Abstract
During sporulation in Bacillus subtilis, the prespore-specific developmental program is initiated soon after asymmetric division of the sporangium by the compartment-specific activation of RNA polymerase sigma factor σF. σF directs transcription of spoIIIG, encoding the late forespore-specific regulator σG. Following synthesis, σG is initially kept in an inactive form, presumably because it is bound to the SpoIIAB anti-sigma factor. Activation of σG occurs only after the complete engulfment of the prespore by the mother cell. Mutations in spoIIIJ arrest sporulation soon after conclusion of the engulfment process and prevent activation of σG. Here we show that σG accumulates but is mostly inactive in a spoIIIJ mutant. We also show that expression of the spoIIIGE155K allele, encoding a form of σG that is not efficiently bound by SpoIIAB in vitro, restores σG-directed gene expression to a spoIIIJ mutant. Expression of spoIIIJ occurs during vegetative growth. However, we show that expression of spoIIIJ in the prespore is sufficient for σG activation and for sporulation. Mutations in the mother cell-specific spoIIIA locus are known to arrest sporulation just after completion of the engulfment process. Previous work has also shown that σG accumulates in an inactive form in spoIIIA mutants and that the need for spoIIIA expression for σG activation can be circumvented by the spoIIIGE155K allele. However, in contrast to the case for spoIIIJ, we show that expression of spoIIIA in the prespore does not support efficient sporulation. The results suggest that the activation of σG at the end of the engulfment process involves the action of spoIIIA from the mother cell and of spoIIIJ from the prespore.
During the early stages of endospore development in Bacillus subtilis, the rod-shaped bacterial cell is divided into a smaller prespore and a larger mother cell. The prespore is engulfed by the mother cell, which lyses at the end of the sporulation process to liberate the mature spore into the environment. Each cell type receives a copy of the bacterial chromosome, and each deploys specific but interdependent genetic programs controlled by the successive appearance of the σF, σE, σG, and σK subunits of RNA polymerase (29, 32, 42, 46). The σF and σG regulators control gene expression during the early and late stages of prespore development, respectively, whereas σE controls the expression of early mother cell genes and is replaced at later stages by σK. The activation of the first compartment-specific sigma factor, σF, involves the action of three regulatory proteins, SpoIIAA, SpoIIAB, and SpoIIE (33, 48). SpoIIAB is an anti-sigma factor that binds to σF as a dimer and inhibits its transcriptional activity (6, 7, 14, 36), whereas SpoIIAA is an anti-anti-sigma factor that in an unphosphorylated state interacts with SpoIIAB and releases σF from the SpoIIAB-σF complex (1, 12, 13). SpoIIE, a septum-bound phosphatase, promotes the preferential dephosphorylation of SpoIIAA-P in the prespore, its binding to SpoIIAB, and consequently the activation of σF (3, 4, 12, 15). Transcription of the spoIIIG gene, encoding the late prespore regulator σG, is driven by the σF form of RNA polymerase (55). However, transcription of spoIIIG is delayed towards the end of the engulfment process relative to transcription of a first class of σF-dependent genes (41). In addition, it requires both the activity of σE in the mother cell and expression of the σF-controlled gene spoIIQ (41, 56). Following synthesis, σG does not become active until engulfment of the prespore by the mother cell is complete (32, 53; reviewed in references 42 and 46). Once active, σG directs expression of its own gene, allowing a rapid increase in the cellular levels of σG (25, 55). Activation of σG at the end of the engulfment sequence demands the expression of the spoIIIA operon (26, 43), which is transcribed in the mother cell by σE RNA polymerase (23). The spoIIIA locus encodes eight proteins predicted to be associated with the prespore outer membrane, all of which are required for the activation of σG (23, 53). σG accumulates in a spoIIIA mutant but is unable to activate transcription from its target promoters (26). Genetic, biochemical, and structural evidence suggests that the anti-sigma factor SpoIIAB negatively regulates σG (7, 8, 17, 26, 28, 44). In particular, during sporulation SpoIIAB tends to disappear from the forespore soon after completion of the engulfment sequence in a spoIIIA-dependent mode (28), and expression of an allele of spoIIIG (spoIIIGE155K) encoding a form of σG that is not efficiently bound by SpoIIAB in vitro permits activation of σG in vivo in the absence of an intact spoIIIA locus (26). These results suggest that the spoIIIA products are required soon after conclusion of the engulfment process to inactivate SpoIIAB, thereby releasing active σG (26).
Mutations in the spoIIIJ locus were also shown to prevent expression of σG-controlled genes but not transcription of the spoIIIG gene (16). spoIIIJ is the first gene of a bicistronic operon transcribed during vegetative growth (16, 39). It encodes a putative lipoprotein that localizes to the cell membrane and septal regions during vegetative growth and sporulation but also to the membranes that surround the developing spore (39). SpoIIIJ is related to the YidC Sec-independent and Sec-dependent membrane protein translocase of Escherichia coli (about 33% identity and 51% conserved residues for a region of overlap between the two proteins of 226 amino acids) (47, 49, 57). SpoIIIJ is also related to the CcfA protein of Enterococcus faecalis, with about 65% similarity and 39% identity between the two proteins (2). Like spoIIIJ, ccfA codes for a putative lipoprotein precursor, but it also encodes the cCF10 peptide pheromone within the signal peptide (2). The cCF10 peptide pheromone is required for the conjugative transfer of the E. faecalis plasmid pCF10. CcfA, like YidC in E. coli, seems to be required for normal growth of E. faecalis (2).
Here we have analyzed the role of spoIIIJ in the activation of σG. We show that σG accumulates in a spoIIIJ mutant but is mostly inactive. We also show that expression of the spoIIIGE155K allele restores transcription of the σG-dependent gene sspE to spoIIIJ mutant cells. Expression of spoIIIJ from a prespore-specific promoter produces a sporulation-proficient strain, whereas its expression from a mother cell-specific promoter results in a block soon after completion of the engulfment process. Conversely, expression of spoIIIA in the prespore does not support efficient sporulation. These results suggest that SpoIIIJ from the prespore and the spoIIIA-encoded products from the mother cell compartment are cell type-specific elements of a signaling pathway that antagonizes the inhibitory action of SpoIIAB and promotes the activation of σG soon after conclusion of the engulfment process.
MATERIALS AND METHODS
Bacterial strains and general methods.
The B. subtilis strains used in this work, most of which are congenic derivatives of the wild-type strain MB24 (trpC2 metC3), are listed in Table 1. Luria-Bertani medium was used for routine growth of E. coli DH5α (BRL) and of B. subtilis strains. Sporulation was induced by exhaustion in Difco sporulation medium (DSM), and the extent of sporulation (expressed as the percentage of heat resistant CFU relative to the total cell count) was assessed as described previously (20, 21). Antibiotics were used as previously described (20, 21).
TABLE 1.
B. subtilis strains
| Strain | Relevant genotype | Origin (reference) |
|---|---|---|
| MB24 | trpC2 metC3 | Laboratory stock |
| AH45 | trpC2 metC3 spoIIIGΔ1 | Laboratory stock |
| AH62 | trpC2 metC3 spoIIIA::Tn917ΩHU24 | Laboratory stock (26) |
| EUE9537 | trpC2 metC3 SPβ::sspE-lacZ spoIIIG::pEK15 (wild-type spoIIIG) | Laboratory stock (26) |
| EUE9538 | trpC2 metC3 SPβ::sspE-lacZ spoIIIG::pEK16 (spoIIIGE155K) | Laboratory stock (26) |
| AH1042 | trpC2 metC3 ΔsspE::sspE-lacZ | Laboratory stock |
| AH2350 | trpC2 metC3(pMK3) | This work |
| AH5008 | trpC2 metC3 ΔamyE::spoIIIG-lacZ | This work |
| AH5009 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::spoIIIG-lacZ | This work |
| AH5011 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIIIJspoIIIJ | This work |
| AH5012 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIIJQ-spoIIIJ | This work |
| AH5013 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIID-spoIIIJ | This work |
| AH5014 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIIIJ-spoIIIJ ΔsspE::sspE-lacZ | This work |
| AH5015 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIIQ-spoIIIJ ΔsspE::sspE-lacZ | This work |
| AH5016 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIID-spoIIIJ ΔsspE::sspE-lacZ | This work |
| AH5017 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIIIJ-spoIIIJ-gfp | This work |
| AH5018 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIIQ-spoIIIJ-gfp | This work |
| AH5019 | trpC2 metC3 ΔspoIIIJ::km ΔamyE::PspoIID-spoIIIJ-gfp | This work |
| AH5020 | trpC2 metC3 spoIIIA::PspoIIQ-spoIIIA | This work |
| AH5024 | trpC2 metC3 ΔspoIIIJ::km ΔsspE::sspE-lacZ spoIIIG::pEK15 | This work |
| AH5025 | trpC2 metC3 ΔspoIIIJ::km ΔsspE::sspE-lacZ spoIIIG::pEK16 | This work |
| AH5029 | trpC2 metC3 spoIIIA::PspoIID-spoIIIA | This work |
| AH5030 | trpC2 metC3 ΔsspE::sspE-lacZ spoIIIG::pEK15 | This work |
| AH5031 | trpC2 metC3 ΔsspE::sspE-lacZ spoIIIG::pEK16 | This work |
| AH5035 | trpC2 metC3 spoIIIJ::sp(pMK3) | This work |
| AH5036 | trpC2 metC3 spoIIIJ::sp(pMS210) | This work |
| AH5037 | trpC2 metC3(pMS210) | This work |
| JOB20 | trpC2 metC3 ΔspoIIIJ::sp | This work |
| JOB34 | trpC2 metC3 ΔspoIIIJ::km ΔsspE::sspE-lacZ | This work |
| JOB44 | trpC2 metC3 ΔspoIIIJ::km | This work |
Construction of a spoIIIJ nonpolar null mutant.
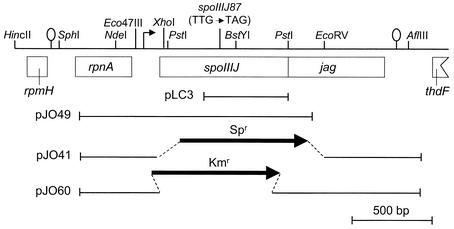
To create a nonpolar insertion into the spoIIIJ gene, we used PCR with Taq DNA polymerase and primers RNPA-75 and JAGD-235R (Table 2) to produce a DNA fragment extending from nucleotide 421 upstream of the spoIIIJ start codon to nucleotide 240 downstream of the jag stop codon. This DNA fragment was cloned into the pCR2.1 TOPO vector (Invitrogen) to create plasmid pJO34. An EcoRI fragment containing the spoIIIJ operon was released from pJO34 and inserted into the EcoRI site of pLitmus38 (New England Biolabs) to create pJO57. Inverse PCR with primers JXMA-1 and JXMA-2 (Table 2) was performed on pJO57 to create a linear fragment that deleted nucleotides +41 to +707 of the spoIIIJ gene (with respect to the transcription start site). This PCR product was ligated with an XmaI restriction fragment containing a promoterless kanamycin resistance (Kmr) determinant resulting from the digestion of pUC18-K (35). The DNA resulting from this ligation was a recircularized plasmid in which the spoIIIJ gene was replaced with the km gene, which was now under control of the spoIIIJ promoter. This plasmid was named pJO60. Strains JOB34 and JOB44 were created by transformation of AH1042 (sspE-lacZ) and MB24 (wild type) (Table 1), respectively, with ScaI-linearized pJO60, selecting for Kmr. Disruption of the spoIIIJ gene by a double-crossover event (marker replacement) was verified by PCR. The resulting strains, JOB34 and JOB44 (Table 1), were tested by reverse transcription-PCR analysis to confirm that the insertion was nonpolar on jag (not shown). Strains JOB34 and AH1042 were transformed with chromosomal DNA from EUE9538 (spoIIIGE155K allele) or EUE9537 (wild-type spoIIIG allele) with selection for spectinomycin resistance (Spr), and appropriate transformants were identified as described previously (Table 1) (26). During the course of this work we also used a spoIIIJ mutant created by insertion of an Spr cassette. pJO34 (see above) was digested with EcoRI, and the resulting fragment was cloned into the EcoRI site of pUC19 to create pJO36. An Spr cassette was then liberated from pAH250 (20) with SmaI and XhoI and cloned between the EcoRV and XhoI sites of pJO36 to create pJO41. ScaI-digested pJO41 was used to transform MB24 to Spr, yielding strain JOB20 (Table 1; Fig. 1).
TABLE 2.
Primers
| Primer | Sequence (5′→3′) |
|---|---|
| RNPA-75 | CCGCCAGTTTGTCTTATATACGC |
| JAGD-235R | CGAAGTGGAAGATTTGTTATCCACA |
| JXMA-1 | CCTTTTCAACAACATTCCCGGGTATAATTAATCTTTACACTC |
| JXMA-2 | GCGATCAACTTCCCGGGGGCTCTTTCTCTTTATTGGG |
| spoIIIJ-460D | CGAAAGCGATGCAGGCTTTACAGCCGG |
| gfp-spoIIIJ-R | CTCCAGTGAAAAGTTCTTCTCCTTTACTCTTTTTCTTTCCTCCGGCTTTTTGCGGC |
| gfp-30D | AGTAAAGGAGAACTTTTCACTGGA |
| gfp-R | GGCGAATTCTTATTTGTATAGTTCATCCAT |
| spoIIIJ-150R | CCAGCTTTACGATCGCAGGC |
| spoIIIJ-spoIIQ | GTTGCTGAGGTGATGAAACAATGTTGTTGAAAAGGAGAATAGGG |
| spoIIIJ-spoIID | CGAGCAGGAGGCAGCTGAATATGTTGTTGAAAAGGAGAATAGGG |
| spoIIIJ-1039R | CAATCCGGATCCTGTACTGCTTCATCGACATTTCGCCC |
| spoIIQ-152D | GTTTCAAAGCTGAATTCCAGGCAGCG |
| spoIIQ-500R | TGTTTCATCACCTCAGCAACATTCTG |
| spoIID-1D | CGGAAGAATTCCGCCGTATGAATGG |
| spoIID-500R | ATTCAGCTGCCTCCTGCTCGGG |
| spoIIIA-spoIIQ | GTTGCTGAGGTGATGAAACATTGAATGAAATCGCTGAGG |
| spoIIIA-spoIID | CGAGCAGGAGGCAGCTGAATTTGAATGAAATCGCTGAGG |
| spoIIIA-5004D | GGAAGCAGCGGATCCGCAATCCCC |
| spoIIIG-392D | GGGAAAAAAGATCTCGAGAAATAAAGTCG |
| spoIIIG-1217R | GCCTTTAAAACAACTCGAGTTAAAGGC |
FIG. 1.
Genetic organization of the spoIIIJ-jag locus of B. subtilis. A partial restriction map and the genetic organization of the spoIIIJ region are shown. The boxes below the line representing a partial restriction map of the region indicate the extents of the different cistrons in the region (30), all of which are transcribed from left to right. The stem-and-loop structures upstream of rpnA and downstream of jag indicate the position of possible transcription terminators. The position of the spoIIIJ promoter is shown by a horizontal arrow. The position of the mutation present in the spoIIIJ87 allele (L138STOP) is indicated. The lines below the restriction map represent DNA fragments present in the indicated plasmids.
Construction of a spoIIIG-lacZ fusion.
To construct a spoIIIG-lacZ translational fusion, we first isolated a 448-bp EcoRI-to-PvuII fragment from pTK4 encompassing the spoIIGB-spoIIIG intergenic region and including the first 30 codons of spoIIIG (27). The 448-bp fragment was introduced between the EcoRI and SmaI sites of pNM480 (37), yielding pMS140. pMS140 was digested with EcoRI and SacI, and the 2,448-bp fragment encompassing the spoIIGB-spoIIIG intergenic region and 2,000 bp of the lacZ gene was cloned between the same sites of the amyE integrational vector pSN32 (38), yielding pMS141. MB24 and JOB44 were both transformed with pMS141 to chloramphenicol resistance (Cmr), and Cmr AmyE− transformants were identified and named AH5008 and AH5009, respectively (Table 1).
Fusions of spoIIIJ to different sporulation promoters.
To insert a copy of the spoIIIJ gene under the control of its promoter at the nonessential amyE locus, we first used PCR amplification with Taq DNA polymerase and primers RNPA-75 and spoIIIJ-150R (Table 2) to produce a DNA fragment extending from nucleotide 421 upstream of the spoIIIJ translational start codon to nucleotide 144 downstream of the end of the spoIIIJ gene. This fragment was cloned into pCR2.1 TOPO (Invitrogen), creating plasmid pJO46. An EcoRI restriction fragment containing the spoIIIJ gene from pJO46 was subcloned into the EcoRI restriction site of pDG364 (10) to create pJO49. Fusions of spoIIIJ to the spoIIQ and spoIID promoters were constructed by the method of splicing by overlay extension (SOE) (22). The spoIIQ and spoIID promoters included regions from −322 to +14 and −464 to +31 relative to the transcriptional start site, respectively. Initially, the spoIIIJ and promoter sequences were amplified separately from chromosomal DNA of MB24 (Table 1). The following primers were used (Table 2): for spoIIIJ, spoIIIJ-spoIIQ or spoIIIJ-spoIID and spoIIIJ-1039R; for spoIIQ, spoIIQ-152D and spoIIQ-500R; and for spoIID, spoIID-1D and spoIID-500R. The 839-bp spoIIIJ fragment was mixed with the 350-bp spoIIQ fragment or with the 500-bp spoIID fragment, and the resulting fragments of 1,189 or 1,339 bp were amplified with primers spoIIQ-152D and spoIIIJ-1039R, to create a PspoIIQ-spoIIIJ fusion, or with primers spoIID-1D and spoIIIJ-1039R, to create a PspoIID-spoIIIJ fusion. The PspoIIQ-spoIIIJ and PspoIID-spoIIIJ fragments were digested with EcoRI and BamHI and ligated to pDG364 (10) digested with the same enzymes, to yield pMS189 and pMS190, respectively. JOB44 was transformed with pJO49, pMS189, and pMS190 to produce the Cmr AmyE− strains AH5011, AH5012, and AH5013, respectively (Table 1). Derivatives of these strains bearing an sspE-lacZ fusion were obtained by transformation with chromosomal DNA from AH1042 (Table 1). To construct a PspoIID-spoIIIJ fusion in a multicopy vector, we isolated a 1,339-bp EcoRI-to-BamHI fragment from pMS190 encompassing the PspoIID-spoIIIJ fusion. The 1,339-bp fragment was introduced between the same sites of pMK3 (54), yielding pMS210. Plasmids pMK3 and pMS210 were introduced in both MB24 and JOB20 (Table 1).
Construction of a spoIIIJ-gfp fusion.
We used the SOE technique (see above) to construct a fusion of spoIIIJ to gfp. Initially, the spoIIIJ and gfp sequences were amplified separately, from chromosomal DNA of a wild-type B. subtilis strain and from pEA18 (a gift from Alan Grossman), respectively. The following primer pairs were used (Table 2): for spoIIIJ, spoIIIJ-460D and gfp-spoIIIJ-R; and for gfp, gfp-30D and gfp-R. The 551-bp spoIIIJ fragment and the 772-bp gfp fragment were purified, mixed together, and amplified with primers spoIIIJ-460D and gfp-R (see above). This produced a 1,245-bp spoIIIJ-gfp fragment which was cloned into pCR2.1 TOPO (Invitrogen) to yield pLC1. The presence and orientation of the insert were confirmed by restriction analysis. An Spr cassette was released from pAH256 (20) with EcoRV and XhoI and cloned in pLC1 that had been cut with the same restriction enzymes. The resulting plasmid, pLC3, was used to transform strains AH5011, AH5012, and AH5013; Spr transformants that were the result of a single reciprocal crossover event (Campbell-type mechanism) were identified and designated AH5017, AH5018, and AH5019, respectively (Table 1).
Fusions of spoIIIA to different sporulation promoters.
Fusions of spoIIIA to the spoIIQ and spoIID promoters were constructed by SOE (see above). Initially, the spoIIIA and promoter sequences were amplified separately from chromosomal DNA of a wild-type B. subtilis strain. The following primers were used (Table 2): for spoIIIA, spoIIIA-spoIIQ, spoIIIA-spoIID, and spoIIIA-5004D; for spoIIQ, spoIIQ-152D and spoIIQ-500R; and for spoIID, spoIID-1D, and spoIID-500R. The 386-bp spoIIIA fragment was mixed with the 350-bp spoIIQ fragment or with the 500-bp spoIID fragment, and the mixture was subjected to PCR amplification with primers spoIIQ-152D and spoIIIA-5004D for spoIIQ or primers spoIID-1D and spoIIIA-5004D for spoIID. The resulting PspoIIQ-spoIIIA and PspoIID-spoIIIA fragments were digested with BamHI and introduced into BamHI- and SmaI-digested pUS19 (5), to yield pMS155 and pMS198. Transformation of MB24 with pMS155 or pMS198 (with selection for Spr cells) produced strains AH5020 and AH5029, respectively (Table 1). In both strains the Campbell-type integration of pMS155 or pMS198 separated spoIIIAA from its native promoter and placed the spoIIIA operon under the control of the spoIIQ or spoIID promoter, respectively. Correct integration was confirmed by PCR.
Overproduction and purification of σG.
The spoIIIG coding region was PCR amplified with primers spoIIIG-392D and spoIIIG-1217R (Table 2). The PCR product was digested with BglII and XhoI and inserted between the BamHI and XhoI sites of pET30c(+) (Novagen), creating pMS112, in which a His6 tag was introduced between the first and second codons of spoIIIG. Plasmid pMS112 was introduced into competent cells of BL21(DE3)/pLysS (Novagen). Growth, induction, and lysate preparation were essentially as described previously (50). The His6-σG fusion protein was partially purified on Hi-Trap chelating columns in the presence of 8 M urea as described by the manufacturer (Amersham Pharmacia Biotech) and used to raise a polyclonal anti-σG antibody in rabbits (Eurogentec, Herstal, Belgium).
Immunoblotting.
Preparation of B. subtilis whole-cell lysates and Western blot analysis were essentially as described previously (50). The anti-σG polyclonal antiserum was incubated with the membranes at a dilution of 1:1,000 in phosphate-buffered saline (8 mM sodium phosphate [pH 7.5], 150 mM NaCl) containing 0.5% low-fat milk. Incubation with an anti-rabbit secondary antibody conjugated to horseradish peroxidase (from Sigma) was for 30 min at a 1:5,000 dilution. A rabbit anti-green fluorescent protein (anti-GFP) antiserum (Clontech) was used according to the manufacturer's protocol.
β-Galactosidase assays.
β-Galactosidase activity was determined with the substrate o-nitrophenol-β-d-galactopyranoside, and enzyme activity was expressed in Miller units as described previously (20, 21).
Transmission electron microscopy.
Samples (5 ml) of DSM cultures of various strains were collected 12 h after the onset of sporulation. The cells were fixed for 2 h with 2.5% glutaraldehyde in 0.1 M cocadylate buffer (pH 7.4) on ice. Samples were washed twice with 0.1 M cocadylate buffer (pH 7.4) and then were treated with 1% osmium tetroxide solution [1.5% K4Fe(CN)6, 1% OsO4, 0.1 M cocadylate buffer]. The cells were dehydrated at room temperature by washing with increasing concentrations of ethanol (up to 100%), followed by a wash with acetone. The cells were embedded in EM bed-812 resin (Electron Microscopy Sciences). Ultrathin sections were processed for observation as described previously (21). Transmission electron microscopy was performed on a Hitachi H7500 transmission electron microscope operated at 75 keV.
Light microscopy and image processing.
Samples (0.5 ml) of DSM cultures were collected throughout growth and sporulation and resuspended in the same volume of phosphate-buffered saline supplemented with DAPI (4′,6′-diamidino-2-phenylindole) (10 μg/ml). Microscope slides were prepared by using an adaptation of the method developed by van Helvoort and Woldringh (58). Images were acquired on a scientific-grade cooled charge-coupled device (Cooke Co.) on a multiwavelength wide-field three-dimensional microscopy system (63x/1.4 OIL Plan Apochromat objective, Zeiss 100 M; Intelligent Imaging Innovation). Samples were imaged in successive 0.2-μm focal planes, and out-of-focus light was removed with a constrained iterative deconvolution algorithm. Standard filters for fluorescein isothiocyanate (for GFP) and DAPI were used. The pattern of nucleoid staining with DAPI was used to as an indication of the sporulation stage. Just after formation of the asymmetric septum, the prespore nucleoid appears highly condensed, whereas soon after conclusion of the engulfment process, the mother cell and prespore nucleoids appear equally condensed (19, 51).
RESULTS
σG accumulates but is mostly inactive in a spoIIIJ mutant.
spoIIIJ is the first gene of a bicistronic operon (16) (Fig. 1). Mutations in the spoIIIJ locus block sporulation after completion of prespore engulfment and abolish σG activity (16). Errington et al. have examined the effect on sporulation of the spoIIIJ87 allele (16), which we found to be a nonsense mutation at codon 138 of SpoIIIJ (Fig. 1). Those authors have also studied the effect of a spoIIIJ insertional mutation on sporulation-specific gene expression and found that neither allele impaired transcription of the σE-dependent phoAIII gene or of the σF-dependent spoIIIG gene (16). However, both spoIIIJ87 and the insertional mutation prevented expression of the σG-dependent spoVA operon (16). Disruption of the second gene of the spoIIIJ operon, jag, did not affect either growth or sporulation (16). However, because both types of spoIIIJ mutants could have a polar effect on the expression of the jag gene, the sporulation phenotype could be caused by the lack of expression of the two genes. Here, we have used a nonpolar mutation in spoIIIJ (ΔspoIIIJ::km; strain JOB44) (Table 1), which permits expression of the jag gene, as verified by reverse transcription-PCR (not shown), to reexamine the individual contribution of this gene to sporulation. In agreement with the results of Errington and coauthors (16), we found that inactivation of spoIIIJ has little effect on the transcription of genes expressed in the mother cell (such as spoIID) or in the prespore (such as spoIIQ) prior to the completion of engulfment (not shown). In contrast, and as also found by Errington and coauthors (16), the ΔspoIIIJ::km mutation reduced the rate of spoIIIG-lacZ expression (Fig. 2A [Fig. 2A is included to allow direct comparison with the immunoblot analysis of σG accumulation; see below]). The ΔspoIIIJ::km mutation also greatly reduced transcription of the σG-dependent gene sspE (34) (data not shown). The ΔspoIIIJ::sp mutant JOB20 (Table 1) behaved like JOB44 (ΔspoIIIJ::km) with respect to spoIIIG- and sspE-lacZ expression (not shown).
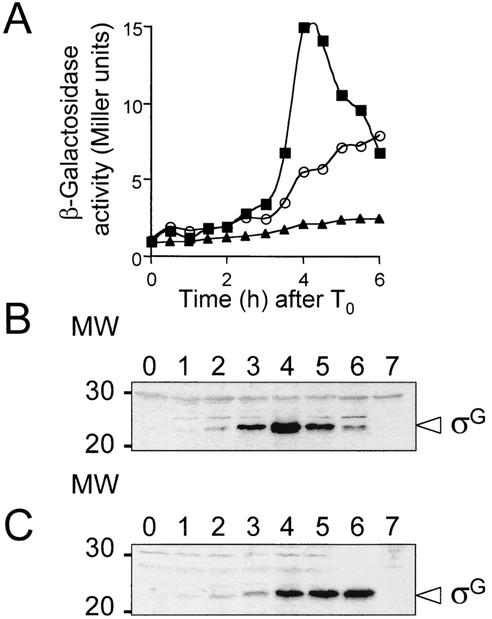
FIG. 2.
Effect of a nonpolar spoIIIJ mutation on the expression of spoIIIG-lacZ and immunoblot analysis of σG accumulation in the spoIIIJ mutant. (A) Expression of a spoIIIG-lacZ fusion was monitored at the indicated times after the onset of sporulation in the spoIIIJ mutant, JOB44 (circles) or in the wild-type strain, MB24 (squares). Endogeneous levels of β-galactosidase activity were determined in the wild-type strain MB24 (triangles). Enzyme activity was measured in Miller units. T0 indicates the end of the exponential phase of growth, defined as the onset of sporulation. (B and C) Immunoblot analysis of σG accumulation in DSM in the wild-type strain MB24 (B) or in the spoIIIJ::km mutant JOB44 (C). Samples from sporulating cultures were collected at the onset of sporulation in DSM (lanes 0) and at hourly intervals thereafter, as indicated. Proteins (30 μg) in each sample were subjected to immunoblot analysis with an anti-σG rabbit polyclonal antibody (see Materials and Methods). Lanes 7, 30 μg of an extract prepared from a culture of a spoIIIG deletion mutant (AH45 [Table 1]) at h 4 of sporulation. The arrowhead indicates the position of σG; other bands represent nonspecific cross-reactive material. The positions of molecular weight (MW) markers are shown on the left.
Since substantial expression of a spoIIIG-lacZ fusion occurs in a spoIIIJ mutant (16) (Fig. 2A), it is likely that SpoIIIJ acts at the level of σG accumulation or activity. To distinguish between these two possibilities, we used an antiserum directed against σG to monitor its accumulation during sporulation in the wild type and in the ΔspoIIIJ::km mutant (JOB44) (Table 1). This σG antiserum detected a species of 35 kDa in immunoblot analysis of extracts prepared from sporulating wild-type cultures of B. subtilis but not from cultures of a spoIIIG deletion mutant (Fig. 2B and C, lanes 7) (26). In the wild-type strain, MB24, σG was first detected at 2 h after the onset of sporulation, and its levels increased during h 3 and 4 (Fig. 2B). In the spoIIIJ mutant, σG accumulated to nearly wild-type levels, although at h 3 the level of σG was reduced about threefold compared to that in the wild type (Fig. 2C). The pattern of σG accumulation in the spoIIIJ mutant probably reflects the decreased rate of spoIIIG transcription (Fig. 2). The somewhat reduced levels of spoIIIG transcription and σG accumulation in the spoIIIJ mutant are probably a result of the autoregulatory effect that σG exerts on transcription of its encoding gene (55). We conclude that the failure to detect expression of σG-dependent genes in the spoIIIJ mutant (16; this work) results from the inactivity of σG.
The E155K form of σG no longer requires SpoIIIJ for activation.
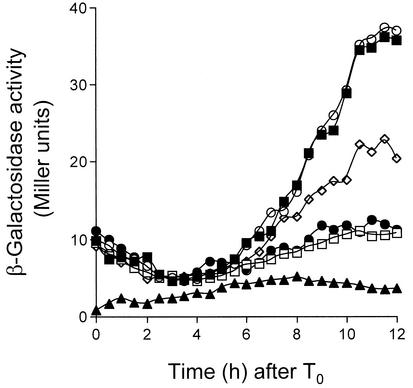
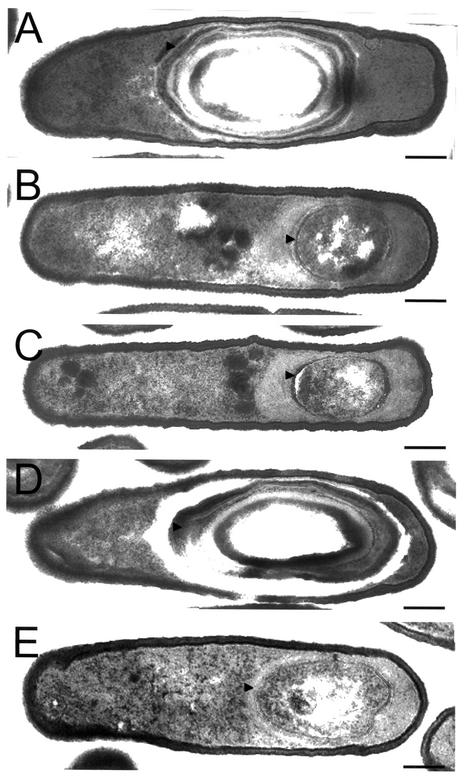
In a spoIIIA mutant, σG also accumulates in an inactive form (26). Activity of σG can be restored to spoIIIA mutant cells by the production of a form of σG (σGE155K) that is not efficiently bound by SpoIIAB in vitro (26). To test whether σGE155K could also bypass the need for spoIIIJ expression for σG activity, we introduced the wild-type or spoIIIGE155K allele of spoIIIG (the later encoding σGE155K) linked to a Spr marker into a strain bearing an sspE-lacZ fusion and either ΔspoIIIJ::km (JOB34 [Table 1]) or the wild-type spoIIIJ allele (AH1042 [Table 1]). Strains EUE9537 and EUE9538 carry Campbell-type insertions at the 3′ end of the spoIIIG gene that recreate either the wild-type spoIIIG sequence or the spoIIIGE155K allele, respectively, and have been described before (26). JOB34 or AH1042 was transformed with chromosomal DNA from EUE9537 or EUE9538, and appropriate transformants were designated AH5030 (spoIIIG wild type), AH5031 (spoIIIGE155K), AH5024 (spoIIIG wild type/ΔspoIIIJ::km), and AH5025 (spoIIIGE155K/ΔspoIIIJ::km) (Table 1). Strains AH5030 and AH5024 were constructed in parallel to control for any expression differences due to strain construction. The activity of σG was then monitored during sporulation by using the sspE-lacZ reporter fusion. We found the profile of sspE-lacZ expression in AH5024 (spoIIIG wild type/ΔspoIIIJ::km) to be very similar to that of the same fusion in the ΔspoIIIJ::km mutant JOB34) (Fig. 3). Strains AH5030 (spoIIIG wild type) and AH5031 (spoIIIGE155K) showed almost identical profiles of sspE-lacZ expression, which seemed to attain a maximum (about 40 Miller units) between h 10 and 12 of sporulation (26) (Fig. 3). Expression of sspE-lacZ in strain AH5025 (spoIIIGE155K/ΔspoIIIJ::km) commenced at the same time as in AH5030 and AH5031 and again seemed to reach a maximum between h 10 and 12 of sporulation (Fig. 3). Expression of sspE-lacZ in AH5025 was greatly reduced compared to that in a wild-type strain such as AH1042 (Table 1; see Fig. 5). However, maximum levels of β-galactosidase production in AH5025 showed a reduction of only about 50% compared to strain AH5030 or AH5031 (Fig. 3), both of which sporulate with wild-type efficiency (26) (Table 3). We also found that the spoIIIGE155K allele permitted expression of a gerE-lacZ fusion in the ΔspoIIIJ::km mutant or in cells carrying the spoIIIA::Tn917ΩHU24 allele, the same used in the study of Kellner et al. (26) (data not shown). gerE-lacZ served as a reporter for σK-dependent transcriptional activity (9), which requires σG function (reviewed in references 29, 32, 42, and 46). We conclude that the absence of σG activity in the spoIIIJ mutant is partially relieved by the spoIIIGE155K allele (Fig. 3). Because the E155K substitution in σG, which reduces its affinity for SpoIIAB, partially bypasses the requirement for the spoIIIA-encoded products (26) and for spoIIIJ for σG activity, we suggest that both loci are required after completion of the engulfment process to antagonize the inhibition exerted by SpoIIAB upon σG. However, even though the spoIIIGE155K allele restores activity of σG and of σK to spoIIIJ mutant cells, the spoIIIGE155K/ΔspoIIIJ::km double mutant is still unable to sporulate (Table 3). Moreover, electron microscopy observations reveal that, as previously reported for the spoIIIJ87 mutant (16), in both JOB44 (ΔspoIIIJ::km) (Table 1) and AH5025 (spoIIIGE155K/ΔspoIIIJ::km) (Table 1) sporulation is arrested soon after completion of the engulfment process (Fig. 4, compare panels B and C with panel A, which depicts a Spo+ strain). Possibly, the level of activity of σG allowed by the spoIIIGE155K allele is not sufficient to restore sporulation to a spoIIIJ mutant. Alternatively, in addition to promoting activation of σG, SpoIIIJ is required in some other way for sporulation.
FIG. 3.
The spoIIIGE155K allele partially bypasses the need for spoIIIJ expression for σG activity. Expression of sspE-lacZ was monitored during sporulation in the following strains (Table 1): AH5030 (spoIIIG::pEK15) (closed squares), AH5031 (spoIIIG::pEK16) (open circles), AH5024 (spoIIIG::pEK15 spoIIIJ::km) (open squares), AH5025 (spoIIIG::pEK16 spoIIIJ::km) (open diamonds), and JOB34 (spoIIIJ::km) (closed circles). Note that integration of pEK15 into the spoIIIG locus recreates a wild-type spoIIIG sequence, whereas integration of pEK16 results in expression of the spoIIIGE155K allele (see Materials and Methods). Strains were grown in DSM, and samples were taken at the indicated times after the onset of sporulation (T0) and assayed for β-galactosidase activity. The endogenous levels of β-galactosidase production were determined in the wild-type strain MB24 (closed triangles). Enzyme activity is indicated in Miller units (see Materials and Methods).
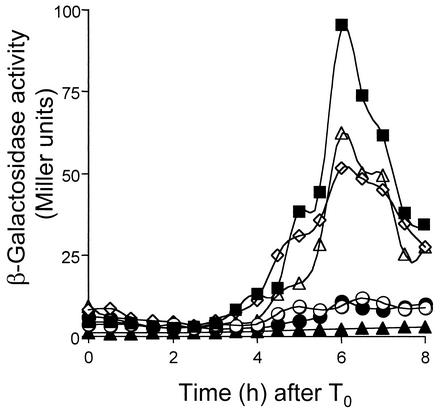
FIG. 5.
Effect of the compartmentalization of spoIIIJ expression on σG-dependent gene expression. An sspE-lacZ gene was introduced into the spoIIIJ mutant JOB44, into the wild-type strain MB24, and into the strains expressing spoIIIJ under the control of its native promoter, the spoIIQ promoter, or the spoIID promoter. The resulting strains (Table 1; see below) were grown in DSM, and samples were taken at 30-min intervals after the onset of sporulation (T0) and assayed for β-galactosidase activity. Closed squares, expression in the wild-type strain (AH1042); open circles, expression in the spoIIIJ::km mutant (JOB34); open diamonds, expression in the PspoIIIJ-spoIIIJ strain (AH5014); open triangles, expression in the PspoIIQ-spoIIIJ strain (AH5015); closed circles, expression in the PspoIID-spoIIIJ strain (AH5016). Endogeneous levels of β-galactosidase production were determined in the wild-type strain MB24 (closed triangles). Enzyme activity is expressed in Miller units (see Materials and Methods).
TABLE 3.
Influence of various mutations on efficiency of sporulation
| Strain | Genotype | Sporulation (CFU/ml)a
|
% Sporu- lation | |
|---|---|---|---|---|
| Viable cells | Heat- resistant cells | |||
| MB24 | Wild type | 5.9 × 108 | 3.5 × 108 | 59.0 |
| JOB44 | spoIIIJ::km | 1.8 × 108 | 1.0 × 102 | <0.001 |
| JOB20 | spoIIIJ::sp | 3.0 × 108 | 1.9 × 103 | <0.001 |
| AH5030 | spoIIIGwtb | 5.4 × 108 | 1.6 × 108 | 30.0 |
| AH5031 | spoIIIGE155K | 9.0 × 108 | 2.9 × 108 | 32.0 |
| AH5024 | spoIIIJ spoIIIGwt | 3.5 × 108 | 1.8 × 103 | <0.001 |
| AH5025 | spoIIIJ spoIIIGE155K | 3.4 × 108 | 2.9 × 103 | <0.001 |
| AH5011 | spoIIIJ PspoIIIJ-spoIIIJ | 5.5 × 108 | 2.2 × 108 | 40.0 |
| AH5012 | spoIIIJ PspoIIQ-spoIIIJ | 4.3 × 108 | 2.6 × 108 | 61.0 |
| AH5013 | spoIIIJ PspoIID-spoIIIJ | 1.2 × 108 | 5.9 × 106 | 5.0 |
| AH2550 | Wild type(pMK3) | 7.9 × 108 | 4.5 × 108 | 57.0 |
| AH5037 | Wild type(pMS210) | 1.5 × 108 | 6.0 × 106 | 4.0 |
| AH5035 | spoIIIJ(pMK3) | 1.8 × 108 | 5.0 × 103 | 0.003 |
| AH5036 | spoIIIJ(pMS210) | 1.5 × 108 | 5.8 × 106 | 3.9 |
| AH62 | spoIIIAΩHU24 | 2.3 × 107 | 9.0 × 102 | <0.001 |
| AH5020 | PspoIIQ-spoIIIA | 3.2 × 108 | 9.0 × 103 | 0.003 |
| AH5029 | PspoIID-spoIIIA | 5.5 × 108 | 3.1 × 108 | 56.0 |
Sporulation was measured 24 h after the onset of the process in liquid sporulation medium as described in Materials and Methods.
spoIIIGwt, wild-type spoIIIG.
FIG. 4.
Electron microscopy of the wild type and the spoIIIJ mutants. The strains were grown in DSM, and samples were taken 12 h after the onset of sporulation and processed for electron microscopy analysis as described in Materials and Methods. (A) Strain AH5011 (spoIIIJ expressed under the control of the spoIIIJ promoter at amyE); (B) JOB44 (spoIIIJ::km); (C) AH5025 (spoIIIJ::km/spoIIIGE155K); (D) AH5012 (spoIIIJ expressed under the control of the spoIIQ promoter at amyE); (E) AH5013 (spoIIIJ expressed under the control of the spoIID promoter at amyE). Strain AH5011 (PspoIIIJ-spoIIIJ) is shown instead of the wild-type MB24 to allow comparison with strains expressing spoIIIJ from the spoIID or spoIIIJ promoter. AH5011 sporulates with wild-type incidence (Table 3) and is indistinguishable from MB24 by electron microscopy (not shown). Both AH5011 (A) and AH5012 (D) form free mature spores like the wild-type MB24 but are represented at a stage prior to lysis of the mother cell to facilitate comparison with the sporulation mutants in panels B, C, and E. Strain JOB44 (B) is blocked just after completion of the engulfment process, as previously reported for the spoIIIJ87 mutant (16). AH5025 (C) and AH5013 (E) are also blocked at the engulfment stage of sporulation. The arrowheads indicate the protective layers that surround the spore (A and D) or the prespore membranes (B, C, and E). Bars, 0.2 μm.
SpoIIIJ does not appear to encode a pheromone peptide.
We also tested whether the SpoIIIJ protein could yield a peptide similar to the one encoded by the ccfA locus of E. faecalis (2). We assayed the effects on sporulation of double alanine substitutions at three different locations of the lipoprotein signal sequence. Each of these strains produced wild-type levels of heat-resistant spores (not shown). We also made substitutions in the signal sequence of spoIIIJ that would be expected to create a peptide identical to the cCF10 peptide from E. faecalis. We tested whether cell-free culture supernatants from this B. subtilis strain would induce wild-type E. faecalis to aggregate, which is a response consistent with the production of the cCF10 peptide (2); however, these supernatant solutions failed to induce clumping of E. faecalis (not shown). Moreover these multiple substitutions in the signal sequence of the SpoIIIJ protein had no effect on sporulation by this B. subtilis strain. Therefore, we found no evidence that spoIIIJ encodes a secreted peptide required for sporulation.
Expression of spoIIIJ in the prespore is sufficient for sporulation.
spoIIIJ is normally expressed during vegetative growth (16), but induction of spoIIIJ transcription from the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Pspac promoter at the onset of sporulation is sufficient for efficient spore formation (39). Because both spoIIIJ and the Pspac-spoIIIJ allele are expressed prior to the asymmetric division of the sporangium, SpoIIIJ could accumulate in the prespore and in the mother cell. To investigate whether expression of spoIIIJ is explicitly required in either the mother cell or the prespore, we fused the coding region of spoIIIJ to the prespore-specific σF-recognized spoIIQ promoter (31) or to the mother cell-specific σE-dependent spoIID promoter (45). The promoter fusions were introduced at the nonessential amyE locus, producing strains AH5012 (PspoIIQ-spoIIIJ) and AH5013 (PspoIID-spoIIIJ) (Table 1). A strain carrying spoIIIJ under the control of its native promoter at amyE (AH5011 [Table 1]) was also constructed in parallel to control for any differences due to ectopic expression of spoIIIJ at amyE. Strain AH5011, which expresses spoIIIJ at amyE under the control of its native promoter, and strain AH5012 (PspoIIQ-spoIIIJ at amyE) both formed spores with the same efficiency as the wild-type strain MB24 (Table 3). In contrast, although expression of spoIIIJ from the spoIID promoter significantly increased the frequency of sporulation of the ΔspoIIIJ::km mutant (JOB44), from 1 × 102 to 5.9 × 106 (Table 3), it did not support wild-type levels of sporulation (AH5013) (Table 3). Electron microscopy revealed that strains AH5011 and AH5012 completed morphogenesis of the spore protective layers (Fig. 4A and D), which relies on the activities of σG and σK (29, 32). In contrast, strain AH5013 was blocked soon after completion of the engulfment process and was indistinguishable from a spoIIIJ null mutant (Fig. 4B and E). In addition, the results in Fig. 5 show that sspE-lacZ was expressed in strains bearing the PspoIIIJ-spoIIIJ allele at amyE (AH5014 [Table 1]) or the PspoIIQ-spoIIIJ fusion (AH5015 [Table 1]) but not the PspoIID-spoIIIJ fusion (AH5016 [Table 1]), indicating that σG was not active in this last strain. Failure of the PspoIID-spoIIIJ fusion to support efficient sporulation may result from low levels of transcription from the spoIID promoter (see below). However, we think that this is unlikely to be the cause of the sporulation phenotype of AH5013. First, translational fusions of the spoIID or spoIIQ promoters to lacZ give rise to similar levels of β-galactosidase activity (52), and even though we used a shorter spoIID promoter fragment (see Materials and Methods), it carries all sequences known to be required for expression of spoIID (45). In addition, a spoIID-lacZ transcriptional fusion is expressed at higher levels than a spoIIQ-lacZ transcriptional fusion in either wild-type or spoIIIJ mutant cells (not shown). We also introduced a replicative plasmid carrying the PspoIID-spoIIIJ fusion (pMS210) into the ΔspoIIIJ::sp mutant JOB20 (Table 1). JOB20 behaves like the ΔspoIIIJ::km mutant with respect to expression of spoIIIJ at amyE from the spoIIIJ, spoIID, or spoIIQ promoter (not shown). The resulting strain (AH5036 [Table 1]) sporulated at the same level as AH5013 (5.9 × 106 spores/ml) (Table 3). The vector used (pMK3 [54]) did not interfere with sporulation or change the spore titer of a spoIIIJ mutant (Table 3). However, we found that pMS210 interfered with sporulation of the wild-type strain MB24 (strain AH5037) (Table 3). Strain AH5037 (multicopy PspoIID-spoIIIJ) sporulated at the same level as AH5013 (ΔspoIIIJ::km/PspoIID-spoIIIJ in single copy) (Table 3). It therefore seems that failure of the PspoIID-spoIIIJ allele to complement a spoIIIJ null mutation is not due to reduced expression of spoIIIJ. Possibly, synthesis of SpoIIIJ is not well tolerated in the mother cell. In any case, our results show that expression of spoIIIJ in the prespore is sufficient for σG activation and for efficient sporulation.
SpoIIIJ-GFP may be degraded in the mother cell.
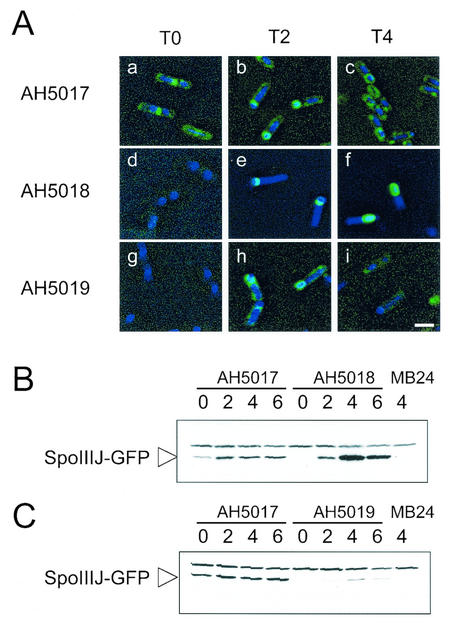
When produced in the mother cell, SpoIIIJ may not localize around the prespore, or if it does, then localization around the prespore is not sufficient for function. To investigate the ability of SpoIIIJ-GFP to localize around the prespore when produced in the mother cell, we fused GFP to the C terminus of SpoIIIJ in cells that express spoIIIJ in single copy at amyE from the spoIID promoter. In addition, we examined the localization of SpoIIIJ-GFP when expressed under the control of its native promoter or from the spoIIQ promoter. In parallel we monitored the accumulation of the SpoIIIJ-GFP fusion protein with an anti-GFP antibody (see Materials and Methods). Strains AH5011, AH5012 and AH5013 (Table 1; see above) were transformed with pLC3 (Fig. 1). Because all three strains bear a deletion of the spoIIIJ locus, transformants were readily obtained that resulted of Campbell-type integration of pLC3 at the amyE region, a recombinational event that fused spoIIIJ to gfp (see Material and Methods). Introduction of the SpoIIIJ-GFP fusion into AH5011 produced a Spo+ strain (AH5017 [Table 1]). Thus, as also found by Murakami et al. (39), the activity of SpoIIIJ does not seems to be inhibited or altered by its fusion to GFP. In strain AH5017, SpoIIIJ-GFP was found associated with the cell membrane and the division septum during vegetative growth (data not shown) and at the onset of sporulation (T0) (Fig. 6A, panel a) (39). Also in confirmation of the observations of Murakami et al. (39), at h 2 of sporulation SpoIIIJ-GFP localized to the asymmetric septum region and around the prespore (Fig. 6A, panel b), and at h 4 the fusion protein was found surrounding the developing spore (Fig. 6A, panel c). Decoration of the cell membrane was also observed at h 2 and persisted at h 4 of sporulation (Fig. 6A, panel c). In AH5017 (PspoIIIJ-spoIIIJ-gfp), SpoIIIJ-GFP accumulated during growth, and its cellular level seemed to increase at about h 2 of sporulation (Fig. 6B). When synthesized in the prespore from the spoIIQ promoter (strain AH5018 [Table 1]), SpoIIIJ-GFP decoration of the asymmetric septum region was first detected 2 h after the onset of sporulation (Fig. 6A, panels d and e), and by h 4 of sporulation SpoIIIJ-GFP was mostly found in a ring-like pattern around the prespore (Fig. 6A, panel f). The results of immunoblot analysis reveal that SpoIIIJ-GFP accumulated from h 2 of sporulation onwards and that its cellular level reached a maximum at around h 4 (Fig. 6B, strain AH5018). This pattern of accumulation coincides with the temporal pattern of expression of a spoIIQ-lacZ fusion (31) (data not shown).
FIG. 6.
SpoIIIJ-GFP localization and accumulation. (A) Typical localization patterns of SpoIIIJ-GFP in the various strains at the indicated times. The strains were grown in DSM, and samples were taken throughout sporulation for observation by fluorescence microscopy with the DNA stain DAPI. Strains AH5017 (PspoIIIJ-spoIIIJ-gfp) (panels a to c), AH5018 (PspoIIQ-spoIIIJ-gfp) (panels d to f), and AH5019 (PspoIID-spoIIIJ-gfp) (panels g to i) were observed at the onset of sporulation (panels a, d, and g), at T2 (panels b, e, and h), and at T4 (panels c, f, and i). Bar, 2 mm. (B and C) Immunoblot analysis of SpoIIIJ-GFP accumulation in DSM in strains AH5017 (PspoIIIJ-spoIIIJ-gfp) and AH5018 (PspoIIQ-spoIIIJ-gfp) (B) and in strains AH5017 (PspoIIIJ-spoIIIJ-gfp) and AH5019 (PspoIID-spoIIIJ-gfp) (C). Results for strain AH5017 are repeated in panels B and C to allow direct comparison. Samples were collected at the end of the exponential phase of growth in DSM, which was defined as the onset of sporulation (lanes 0) and at 2, 4, and 6 h thereafter. Proteins (30 μg) were electrophoretically resolved on sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose membranes, and GFP was detected with a polyclonal rabbit antibody (see Materials and Methods). The arrowhead indicates the position of the SpoIIIJ-GFP fusion protein. Another band with slower migration that is also found in MB24 represents a cross-reactive species. The last lane in panels B and C contains 30 μg of an extract prepared from a culture of the wild-type strain (MB24) at h 4 of sporulation in DSM.
When synthesized in the mother cell under the control of the spoIID promoter (strain AH5019 [Table 1]), SpoIIIJ-GFP was again first detected 2 h after the onset of sporulation by fluorescence microscopy (Fig. 6A, panels g and h) and by immunoblot analysis (Fig. 6C). At this time in spore development, the fusion protein decorated the cell membrane region and the septal region, and it also tended to localize around the prespore (Fig. 6A, panels g and h). Although the immunoblot analysis of AH5019 reveals that SpoIIIJ-GFP accumulated to reduced levels compared to the strain (AH5017) expressing the PspoIIIJ-spoIIIJ-gfp fusion at amyE (Fig. 6C), the intensities of the fluorescence signal in the cell membrane region, the septal region, or around the prespore were equivalent in the two strains (Fig. 6A, compare panels b and h). Moreover, by h 2 of sporulation, about 22% of the cells in AH5017 or AH5019 showed condensed prespore chromosomes (>100 cells counted), as judged from the analysis of DAPI-stained images (see Materials and Methods). These cells are presumed to have completed the asymmetric division of the sporangium. Of these, about 80% in either AH5017 (PspoIIIJ-spoIIIJ-gfp) or AH5019 (PspoIID-spoIIIJ-gfp) showed decoration of the septum or prespore regions by SpoIIIJ-GFP (see also below). It appears that the PspoIID-spoIIIJ-gfp allele results in sufficient product to decorate the prespore region in most of the sporulating cells at h 2 of sporulation. About 17% of the cells (over 100 cells counted) of both AH5017 or AH5019 appeared to have completed the engulfment process by h 4 of sporulation, based on the analysis of DAPI images (see Materials and Methods). However, at this time in sporulation, SpoIIIJ-GFP decoration of both the cell membrane and the prespore region was considerably reduced in AH5019 compared to AH5017 (Fig. 6A, compare panels c and i). This reduction in the decoration of the prespore was not accompanied by an increase in cytoplasmic fluorescence, even though the accumulation of SpoIIIJ-GFP as monitored by immunoblot analysis appeared to increase (Fig. 6C). The immunoblot analysis did not reveal signs of proteolysis of SpoIIIJ-GFP (Fig. 6C). Nevertheless, even though a spoIID-lacZ fusion appears to be expressed at higher levels in both wild-type and spoIIIJ mutant cells than is a spoIIQ-lacZ fusion (see above), our PspoIID-spoIIIJ-gfp fusion resulted in reduced cellular levels of SpoIIIJ-GFP. The observation that a PspoIID-spoIIIJ multicopy allele interfered with sporulation of a wild-type strain (see above) suggests that accumulation of SpoIIIJ in the mother cell is not well tolerated. Together, the results suggest that when produced in the mother cell, the SpoIIIJ-GFP protein may be subjected to degradation.
Expression of spoIIIA in the prespore does not permit activation of σG
The results described above suggest that SpoIIIJ and the spoIIIA-encoded products may function at the end of the engulfment process to antagonize the action exerted by SpoIIAB upon σG. However, while expression of spoIIIJ is sufficient in the prespore (this work), earlier studies have shown that the spoIIIA operon, which is transcribed from a promoter recognized by the σE form of RNA polymerase, is specifically required in the mother cell (23, 24). To determine whether expression of spoIIIA in the mother cell is an absolute requirement or whether the spoIIIA-encoded products could promote σG activation and sporulation if produced only in the prespore, we fused the spoIIIA operon to the prespore-specific spoIIQ promoter (strain AH5020 [Table 1]). The results in Table 3 indicate that expression of the spoIIIA operon from the spoIIQ promoter resulted in very poor sporulation (about 104 spores/ml). We also found that expression of the spoIIIA operon solely from the spoIIQ promoter (in strain AH5020, bearing the PspoIIQ-spoIIIA allele) did not support expression of the σG-dependent sspE-lacZ fusion (not shown). Moreover, electron microscopy observations revealed that strain AH5020 is blocked soon after completion of the engulfment sequence (data not shown). In contrast, expression of the spoIIIA operon from the similarly expressed mother cell-specific spoIID promoter (45, 52) resulted in wild-type levels of sporulation (Table 3). We suggest that at the end of the engulfment process, the spoIIIA-encoded products from the mother cell and SpoIIIJ from the prespore are both required to antagonize the inhibitory action imposed by SpoIIAB upon σG in the prespore (Fig. 7).
FIG. 7.
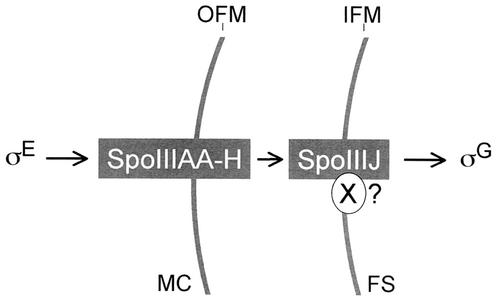
Model for the signaling pathway that controls σG activation. Shown is the proposed sequence of events that originate in the mother cell leading to σG activation in the prespore at engulfment. The eight membrane-associated proteins encoded by the spoIIIA locus (SpoIIIAA to SpoIIIAH) and SpoIIIJ participate in conveying the signal that activates σG. The spoIIIA-encoded products act from the mother cell across the space delimited by the outer forespore membrane (OFM) and the inner forespore membrane (IFM). SpoIIIJ in turn acts from the prespore. SpoIIIJ may be required for localizing a prespore-specific component (X) of the spoIIIA signaling pathway. The signal conveyed through the spoIIIA-encoded proteins and SpoIIIJ results in the activation of σG; it appears to serve in part to antagonize the inhibitory action of SpoIIAB upon σG, but it may also involve other, as-yet-unidentified factors. MC, mother cell; FS, engulfed prespore.
DISCUSSION
Here we have analyzed the role of spoIIIJ in the activation of σG. Mutations in spoIIIJ arrest development soon after engulfment of the prespore by the mother cell, and greatly diminish the activity of σG (16). However, mutations in spoIIIJ do not prevent transcription of the spoIIIG gene (16; this work), and as shown here they allow the accumulation of σG to nearly wild-type levels (Fig. 2). Therefore, most of the σG that accumulates in spoIIIJ mutant cells is inactive. This is reminiscent of the situation in a spoIIIA mutant, in which σG also accumulates but is inactive (26). Activation of σG at the end of the engulfment process then relies on spoIIIA- and spoIIIJ-dependent events. The available evidence suggests that the inhibitory action of SpoIIAB is responsible for the absence of σG activity in a spoIIIA mutant. SpoIIAB tends to disappear from the prespore compartment after conclusion of the engulfment process in a spoIIIA-dependent manner (28). More importantly, a single amino acid substitution in σG that results in inefficient binding by SpoIIAB in vitro bypasses the requirement for spoIIIA expression for σG activity (26). This implies that SpoIIIA acts after engulfment of the prespore by the mother cell to antagonize the inhibition of σG activity by SpoIIAB (26). We found that expression of the spoIIIGE155K allele (encoding the σGE155K form of σG) also allows σG activity in the absence of SpoIIIJ (Fig. 3). Therefore, our results suggest that both SpoIIIJ and the spoIIIA-encoded products are involved in relieving the inhibition imposed by SpoIIAB upon σG in the prespore. The spoIIIGE155K mutation does not causes premature expression of sspE-lacZ and in that sense does not uncouple the activity of σG from the conclusion of the engulfment sequence (Fig. 3) (26). One possible explanation is that SpoIIAB is important to antagonize σG only towards the end of the engulfment sequence and that another, as-yet-unidentified factor contributes to the negative regulation of σG activity during the process of engulfment.
We also note that even though the spoIIIGE155K allele bypasses the requirement for SpoIIIJ for both σG and σK activities, it did not restore formation of heat-resistant spores (Fig. 4C; Table 3). Interestingly, the spoIIIGE155K allele does not restore sporulation to a spoIIIA mutant either (26). There are several possible explanations for these observations. First, because σG is autoregulatory (25, 55), a mutation that would make it less susceptible to SpoIIAB could in principle result in increased levels of σG activity in the mother cell. However, ectopic activation of σG in the mother cell is unlikely to be the cause of the sporulation phenotype of the spoIIIGE155K/ΔspoIIIJ::km double mutant, since a strain carrying the spoIIIGE155K allele in an otherwise wild-type background produces heat-resistant spores with wild-type efficiency (26) (not shown). The block in sporulation of the spoIIIGE155K/ΔspoIIIJ::km double mutant could also result from insufficient activity of σG, even though peak levels of sspE-lacZ expression in the spoIIIGE155K/ΔspoIIIJ::km double mutant are only half of what is found in a strain carrying only the spoIIIGE155K allele and which is Spo+ (Fig. 3). The E155K substitution was introduced in σG (26) on the basis of the finding that a glutamic acid-to-lysine substitution was found at an equivalent position (E149K) in a screen for σF mutants with reduced affinity for SpoIIAB (11). The structure of a complex between a dimer of the Bacillus stearothermophilus SpoIIAB protein and σF helps to explain the specificity of SpoIIAB for σF and for σG (7). In the B. stearothermophilus σF protein, the residue equivalent to E149 of B. subtilis σF, as well as three other residues found in genetic screens for mutants resistant to inhibition by SpoIIAB (11), is located within a region that contains 17 amino acids found to interact with SpoIIAB in the crystal structure (7). Of those residues, 15 are either identical or homologous in σG and 3 are uniquely conserved between σF and σG (7). However, the structure also suggests that even though σG appears to contact SpoIIAB through the same region that mediates the interaction of σF with SpoIIAB, the details of the interaction of σG with SpoIIAB may differ (7). It is possible that the E155K substitution makes σG less refractory to SpoIIAB than the corresponding mutation (E149K) in σF. Lastly, another possibility for the incapacity of spoIIIGE155K/spoIIIA or spoIIIGE155K/ΔspoIIIJ::km double mutants to sporulate is that in addition to the activation of σG, SpoIIIJ and the eight products of the spoIIIA operon play other roles in sporulation (26).
In any case, SpoIIIJ and the products of the spoIIIA locus function at least in part in the same signaling pathway that is involved in the activation of σG in the prespore after completion of the engulfment sequence. This signaling pathway appears to originate in the mother cell with the σE-dependent transcription of the spoIIIA operon (23). Because their synthesis is restricted to the mother cell compartment, it seems unlikely that the spoIIIA-encoded proteins interact directly with SpoIIAB in the prespore (26). However, other than SpoIIAB and σG, no prespore-specific components of the σE-to-σG signaling pathway have been identified. We tested whether the function of SpoIIIJ is specifically required in either the prespore or the mother cell by expressing spoIIIJ under the control of a promoter utilized by the σF (spoIIQ) or σE (spoIID) form of RNA polymerase. We found that expression of spoIIIJ is sufficient in the prespore both for the activation of σG and for sporulation (Fig. 6; Table 3). Nevertheless, expression of a PspoIID-spoIIIJ fusion in single copy significantly increases the sporulation efficiency of a spoIIIJ null mutant, and thus it appears that SpoIIIJ retains some functionality when expressed in the mother cell (Table 3). The lack of full complementation of a spoIIIJ null mutant by PspoIID-spoIIIJ does not seem to be due to reduced expression of spoIIIJ, because the presence of the same fusion in a multicopy plasmid does not further increase the efficiency of sporulation. Since the multicopy PspoIID-spoIIIJ allele interferes with sporulation of the wild-type strain, we speculate that SpoIIIJ may promote the incorporation into the mother cell side of the prespore membranes of a protein that interferes with the signaling events that lead to σG activation and that the mother cell has mechanisms to prevent SpoIIIJ accumulation to high levels. Preferential degradation of SpoIIIJ in the mother cell could be another example of proteolysis playing a role in the reinforcement of the correct compartmentalization on σ factor activity during sporulation (see, for example, references 18 and 40).
In contrast to the case for spoIIIJ, we found that expression of spoIIIA from the prespore-specific spoIIQ promoter does not support efficient sporulation (Fig. 6; Table 3). Like SpoIIIJ (39; this work), the spoIIIA-encoded products also localize around the developing spore (46; W. Blaylock and C. P. Moran, Jr., unpublished results). Together, our results suggest that the spoIIIA-encoded products from the mother cell and SpoIIIJ from the prespore act together to convey a signal that results in the activation of σG in the prespore after completion of the engulfment process (Fig. 7). We favor the idea that the spoIIIA-encoded proteins and SpoIIIJ function in the same pathway as suggested in Fig. 7, because mutations in either loci respond similarly to the spoIIIGE155K allele. However, we cannot presently exclude that spoIIIA and spoIIIJ act independently of each other. SpoIIIJ is related to the YidC membrane protein translocase of E. coli (47, 49) and may have a similar function in B. subtilis (57). Since its function is sufficient in the prespore, SpoIIIJ may be required for the membrane localization of an as-yet-unknown forespore-specific component in the σE to σG signaling pathway (Fig. 7). It remains unclear whether SpoIIAB is the only factor responsible for keeping σG inactive prior to engulfment and, if this is the case, how σF can escape from SpoIIAB while σG is held inactive. It is also unknown whether SpoIIAA and SpoIIE, which are required for σF activation (1, 3, 4, 12, 13, 15), play a role in the activation of σG.
Acknowledgments
We thank Bill Blaylock and Gonçalo Real for helpful discussions and for critically reading the manuscript, Patrick Piggot and Alan Grossman for the gift of strains, Daniel Kalman for help with the fluorescence microscope and for helpful discussions, and Hong Hy (Emory Microscopy Core Facility) and Jan Pohl (Emory Microchemical Facility) for technical assistance.
This work was supported by grants Praxis XXI/PCNA/C/BIO/13201/98 and PRAXIS/BIO/35109/99 from the Fundação para a Ciência e a Tecnologia to A.O.H. and by grant GM54395 from the National Institutes of Health to C.P.M. M.S. is the recipient of a Ph.D. fellowship (PRAXIS XXI/BD 18 251/98) from the Fundação para a Ciência e a Tecnologia.
REFERENCES
- 1.Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77:195-205. [DOI] [PubMed] [Google Scholar]
- 2.Antiporta, M. H., and G. M. Dunny. 2002. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 184:1155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arigoni, F., K. Pogliano, C. D. Webb, P. Stragier, and R. Losick. 1995. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science 270:637-640. [DOI] [PubMed] [Google Scholar]
- 4.Arigoni, F., L. Duncan, S. Alper, R. Losick, and P. Stragier. 1996. SpoIIE governs the phosphorylation state of a protein regulating transcription factor sigma F during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:3238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of σB levels and activity in Bacillus subtilis. J. Bacteriol. 175:2347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, E. A., and S. A. Darst. 2000. The anti-σ factor SpoIIAB forms a 2:1 complex with σF, contacting multiple conserved regions of the σ factor. J. Mol. Biol. 300:17-28. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, E. A., S. Masuda, J. L. Sun, O. Muzzin, C. A. Olson, S. Wang, and S. A. Darst. 2002. Crystal structure of the Bacillus stearothermophilus anti-σ factor SpoIIAB with the sporulation factor σF. Cell 108:795-807. [DOI] [PubMed] [Google Scholar]
- 8.Coppolechia, R., H. DeGrazia, and C. P. Moran, Jr. 1991. Deletion of spoIIAB blocks endospore formation in Bacillus subtilis at an early stage. J. Bacteriol. 173:6678-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting, S., S. Panzer, and R. Losick. 1989. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J. Mol. Biol. 207:393-404. [DOI] [PubMed] [Google Scholar]
- 10.Cutting, S. M., and P. B. V. Horn. 1990. Genetics analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 11.Decatur, A., and R. Losick. 1996. Multiple sites of contact between the Bacillus subtilis developmental transcription factor σF and its anti-sigma factor SpoIIAB. Genes Dev. 10:2348-2358. [DOI] [PubMed] [Google Scholar]
- 12.Diederich, B., J. F. Wilkinson, T. Magnin, M. Najafi, J. Errington, and M. D. Yudkin. 1994. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor sigma F of Bacillus subtilis. Genes Dev. 8:2653-2663. [DOI] [PubMed] [Google Scholar]
- 13.Duncan, L., A. Alper, and R. Losick. 1996. SpoIIAA governs the release of the cell-type specific transcription factor σF from its anti-sigma factor SpoIIAB. J. Mol. Biol. 260:147-164. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, L., and R. Losick. 1993. SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein sigma F from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 90:2325-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan, L., S. Alper, F. Arigoni, R. Losick, and P. Stragier. 1995. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science 270:641-644. [DOI] [PubMed] [Google Scholar]
- 16.Errington, J., L. Appleby, R. A. Daniel, H. Goodfellow, S. R. Partridge, and M. D. Yudkin. 1992. Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sigma G activity at an intermediate stage of sporulation. J. Gen. Microbiol. 138:2609-2618. [DOI] [PubMed] [Google Scholar]
- 17.Foulger, D., and J. Errington. 1993. Effects of new mutations in the spoIIAB gene of Bacillus subtilis on the regulation of σF and σG activities. J. Gen. Microbiol. 139:3197-3203. [DOI] [PubMed] [Google Scholar]
- 18.Fujita, M., and R. Losick. 2002. An investigation into the compartmentalization of the sporulation transcription factor sigma E in Bacillus subtilis. Mol. Microbiol. 43:27-38. [DOI] [PubMed] [Google Scholar]
- 19.Harry, E. J., K. Pogliano, and R. Losick. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henriques, A. O., L. R. Melsen, and C. P. Moran, Jr. 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 23.Illing, N., and J. Errington. 1991. The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the sigma E form of RNA polymerase. Mol. Microbiol. 5:1927-1940. [DOI] [PubMed] [Google Scholar]
- 24.Illing, N., M. Young, and J. Errington. 1990. Use of integrational plasmid excision to identify cellular localization of gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 172:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karmazyn-Campelli, C., C. Bonamy, B. Savelli, and P. Stragier. 1989. Tandem genes encoding σ-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 3:150-157. [DOI] [PubMed] [Google Scholar]
- 26.Kellner, E. M., A. Decatur, and C. P. Moran, Jr. 1996. Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol. Microbiol. 21:913-924. [DOI] [PubMed] [Google Scholar]
- 27.Kenney, T. J., and C. P. Moran, Jr. 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchman, P. A., H. DeGrazia, E. M. Kellner, and C. P. Moran, Jr. 1993. Forespore-specific disappearance of the sigma-factor antagonist spoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol. Microbiol. 8:663-671. [DOI] [PubMed] [Google Scholar]
- 29.Kroos, L., B. Zhang, H. Ichikawa, and Y.-T. N. Yu. 1999. Control of σ factor activity during Bacillus subtilis sporulation. Mol. Microbiol. 31:1285-1294. [DOI] [PubMed] [Google Scholar]
- 30.Kunst, F., et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 31.Londoño-Vallejo, J.-A., C. Fréhel, and P. Stragier. 1997. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 24:29-39. [DOI] [PubMed] [Google Scholar]
- 32.Losick, R., and P. Stragier. 1992. Crisscross regulation of cell-type-specific gene expression during development in Bacillus subtilis. Nature 355:601-604. [DOI] [PubMed] [Google Scholar]
- 33.Margolis, P., A. Driks, and R. Losick. 1991. Establishment of cell type by compartmentalized activation of a transcription factor. Science 254:562-565. [DOI] [PubMed] [Google Scholar]
- 34.Mason, J. M., R. H. Hackett, and P. Setlow. 1988. Regulation of transcription of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ fusions. J. Bacteriol. 170:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min, K.-T., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. σF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-σ factor that is also a protein kinase. Cell 74:735-742. [DOI] [PubMed] [Google Scholar]
- 37.Minton, N. P. 1984. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene 31:269-273. [DOI] [PubMed] [Google Scholar]
- 38.Mota, L. J., P. Tavares, and I. Sá-Nogueira. 1999. Mode of action of AraR, the key regulator of l-arabinose metabolism in Bacillus subtilis. Mol. Microbiol. 33:476-489. [DOI] [PubMed] [Google Scholar]
- 39.Murakami, T., K. Haga, M. Takeuchi, and T. Sato. 2002. Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J. Bacteriol. 184:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan, Q., D. A. Garsin, and R. Losick. 2001. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-σ factor in B. subtilis. Mol. Cell 8:873-883. [DOI] [PubMed] [Google Scholar]
- 41.Partridge, S., and J. Errington. 1993. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8:945-955. [DOI] [PubMed] [Google Scholar]
- 42.Piggot, P. J., and R. Losick. 2001. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives from genes to cells. ASM Press, Washington, D.C.
- 43.Rather, P. N., and C. P. Moran, Jr. 1988. Compartment-specific transcription in Bacillus subtilis: identification of the promoter gdh. J. Bacteriol. 170:5086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rather, P. N., R. Coppolechia, H. DeGrazia, and C. P. Moran, Jr. 1990. Negative regulator of σG-controlled gene expression in stationary-phase Bacillus subtilis. J. Bacteriol. 172:709-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rong, S., M. S. Rosenkrantz, and A. L. Sonenshein. 1986. Transcriptional control of the Bacillus subtilis spoIID gene. J. Bacteriol. 165:771-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudner, D. Z., and R. Losick. 2001. Morphological coupling in development: lessons from prokaryotes. Dev. Cell 1:733-742. [DOI] [PubMed] [Google Scholar]
- 47.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt, R., P. Margolis, L. Duncan, R. Coppolecchia, C. P. Moran, Jr., and R. Losick. 1990. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:9221-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scotti, P. A., M. L. Urbanus, J. Brunner, J. W. de Gier, G. von Heijne, C. van der Does, A. J. Driessen, B. Oudega, and J. Luirink. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano, M., R. Zilhão, A. J. Ozin, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 1999. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J. Bacteriol. 181:3632-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Setlow, B., N. Magill, P. Febbroriello, L. Nakhimovsky, D. E. Koppel, and P. Setlow. 1991. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J. Bacteriol. 173:6270-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharp, M. D., and K. Pogliano. 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295:137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stragier, P. 1992. Establishment of forespore-specific gene expression during sporulation of Bacillus subtilis, p. 297-310. In J. A. Cole, F. Mohan, and C. Dow (ed.), Prokaryotic structure and function. Society for General Microbiology, Cambridge, United Kingdom.
- 54.Sullivan, M. A., R. E. Yasbin, and F. E. Young. 1984. New shuttle vectors for Bacillus subtilis and E. coli which allow rapid detection of inserted fragments. Gene 29:21-26. [DOI] [PubMed] [Google Scholar]
- 55.Sun, D., R. M. Cabrera-Martinez, and P. Setlow. 1991. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor σG. J. Bacteriol. 173:2977-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun, Y.-L., M. D. Sharp, and K. Pogliano. 2000. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J. Bacteriol. 182:2919-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tjalsma, H., S. Bron, and J. M. van Dijl. 2003. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem., 278:15622-15632. [DOI] [PubMed]
- 58.van Helvoort, J. M., and C. L. Woldringh. 1994. Nucleoid partitioning in Escherichia coli during steady-state growth and upon recovery from chloramphenicol treatment. Mol. Microbiol. 13:577-583. [DOI] [PubMed] [Google Scholar]