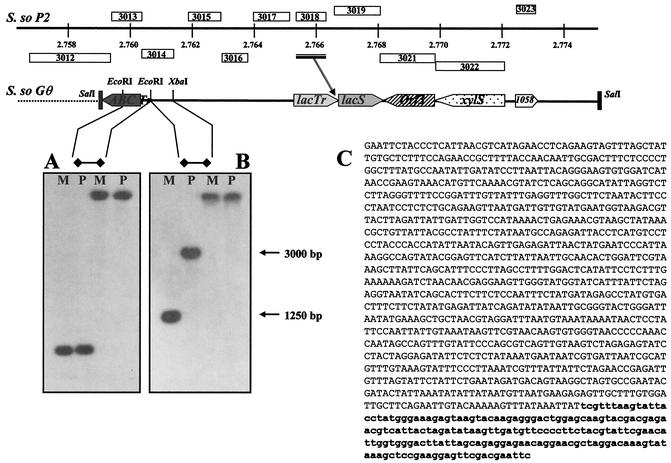
FIG. 4.
Fine mapping of the deletion join end. On top is shown the schematic alignment of the lacS loci, considered in this study, on the P2 (S.so P2; positions are indicated in mega-base pairs) and Gθ (S. so Gθ) Sulfolobus chromosomes. The main difference resulted from the insertion of an ISC element (sso3018) in the 3′ end of ORF sso3017 (the bar and arrow indicate the position of the insertion), namely, the lacTr gene, encoding a putative lactose transporter. The extreme 5′ DNA fragment of the 15-kbp region used for Southern analysis was dissected to produce two probes that showed one-band cross-hybridization patterns (A and B), on both the wild-type (P) and mutant (M) genomic DNAs, digested with EcoRI (two left lanes of each gel) and XbaI (two right lanes of each gel). The EcoRI DNA fragment from the mutant genomic DNA corresponding to the 1,250-bp band was cloned, sequenced (C), and compared with the full sequence of the S. solfataricus P2. The larger region of the fragment was identical to part of ORF sso3012, encoding a putative ABC transporter (uppercase letters), abruptly interrupted by a shorter DNA element (lowercase boldface letters) showing identity with multiple repetitive sequences found on the P2 genome and indicated as transposase-coding ICS1058 elements (sso3023).
A 1,250-bp EcoRI fragment from the mutant GθW chromosome, supposed to contain the 5′ joining end of the deletion, was isolated from a plasmid sublibrary of EcoRI genomic fragments in the size range of 1,100 to 1,300 bp, established in the pGEM7Zf(+) vector. Three thousand recombinant clones were screened by colony hybridization under standard procedures, using the same XbaI/EcoRI 32P-labeled fragment which produced the 1,250-bp band in Southern analysis (see Fig. 4). DNA inserts from three positive clones were sequenced by MWG-BIOTECH AG and analyzed for matching sequences on the whole P2 strain genome (http://www-archbac.u-psud.fr/projects/sulfolobus/).
Construction of β-glycosidase expression vectors and complementation of the ΔlacS mutation.
Two genomic fragments containing the lacS gene from S. solfataricus MT-4 (12) were inserted, after suitable modification, into the NsiI site of the pEXSs plasmid (9) to produce the expression vectors pEXlacS and pEXlacOP (see Fig. 5). The first carried a genomic XbaI fragment derived from the pD22 (12) plasmid and contained the lacS coding sequence and 5′ and 3′ flanking regions of 653 and 648 bp, respectively. The second contained a larger XhoI/XbaI genomic region (lacOP) also comprising the lacTr gene with its own regulatory sequence up to 632 bp upstream from the TTG start codon; this gene encodes a putative membrane protein homologous to the major facilitator superfamily (27). The DNA constructs were transferred into S. solfataricus GθW competent cells by electroporation, following the procedure described by Schleper et al. for transferring the SSV1 virus DNA from Sulfolobus shibatae into S. solfataricus P2 (32). After a 3-h recovery at 75°C, 50% of the electroporated cells were plated onto solid mineral medium supplemented with lactose as the only nutrient; as a control for transformation, the remaining cells were plated onto medium A-gelrite plates containing 150 μg of hygromycin B/ml. For the X-Gal staining test, performed as described above, transformants from the lactose plates were propagated up to late exponential phase (A600 = 0.7 OD unit) in 10 ml of medium A containing 200 μg of hygromycin B/ml; 10-μl aliquots were spotted onto fresh medium A plates without the antibiotic; cultures of the parental Gθ and untransformed GθW were spotted as a positive and a negative control for staining, respectively.
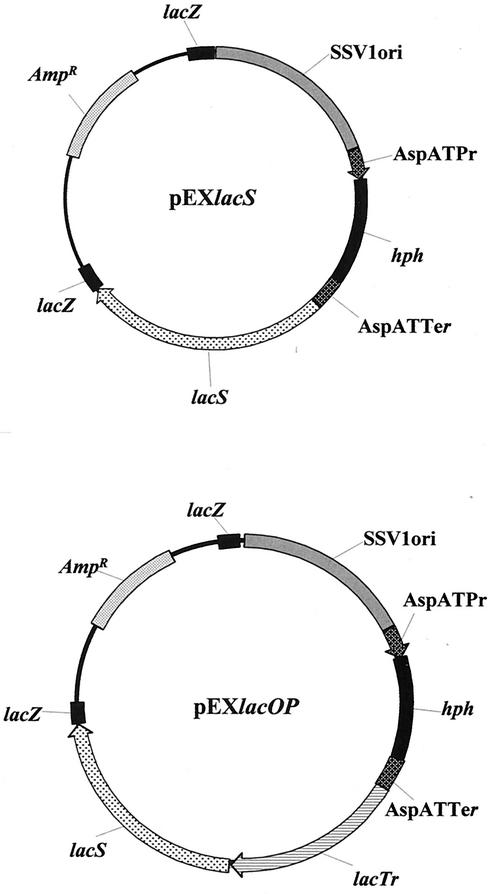
FIG. 5.
Plasmid map of the pEXSs expression vector derivatives pEXlacS and pEXlacOP. The lacS gene and a genomic fragment, encompassing both lacS and lacTr genes, were inserted into the polycloning site of the pEXSs plasmid to produce the pEXlacS and pEXlacOP DNA constructs. SsV1ORI indicates the 1,700-bp fragment carrying the autonomous replication sequence of the S. shibatae SSV1 viral genome. AspATPr and AspATTer are the promoter and terminator sequences of the S. solfataricus aspartate aminotransferase gene, respectively. hph is the E. coli randomly mutagenized hygromycin phosphotransferase gene. The E. coli pGEM5Zf(−) plasmid moiety lies between the two lacZ gene fragments and comprises the sequences necessary for propagation (ORI) and transformant selection for ampicillin resistance (Ampr) in E. coli.
Isolated transformants were also checked for quantification of β-glycosidase activity. Protein extracts were prepared according to the method of Cannio et al. (10) in 25 mM Tris-HCl- 200 mM sodium chloride, pH 7.0, from cells grown either in medium A containing 200 μg of hygromycin B/ml or in medium B without the antibiotic and harvested at early stationary phase. The enzyme assay was performed spectrophotometrically at 405 nm and 75°C on the cytosolic extracts, using p-nitrophenyl-β-d-glucopyranoside as described by Moracci et al. (26).
RESULTS
Selection and characterization of β-glycosidase-deficient mutants.
Three different spontaneous mutants of S. solfataricus Gθ defective for β-glycosidase activity were selected by screening 5 × 105 clones on lactose-tryptone plates with an in situ assay based on direct staining of colonies with X-Gal. It has been shown that wild-type S. solfataricus strain P2 colonies develop a dark-blue color when exposed to aerosols of the chromogenic substrate X-Gal (33). This color results from the activity of the S. solfataricus β-glycosidase that is encoded by lacS (12).
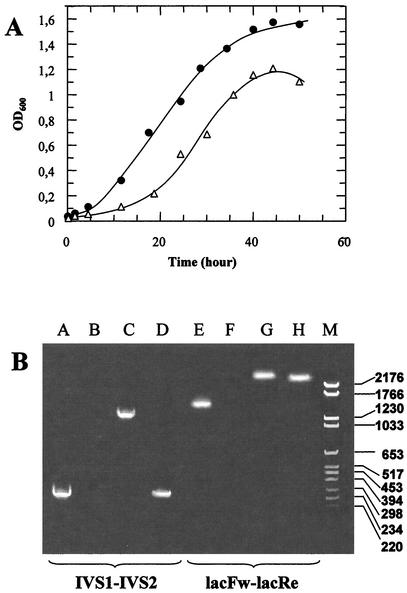
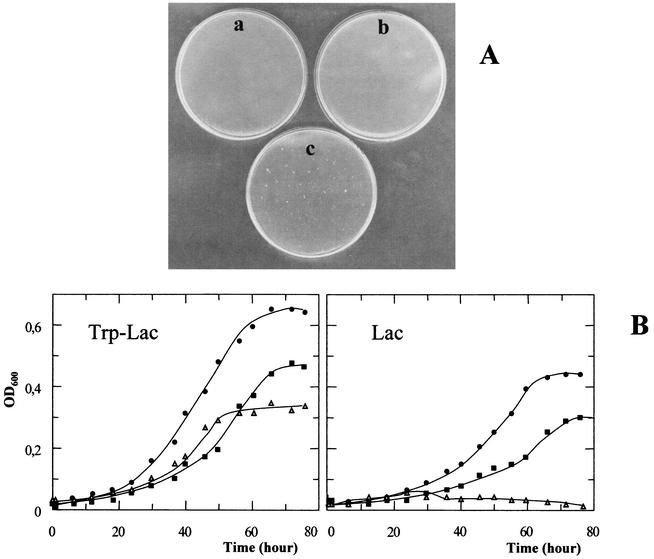
Only one clone (GθW) out of the three isolates showed a stable LacS− phenotype, i.e., even after several generations and plating of the mutant cells, the mutation remained totally nonpermissive for growth on lactose minimal medium; furthermore, all colonies generated by this mutant were colorless when screened by X-Gal staining. Reversion was observed in the cases of the other two mutants (Gθwb1 and Gθwb2), which formed colonies, 10 to 20% of which were pale to dark blue when plated and stained after isolation. The percentage of revertants for both Gθwb1 and Gθwb2 remained constant during each of three subsequent replating and reisolation cycles of colorless clones and also when tested for the ability to grow on lactose minimal medium. Interestingly, the GθW mutant was faster growing in rich medium than the wild-type Gθ, with a doubling time of 4 versus 6 h (Fig. 1A), whereas Gθwb1 and Gθwb2 did not show any difference from the parental strain.
FIG. 1.
Growth rate and PCR amplification analysis of the lacS gene in S. solfataricus Gθ and LacS− mutant derivatives. (A) Cell density was monitored at 600 nm in mineral medium supplemented with 0.1% yeast extract, 0.1% Casamino Acids, and 0.1% glucose for cultures of the wild-type Gθ (▵) and mutant GθW (•). Growth trends for the mutants Gθwb1 and Gθwb2 and the parental Gθ matched perfectly. (B) Amplicons were obtained with two pairs of primers, IVS1-IVS2 and lacFw-lacRe, designed on the wild-type DNA sequence to amplify a 248-bp internal region (lane A) or the entire 1,470-bp coding sequence (lane E). Corresponding amplification products from mutant GθW (lanes B and F), as well as Gθwb1 (lanes C and G) and Gθwb2 (lanes D and H), genomic DNAs were analyzed by agarose gel electrophoresis with reference to a mixture of pBR328 DNA cleaved with BglI and pBR328 DNA cleaved with HinfI used as a molecular weight marker (lane M). OD600, optical density at 600 nm.
The chromosomes of the three mutant isolates were analyzed for the presence of mobile elements or deletion of the lacS gene by PCR amplification of a 248-bp internal region and of the entire 1,470-bp coding sequence (Fig. 1B). The amplicons obtained from the DNAs of the Gθwb1 (internal region) and Gθwb2 (entire coding sequence) mutants were ∼1,000 bp larger than those expected for the wild-type Gθ strain, indicating the presence of IS elements, whereas no amplification was detectable for both DNA fragments from the mutant GθW chromosome. Interestingly, the insertion region of the extra DNA element in the Gθwb1 lacS gene is identical to that determined for the PH1 mutant of S. solfataricus P1 by Schleper et al. (33).
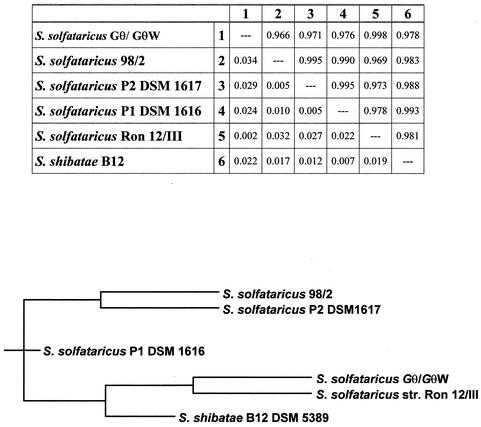
Because of the stability of the defective phenotype, the mutant GθW was chosen for further and more detailed study. As a first genetic characterization, rDNA was isolated and sequenced in order to ascertain the real derivation of the mutant GθW from the Gθ strain and to determine theevolutionary distance of the strains from other closely related Sulfolobales species. The pairwise alignment of the sequences produced the distance matrix of weighted neighbors and the derived phylogenetic tree shown in Fig. 2, where only one rDNA sequence was included. In fact, the wild-type Gθ strain sequence produced a 100% identity score in the direct alignment with that of the mutant strain. The analysis confirmed that the mutant phenotype was distinguishable and occurred spontaneously in the Gθ strain population and was not the consequence of laboratory strain contamination or coculture. Moreover the multiple alignment indicated significant nucleotide substitutions and deletions that allowed the identification of Gθ as a novel strain distinct from other well-characterized strains of S. solfataricus, such as P1 and P2, and very close but not identical to the isolate Ron 12/III isolated and characterized by Fuchs et al. (15).
FIG. 2.
Sequence analysis of Gθ-GθW rDNAs. The identical rDNA sequences amplified from both the wild-type and deletion mutant chromosomes were analyzed for distance evaluation in comparison with the five closest Sulfolobus type strains as indicated. The distance matrix of weighted neighbors and the derived phylogenetic tree were produced in the analysis performed with the Ribosomal Database Project Phylip interface program. In the matrix, values above and below the diagonal are similarity and distance indices, respectively. A similarity of 1.000 and a distance of 0.000 correspond to 100% sequence identity.
Map of the deletion in the GθW mutant.
Southern blot analysis performed in comparison with the wild-type chromosomal DNA, using an XbaI genomic fragment as the labeled probe, confirmed the complete absence, and hence deletion, of the lacS gene in the GθW strain (Fig. 3).
The mapping of the deletion in the lacS gene locus of GθW was performed by Southern blot- genome-walking analysis with contiguous probes covering a larger 15-kbp chromosomal region, comprising the lacS coding sequences and matching the corresponding locus on the P2 genome, with the only difference being a transposase ORF inserted into the sso3017/lacTr sequence (Fig. 4 shows a schematic alignment of the regions). As shown in Fig. 3, only the extreme 5′ and 3′ DNA fragments used as probes were able to cross-hybridize with distinct patterns for the wild-type and mutant genomic DNAs. This analysis revealed that GθW lacks a DNA region of ∼13 kbp that comprises (among other ORFs not characterized previously) not only lacS but also xylS (sso3022) and lacTr (sso3017) genes encoding an α-xylosidase (25, 40) and a putative membrane protein involved in sugar transport (27), respectively.
The extreme XbaI/SalI 5′ fragment (∼1,600 bp) was further dissected into three different restriction subfragments that were used as probes in independent Southern blot experiments in order to find single-band and distinguishable signals on the two genomic DNAs. The 550-bp probe obtained by restriction with EcoRI was able to reveal and distinguish single bands appearing on EcoRI digests of the parental (∼3,000-bp) and mutant (∼1,250-bp) genomic DNAs (Fig. 4B). The 1,250-bp region from the mutant chromosome was sequenced (Fig. 4C) after isolation from a plasmid sublibrary in E. coli. The search for matching sequences on the S. solfataricus P2 genome revealed that this DNA fragment contained a 1,018-bp sequence located, as expected, in the lacS locus and identical to part (positions 511 to 1528) of an ORF encoding a putative ABC transporter-ATPase (sso3012); this region was found to be fused to a shorter sequence (234 bp) with high identity (98 to 100%) to repetitive sequences spread on the P2 genome and indicated as transposase coding sequences in the insertion elements belonging to the ISC1058 family (7, 24, 28). The closest of these elements (the second ORF of the transposase element) is found upstream of the xylS gene at a distance of 13.3 kbp from the insertion point of the ISC1058 element in the ABC transporter ORF; this extension indicates a very large deletion and is in good agreement with the approximate value of 13 kb estimated for the missing sequence by Southern analysis.
lacS gene transfer and complementation of the defective β-glycosidase ΔlacS-ΔlacTr mutation.
The lacS gene was tested for the capability to restore β-glycosidase activity of the mutant GθW, namely, to complement the defective deletion genotype. The pEXSs vector (9) was used as a transfer and expression vehicle for the lacS gene and for a genomic fragment (lacOP), carrying lacS and the lacTr gene, supposed to be necessary for sugar uptake and named ORF2 by Prisco et al. (27). This gene from strain Gθ corresponds to the ORF sso3017 on the P2 strain genome, with the only difference being the insertion of a transposase ISC1439 element in the 3′ end of the latter (see the schematic alignment in Fig. 4).
The pEXSs derivative vectors obtained, pEXlacS and pEXlacOP (Fig. 5), were transferred by electroporation into Sulfolobus cells, and transformants were selected on gelrite plates containing only lactose as the nutrient or on rich medium (yeast extract, Casamino Acids, and glucose) containing the selective agent hygromycin B. Unlike the wild-type Gθ, the mutant GθW and its lacS transformant derivatives did not produce any colonies when plated onto lactose medium, demonstrating that lacS alone was unable to correct the metabolic defect. In fact, the transfer of this DNA construct was able to confer resistance to hygromycin B, and although all the drug-resistant clones analyzed expressed β-glycosidase activity (∼20-fold lower than the activity measured in the wild-type Gθ strain), they were still unable to grow on lactose.
The presence of both lacS and lacTr was necessary to sustain growth on the specific sugar and hence to complement the mutation (Fig. 6A); the average transformation efficiency calculated was ∼5 × 102 per μg of plasmid DNA for both minimal medium and rich medium-hygromicin B plates, namely, the same value (9) reported for the pEXSs vector alone. Moreover the transforming capability of the pEXSs vector was confirmed to be unaltered by insertion of larger extra sequences, since all 10 of the clones tested for transformation with both pEXlacS and pEXlacOP had acquired resistance to hygromycin B and were able to propagate when exposed to the antibiotic.
FIG. 6.
lac complementation and growth analysis of transformants. The pEXSs derivative vectors pEXlacS and pEXlacOP were transferred into the mutant GθW cells by electroporation, and transformants were selected on gelrite plates containing only lactose as a nutrient. (A) The LacS− GθW did not produce any colonies when plated onto lactose medium (negative control; plate a), and lacS transformants failed to grow, maintaining the lactose metabolic defect (plate b), whereas transformation with the construct containing both lacS and lacTr restored the wild-type phenotype and sustained growth on minimal medium (plate c). (B) Growth of four independent transformants was monitored in both rich tryptone-lactose (Trp-Lac) and minimal lactose (Lac) media and resulted in nearly identical overlapping curves (solid squares) compared to cultures of the wild-type Gθ (•) and untransformed GθW (▵) strains under the same conditions. OD600, optical density at 600 nm.
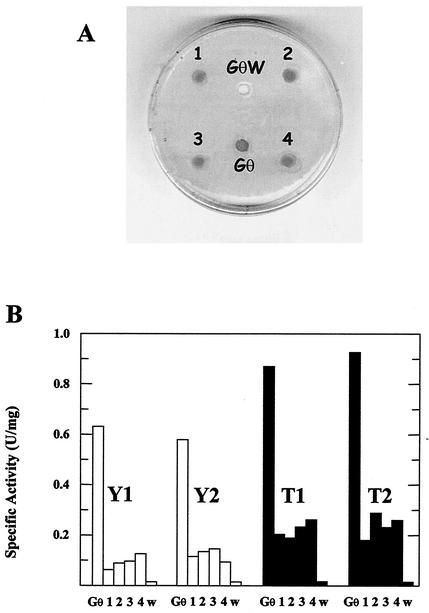
Independent pEXlacOP transformants were further characterized for the restoration of β-glycosidase activity; they were picked from the lactose plates, propagated in hygromycin selective medium, and seeded onto fresh solid medium. After growth, X-Gal solutions were overlaid dropwise onto the colonized areas, and β-glycosidase activity was detectable after incubation for 4 to 5 h at 75°C. Figure 6 shows the analysis of four clones (1, 2, 3, and 4) that all developed blue color upon being stained with the chromogenic substrate X-Gal (Fig. 6B), thus demonstrating the complementing capability of the genomic fragment used for transformation. The specific spectrophotometric assay performed on cell extracts of different clones revealed the persistence of fairly constant enzyme activity levels after prolonged growth (Fig. 7); nevertheless, the activity displayed by the transformant cell extracts was 5 to 20% of the activity of wild-type Gθ, with all single absolute values higher in lactose-supplemented cultures.
FIG. 7.
β-Glycosidase activity test of isolated clones transformed with pEXlacOP. (A) Four independent transformants were picked from lactose plates, propagated in hygromycin selective medium, and seeded onto fresh plates (1, 2, 3, and 4). After growth, the colonized areas showed β-glycosidase activity, developing blue color upon being stained with the chromogenic substrate X-Gal. Wild-type Gθ and the deficient mutant GθW were used as positive and negative controls for enzyme detection, respectively. Cultures of the same clones were grown to early stationary phase and reinoculated for a subsequent four-step scaling up. Cells were harvested at the early stationary phase of growth for each step, and extracts were assayed for cytosolic β-galactosidase activity (in enzyme units per milligram of total cytosolic proteins). (B) Results for cultures (1, 2, 3, and 4) in both yeast extract-Casamino Acids-glucose (open bars) and tryptone-lactose (solid bars) media at the first (Y1 and T1) and the fourth (Y2 and T2) step of propagation. Activities in the cell extracts of Gθ and GθW (w) cultures were used as reference points for 100 and 0% activity, respectively, in every set of measures.
DISCUSSION
The results of this study strongly support the hypothesis that transposon mutagenesis (22) is a prominent mechanism of mutation in the hyperthermophile S. solfataricus and that IS elements play a central role in the genome dynamics of this organism. We found two different active insertions into the lacS locus in two independent transposition events, which were demonstrated to be unstable and reversible. Reversible insertions able to restore wild-type phenotypes but with higher reversion frequencies have been reported before (17), although the apparent mutational frequency (10−4 to 10−5 per plated cell) of lacS in the Gθ strain was comparable to that calculated for both lacS (17, 33) and pyrE (24) genes in the 98/2 and P1 strains. Spontaneous S. solfataricus deletion mutants have been isolated before (17); nevertheless, the precise mapping of the extended deletions described, as well as the indication of transposable elements as mediators of dramatic chromosome rearrangements, has never been reported before. The stable mutant phenotype of the GθW isolate was indeed caused by such an event driven by a transposable element of the ISC1058 family. This element, which is present in 14 full-length copies on the S. solfataricus P2 genome, belongs to the IS5 family phylogenetically clustered with proteobacterial elements (23). Its activity of “jumping” from one site on the chromosome to another has already been demonstrated, even in the brief time range of a single culture (24), and hence, the presence of increased numbers of identical copies suggests that others have been duplicated only recently. The different behavior of distinct S. solfataricus strains in the conservation or loss of genomic sequences (36a) is probably due to differences in the recombination and repair systems of the host cells. The horizontal spread of IS elements among different strains with differentiated regulated control is a quite reasonable explanation of the event, if one considers inter- and intraspecies conjugation (29, 39) and/or the wide host range of a number of natural IS “carriers,” such as some Sulfolobus viruses and plasmids that also contain ISs (34, 35).
To our knowledge, this is the first report of the detailed characterization of an extended nonlethal deletion on the chromosome of S. solfataricus. From an evolutionary perspective (5), this finding highlights the potential of dynamic genomes as a means of growth under highly specialized environmental conditions, such as occurs in hot springs. Rapid deletion of nonessential genes, leading to a reduction of the genome to a minimum number of genes, required in a specialized ecological niche, may confer a competitive advantage over microbes maintaining complex genomes. Our results demonstrate that the parental Sulfolobus species and its derivative mutant, able to propagate at a higher doubling time under the laboratory conditions tested, could also serve as a model system to study this phenomenon.
So far, the major attention devoted to the applied biotechnology of hyperthermophilic archaea (6) has been on the elucidation of their basic molecular cell biology, which requires transformation strategies for in vivo studies (21). Despite the increasing amount of information in this field, which indicates that S. solfataricus is an interesting organism among the characterized representatives of crenarchaea, the art of manipulative genetics for this class of archaea is still in its infancy. Genetic tools could make possible still unattempted investigations of the effect of genotype on phenotype and of the relationship between environmental factors and gene function in archaea; they represent the necessary base on which to build strategies, such as RNA interference and induction of differential expression, aimed at the reconstruction of functional networks or the remodeling of biological pathways.
Therefore, the isolation of stable complementable mutations from S. solfataricus opens new possibilities for the development of genetic tools. So far, three selectable markers that confer resistance to specific inhibitors have been used in Sulfolobus: an alcohol dehydrogenase (3), a hygromycin phophotransferase (9, 10), and wild-type pyrEF genes usable with uracil-auxotrophic mutant hosts (24). Similarly, the lacS gene, together with the metabolically related lacTr, can now be used as an even more stringent selectable marker for growth on lactose when transferred to suitable hosts such as the GθW mutant, despite the lower β-galactosidase activity measured in the transformant clones compared to the wild-type strain Gθ. This lower gene expression could be explained by the absence of additional and important ORFs (among those indicated as coding for proteins of unknown function on the P2 genome annotations), probably encoding protein regulators, present in the missing genomic region and not comprised in the lacOP DNA fragment; it might also be due to the different structure of the episomal lac DNA compared to its counterpart on the wild-type chromosome, which could impair gene transcription. The lacS gene has already been shown to possess autonomous regulatory sequences in the 5′ flanking region and to be cotranscribed with the lacTr mRNA only in barely detectable amounts (27), i.e., putatively common regulatory sequences upstream of the lacTr gene should not be necessary for lacS expression. Moreover, the lacS gene with limited flanking regions has already been proven effective in the complementation of insertional mutations, disrupting only the lacS gene in vivo (41).
This marker was shown to be effectively expressed in a low-copy-number and autonomously replicating vector like pEXSs, but it has recently been demonstrated to also function as a single copy in a chromosome-integrated fashion (41). Therefore, it should work even more effectively in recently discovered high-copy-number extrachromosomal elements (4).
Finally, the efficacy of the host-vector system described and its application in the complementation of conditionally lethal mutations, such as the lack of some sugar-metabolizing enzymes, contributes to the boost in the “postgenomic” phase in S. solfataricus studies. In fact, similarly to the lacTr gene in this work, it may help in the identification in vivo of other still uncharacterized genes on the recently fully sequenced genome and in gaining insights into fundamental matters concerning the evolution of thermophiles and their adaptation to their extreme environments.
Acknowledgments
We are grateful to Patrizia Contursi and Gabriella Fiorentino for helpful discussion and to Giuseppe Ruggiero for technical assistance for S. solfataricus cell culture.
This work was supported by grants from the European Union in the framework of the projects “Screen” (contract QLK3-CT-2000-00649) and “Hypersolutes” (QLRT-2000-00640).
REFERENCES
- 1.Aagaard, C., I. Leviev, R. N. Aravalli, P. Forterre, D. Prieur, and G. A. Garrett. 1996. General vectors for archaeal hyperthermophiles: strategies based on a mobile intron and a plasmid. FEMS Microbiol. Rev. 18:93-104. [DOI] [PubMed] [Google Scholar]
- 2.Ammendola, S., L. Politi, and R. Scandurra. 1998. Cloning and sequencing of ISC1041 from the archaeon Sulfolobus solfataricus MT-4, a new member of the IS30 family of insertion elements. FEBS Lett. 428:217-223. [DOI] [PubMed] [Google Scholar]
- 3.Aravalli, R. N., and R. A. Garrett. 1997. Shuttle vectors for hyperthermophilic Archaea. Extremophiles 1:183-191. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, H. P., Q. She, H. Phan, K. Stedman, D. Prangishvili, I. Holz, J. K. Kristjansson, R. Garrett, and W. Zillig. 1999. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 34:217-226. [DOI] [PubMed] [Google Scholar]
- 5.Bansal, A. K., and T. E. Meyer. 2002. Evolutionary analysis by whole-genome comparisons. J. Bacteriol. 184:2260-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breithaupt, H. 2001. The hunt for living gold. The search for organisms in extreme environments yields useful enzymes for industry. EMBO Rep. 2:968-971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brügger, K., P. Redder, Q. She, F. Confalonieri, Y. Zivanovic, and R. A. Garrett. 2002. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 206:131-141. [DOI] [PubMed] [Google Scholar]
- 8.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, et al. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 9.Cannio, R., P. Contursi, M. Rossi, and S. Bartolucci. 1998. An autonomously replicating transforming vector for Sulfolobus solfataricus. J. Bacteriol. 180:3237-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannio, R., P. Contursi, M. Rossi, and S. Bartolucci. 2001. Thermoadaptation of a mesophilic hygromycin B phosphotransferase by directed evolution in hyperthermophilic Archaea: selection of a stable genetic marker for DNA transfer into Sulfolobus solfataricus. Extremophiles 5:153-159. [DOI] [PubMed] [Google Scholar]
- 11.Charlebois, R. L., and W. F. Doolittle. 1989. Transposable elements and genome structure in halobacteria, p. 297-307. In D. E. Berg, and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 12.Cubellis, M. V., C. Rozzo, P. Montecucchi, and M. Rossi. 1990. Isolation and sequencing of a new beta-galactosidase-encoding archaebacterial gene. Gene 94:89-94. [DOI] [PubMed] [Google Scholar]
- 13.Elferink, M. G., C. Schleper, and W. Zillig. 1996. Transformation of the extremely thermoacidophilic archaeon Sulfolobus solfataricus via a self-spreading vector. FEMS Microbiol. Lett. 137:31-35. [DOI] [PubMed] [Google Scholar]
- 14.Fitz-Gibbon, S. T., H. Ladner, K. Ung-Jin, K. O. Stetter, M. I. Simon, and J. H. Miller. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99:984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, T., H. Huber, S. Burggraf, and K. O. Stetter. 1996. 16S rDNA-based phylogeny of the archaeal order Sulfolobales and reclassification of Desulfurolobus ambivalens as Acidianus ambivalens comb. nov. Syst. Appl. Microbiol. 19:56-60. [Google Scholar]
- 16.Grogan, D. W. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haseltine, C., R. Montalvo-Rodriguez, A. Carl, E. Bini, and P. Blum. 1999. Extragenic pleiotropic mutations that repress glycosyl hydrolase expression in the hyperthermophilic archaeon Sulfolobus solfataricus. Genetics 152:1353-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurka, J. 1998. Repeats in genomic DNA: mining and meaning. Curr. Opin. Struct. Biol. 8:333-337. [DOI] [PubMed] [Google Scholar]
- 19.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, et al. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 20.Klenk, H. P., R. Clayton, J. Tomb, O. White, K. E. Nelson, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 21.Luo, Y., and A. Wasserfallen. 2001. Gene transfer systems and their applications in Archaea. Syst. Appl. Microbiol. 24:15-25. [DOI] [PubMed] [Google Scholar]
- 22.Mahillon, J. 1998. Transposons as gene haulers. APMIS Suppl. 84:29-36. [DOI] [PubMed] [Google Scholar]
- 23.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martusewitsch, E., C. W. Sensen, and C. Schleper. 2000. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J. Bacteriol. 182:2574-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moracci, M., B. Cobucci Ponzano, A. Trincone, S. Fusco, M. De Rosa, J. van Der Oost, C. W. Sensen, R. L. Charlebois, and M. Rossi. 2000. Identification and molecular characterization of the first alpha-xylosidase from an archaeon. J. Biol. Chem. 275:22082-22089. [DOI] [PubMed] [Google Scholar]
- 26.Moracci, M., M. Ciaramella, and M. Rossi. 2001. Beta-glycosidase from Sulfolobus solfataricus. Methods Enzymol. 330:201-215. [DOI] [PubMed] [Google Scholar]
- 27.Prisco, A., M. Moracci, M. Rossi, and M. Ciaramella. 1995. A gene encoding a putative membrane protein homologous to the major facilitator superfamily of transporters maps upstream of the beta-glycosidase gene in the archaeon Sulfolobus solfataricus. J. Bacteriol. 177:1614-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redder, P., Q. She, and R. A. Garrett. 2001. Non-autonomous mobile elements in the crenarchaeon Sulfolobus solfataricus. J. Mol. Biol. 306:1-6. [DOI] [PubMed] [Google Scholar]
- 29.Reilly, M. S., and D. W. Grogan. 2001. Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J. Bacteriol. 183:2943-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rezsöhazy, R., B. Hallet, J. Delcour, and J. Mahillon. 1993. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 9:1283-1295. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schleper, C., K. Kubo, and W. Zillig. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. USA 89:7645-7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schleper, C., R. Röder, T. Singer, and W. Zillig. 1994. An insertion element of the extremely thermophilic archaeon Sulfolobus solfataricus transposes into the endogenous β-galactosidase gene. Mol. Gen. Genet. 243:91-96. [DOI] [PubMed] [Google Scholar]
- 34.Schleper, C., I. Holz, D. Janekovic, J. Murphy, and W. Zillig. 1995. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 177:4417-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.She, Q. X., H. E. Phan, R. A. Garrett, S. V. Albers, K. Stedman, and W. Zillig. 1998. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles 2:417-425. [DOI] [PubMed] [Google Scholar]
- 36.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, et al. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.She, Q., S. Peng, W. Zillig, and R. A. Garrett 2001. Gene capture in archaeal chromosomes. Nature 409:478. [DOI] [PubMed] [Google Scholar]
- 37.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J.-I. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum H: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stedman, K. M., C. Schleper, E. Rumpf, and W. Zillig. 1999. Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: construction and testing of viral shuttle vectors. Genetics 152:1397-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stedman, K. M., Q. She, H. Phan, I. Holz, H. Singh, D. Prangishvili, R. A. Garrett., and W. Zillig. 2000. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J. Bacteriol. 182:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trincone, A., B. Cobucci-Ponzano, B. Di Lauro, M. Rossi, Y. Mitsuishi, and M. Moracci. 2001. Enzymatic synthesis and hydrolysis of xylogluco-oligosaccharides using the first archaeal alpha-xylosidase from Sulfolobus solfataricus. Extremophiles 5:277-282. [DOI] [PubMed] [Google Scholar]
- 41.Worthington, P., V. Hoang, F. Perez-Pomares, and P. Blum. 2003. Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 185:482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zillig, W., D. Prangishvilli, C. Schleper, M. Elferink, I. Holz, et al. 1996. Viruses, plasmids and other genetic elements of thermophilic and hyperthermophilic Archaea. FEMS Microbiol. Rev. 18:225-236. [DOI] [PubMed] [Google Scholar]