Abstract
The functions of the serpin PAI-1 (plasminogen activator inhibitor-1) are based on molecular interactions with its target proteases uPA and tPA (urokinase-type and tissue-type plasminogen activator respectively), with vitronectin and with endocytosis receptors of the low-density-lipoprotein family. Understanding the significance of these interactions would be facilitated by the ability to block them individually. Using phage display, we have identified the disulfide-constrained peptide motif CFGWC with affinity for natural human PAI-1. The three-dimensional structure of a peptide containing this motif (DVPCFGWCQDA) was determined by liquid-state NMR spectroscopy. A binding site in the so-called flexible joint region of PAI-1 was suggested by molecular modelling and validated through binding studies with various competitors and site-directed mutagenesis of PAI-1. The peptide with an N-terminal biotin inhibited the binding of the uPA–PAI-1 complex to the endocytosis receptors low-density-lipoprotein-receptor-related protein 1A (LRP-1A) and very-low-density-lipoprotein receptor (VLDLR) in vitro and inhibited endocytosis of the uPA–PAI-1 complex in U937 cells. We conclude that the isolated peptide represents a novel approach to pharmacological interference with the functions of PAI-1 based on inhibition of one specific molecular interaction.
Keywords: endocytosis, ligand docking, lipoprotein receptor, phage display, plasminogen activator inhibitor-1 (PAI-1), vitronectin
Abbreviations: bis-ANS, 4,4′-dianilino-1,1′-binaphthyl-5,5-disulfonic acid; cfu, colony-forming unit; CMC, critical micelle concentration; h, α-helix; HBS, Hepes-buffered saline; HEK-293T cells, human embryonic kidney 293T cells; HRP, horseradish peroxidase; LDLR, low-density-lipoprotein receptor; LRP-1A, LDLR-related protein; MBS, Mes-buffered saline; MC, Monte Carlo; NOE, nuclear Overhauser effect; PAI-1, plasminogen activator inhibitor type 1; R, relaxed; RAP, receptor-associated protein; RCL, reactive centre loop; RMSD, root mean square deviation; RU, response units; s, β-strand; S, stressed; SPD, serine protease domain; SPR, surface plasmon resonance; tPA, tissue-type plasminogen activator; uPA, urokinase-type plasminogen activator; uPAR, uPA receptor; VLDLR, very-low-density-lipoprotein receptor
INTRODUCTION
The plasminogen activators tPA (tissue-type) and uPA (urokinase-type) are serine proteases catalysing the conversion of the zymogen plasminogen into the active serine protease plasmin which can degrade fibrin and other extracellular proteins. The activities of tPA and uPA are regulated by PAI-1 (plasminogen activator inhibitor-1) (for reviews, see [1,2]). In tissues, plasminogen activation is concentrated on cell surfaces where the glycosylphosphatidylinositol-anchored uPAR (uPA receptor) recruits uPA (for a review, see [3]). Plasminogen activation is implicated in the spread of cancers in which uPA-catalysed plasmin generation in tumours leads to degradation of basement membranes, thereby supporting the cancer cell invasiveness. It was therefore initially surprising that a high tumour level of PAI-1 is an even better marker for a poor prognosis. The functions of PAI-1 in the spread of malignant tumours are not completely known, but one important aspect seems to be a pro-angiogenic effect of PAI-1 (for a review, see [2]).
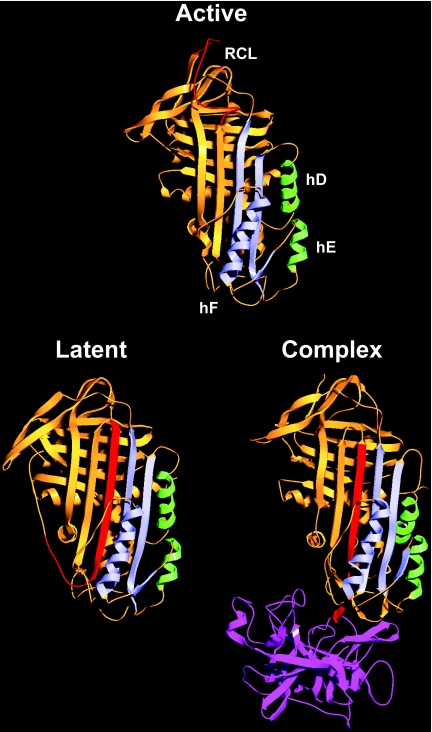
PAI-1 belongs to the serpin family of protease inhibitors (for a review, see [4]). During the reaction of a serpin with its target serine protease, the P1–P1′ bond in the RCL (reactive centre loop) of the serpin is cleaved, and the protease becomes attached to the serpin by an ester bond between the active-site serine and the P1 residue [5]. At this point, the N-terminal part of the RCL is inserted as s4A (β-strand 4A) in the serpin (Figure 1). The protease is thereby pulled to the opposite pole of the serpin, its active site is distorted, and hydrolysis of the ester bond prevented, resulting in a trapped covalent serpin–protease complex [6,7]. Insertion of the RCL changes the serpin from a metastable S (stressed) conformation to a more stable R (relaxed) conformation, and the released energy is believed to fuel protease distortion [8,9]. For some serpins, including PAI-1, the N-terminal part of the RCL can be inserted without P1–P1′ bond cleavage, resulting in formation of the latent state [10]. The S-to-R transition includes conformational changes around hD (α-helix D) and hE which serve as a flexible joint region when the small and the large serpin fragments slide apart to allow RCL insertion [11] (Figure 1). In PAI-1, a hydrophobic cavity in this region serves as the binding site for multiple PAI-1-neutralizing compounds and is rearranged during the S-to-R transition [12–14]. Also, a high-affinity binding site for vitronectin exists in the flexible joint region of the S-form of PAI-1, but not in the R-form [15,16].
Figure 1. Serpin structures.
Ribbon diagrams of active PAI-114–1B (a stable active PAI-1 variant carrying four mutations [42], PDB code 1DVM [43]), latent PAI-1 (PDB code 1LJ5), and the complex between the serpin α1-proteinase inhibitor and the protease elastase (PDB code 2D26 [6]). For the serpins, the RCL is in red, the large serpin fragment is in orange, the small serpin fragment including hF is in blue and hD and hE from the flexible joint region are in green. Elastase is shown in purple.
PAI-1's complexes with its target proteases bind to and are endocytosed by certain receptors of the LDLR (low-density-lipoprotein receptor) family, including LRP-1A (LDLR-related protein 1A), megalin and VLDLR (very-low-density-lipoprotein receptor). These receptors have also been implicated in initiating intracellular signalling events (for a review, see [17]). Binding of the uPA–PAI-1 complex to LRP-1A and VLDLR was shown previously to require basic residues in hD and hE in the flexible joint region of the inhibitor [18–20].
Various attempts have been made to design compounds which could be used to interfere pharmacologically with the functional molecular interactions of PAI-1. The studied types of compounds include monoclonal antibodies and organochemicals (reviewed in [2]). In order to isolate PAI-1binding agents with specificity comparable with that of monoclonal antibodies and a size eventually allowing for chemical synthesis and modification, we screened phage display peptide libraries with PAI-1 as bait and characterized the effects of one isolated peptide sequence on the various molecular interactions of PAI-1.
EXPERIMENTAL
Cell culture
U937 cells (ATCC CR1 1593) were cultured in RPMI 1640 medium. HEK-293T (human embryonic kidney 293T) cells were cultured in Dulbecco's modified Eagle's medium. Both media were supplemented with 10% (v/v) foetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin. The medium for HEK-293T cells additionally contained 1% non-essential amino acids. Cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C.
PAI-1
Natural glycosylated wild-type human PAI-1 was purified from serum-free conditioned medium of dexamethasone-treated HT-1080 fibrosarcoma cells [21]. Murine PAI-1 cDNA was a gift from Dr Ann Gills (Katholieke Universiteit Leuven, Leuven, Belgium). A DNA fragment encoding His6-tagged murine PAI-1 was constructed by PCR and cloned for expression in Escherichia coli cells as described previously for human PAI-1 [15,22]. Site-directed mutagenesis, expression of His6-tagged PAI-1 variants in E. coli and purification by metal-affinity chromatography and size-exclusion chromatography was performed as described in [15,22]. The specific inhibitory activity of PAI-1, i.e. the fraction of PAI-1 capable of inhibiting the proteolytic activity of uPA, was determined in a chromogenic assay as described previously [22].
Peptides
The following peptides were purchased from Eurogentec, from Biopeptide or from Peptide Protein Research Ltd: NH2-DVPCFGWCQDA-CO2H; biotin–DVPCFGWCQDA-CONH2; biotin–DVPCFGWCQDAGAKK-CONH2, all with a disulfide-bridge-constrained conformation. Linear forms with the two cysteine residues replaced by serines were obtained for control experiments. We predominantly used freshly prepared stock solutions of less than 500 μM biotin–paionin-1 in HBS (Hepes-buffered saline: 140 mM NaCl and 10 mM Hepes, pH 7.4). If storage was required, stock solutions were snap-frozen in liquid N2 and kept at −80 °C.
Miscellaneous materials
Vitronectin in a multimerized form was purchased from BD Biosciences, chicken ovalbumin and heparin from porcine intestinal mucosa was purchased from Sigma–Aldrich, rat PAI-1 was purchased from Molecular Innovations, and uPA was purchased from Wakamoto Pharmaceutical Company. α1-Proteinase inhibitor was a gift from Professor J. Enghild (Department of Molecular Biology, University of Aarhus, Aarhus, Denmark). Antithrombin III was a gift from the late Professor S. Magnusson (Department of Molecular Biology, University of Aarhus, Aarhus, Denmark). RAP (receptor-associated protein) was prepared as described previously [23]. Antibodies were described previously as follows: monoclonal anti-PAI-1 antibodies Mab-1, Mab-2, Mab-3, Mab-5, Mab-6 and Mab-7 [24–26]; polyclonal rabbit anti-PAI-1 antibody [27]; and monoclonal anti-uPA Mab-2 [28].
Receptors
A stop codon following the codon for amino acid 772 was introduced into the human VLDLR type-I cDNA [29]. This cDNA, encoding soluble VLDLR (residues 1–772) without the cytosolic domain and transmembrane region, was then cloned into the plasmid pcDNA3.1(−) (Invitrogen) and was authenticated by DNA sequencing. E. coli DH5α cells (Invitrogen) were used for plasmid propagation, and transient transfection of HEK-293T cells was carried out using standard methods [30]. Soluble VLDLR was purified from serum-free conditioned medium from transfected cells by affinity chromatography at 4 °C on a 5 ml RAP-coupled Sepharose column. HBS supplemented with 2 mM CaCl2 and 1 mM MgCl2 was used for equilibration and washing of the column, while elution was carried out using 0.1 M ethanoic acid, pH 3.5, 0.5 M NaCl and 10 mM EDTA. Fractions were neutralized immediately with 0.1 vol. 1 M Tris/HCl, pH 9, and 25 mM CaCl2, and those containing soluble VLDLR were selected on the basis of 125I-RAP ligand blots [31]. Purified soluble VLDLR was concentrated with a Centriplus YM-10 spin column, dialysed into PBS and stored at −80 °C.
Full-length LRP-1A purified from human placentas was a gift from Dr J. Gliemann (Institute of Medical Biochemistry, University of Aarhus, Aarhus, Denmark) [32].
Isolation of PAI-1-binding peptides from a phage display peptide library
Phage display random peptide libraries in the formats X7, CX7C, CX10C and CX3CX3CX3C (where C is cysteine and X is any residue) were obtained from Dr Erkki Koivunen (Division of Biochemistry, University of Helsinki, Helsinki, Finland) [33]. Selection of PAI-1-binding peptides was performed using standard procedures [34]. Phage and PAI-1 expressed from HT-1080 cells (100 nM) were incubated in MBS (Mes-buffered saline: 140 mM NaCl and 10 mM Mes, pH 6) supplemented with 1% (w/v) BSA and 5% (v/v) glycerol for 1 h and formed phage–PAI-1 complexes were captured on polystyrene-adsorbed monoclonal anti-PAI-1 antibody Mab-7 (first and fourth rounds of panning), Mab-5 (second and fifth rounds of panning) or Mab-3 (third round of panning). After washing with MBS, bound phage was eluted with 0.1 M glycine, pH 2.2. For all rounds of panning, enrichment of binders was monitored by determining the amount of input and output phage, measured as cfu (colony-forming units).
Phage was prepared from 16 individual clones eluted from each of the panning rounds 2–5 and tested for PAI-1 binding in ELISA format 1 (see below). PAI-1-binding peptides were identified by sequencing the corresponding phage [25]. Concentrated phage (∼1012 cfu/ml) was prepared by precipitation with NaCl and PEG [poly(ethylene glycol)] 6000 [34], followed by resuspension in MBS supplemented with 10% glycerol and 0.02% NaN3.
The binding of the phage to PAI-1 from HT-1080 cells was compared by incubating ∼109 cfu/ml phage with PAI-1 (300–0.2 nM) followed by detection of any formed phage–PAI-1 complexes in ELISA format 1. The EC50 value for each phage was determined as the PAI-1 concentration required for half-maximal saturation in this ELISA.
NMR structure determination
NMR spectra were recorded on a Bruker NMR spectrometer operating at 600.13 MHz at 10 °C. The sample was 1 mM NH2-DVPCFGWCQDA-CO2H in 20% (v/v) 2H6-DMSO, 10 mM NaH2PO4 and 75 μM NaN3 in water, pH 4.7. COSY [35], and NOESY spectra (400 ms) [36] were recorded. Water was suppressed using pre-saturation. NMR data was processed using NMRPipe [37]. Complete assignment of 1H chemical shifts [38] was carried out using NMRVIEW [39] (see Supplementary Table S1 at http://www.BiochemJ.org/bj/399/bj3990387add.htm). A total of 1000 structures were calculated using the CNS (Crystallographic and NMR system) program package version 1.1 [40] using a total of 51 NOE (nuclear Overhauser effect)-derived distance constraints (see Supplementary Table S2 at http://www.BiochemJ.org/bj/399/bj3990387add.htm). The obtained structures were visualized using the program MOLMOL [41]. Based on the orientations of the aromatic Phe5 and Trp7 side chains, 88 of the 100 structures with the lowest energy could be divided into nine distinct conformational groups.
Molecular modelling
The molecular modelling studies were carried out using the Cerius2 suite of programs (version 4.8, Accelrys Software) and the PDB files 1DVM (active PAI-1 carrying the stabilizing mutations N152H/K156T/Q321L/M356I, also known as PAI-114–1B [42,43]) and 1LJ5 (latent PAI-1). All waters of crystallization were deleted and hydrogen atoms were added in Cerius2 using standard protonation states. Cavity search was carried out with a 1.4 Å (1 Å=0.1 nm) probe on a 0.5 Å grid, saving only sites with an opening of less than 8.0 Å and a volume of more than 62.5 Å3. Docking of the paionin-1 peptide with charged termini was performed using the lowest-energy structures from each of the nine conformations identified in the NMR study and docking these was done using the LigandFit module of Cerius2. Flexible docking with the cff1.01 force field was performed by allowing MC (Monte Carlo) conformational search of the peptide structure inside the protein (10000 MC trials, maximum ten conformations saved). Soft potentials were included to model some plasticity into the protein. A rigid body minimization was included for optimal placement of the peptide inside the cavity (ten iterations, final minimization with 100 iterations). The generated peptide conformations were clustered using the complete linkage option and three ligand–protein structures for each of the nine original paionin-1 conformations were saved. In these poses, the peptide structure was fully minimized (maximum 10000 iterations using the Cerius2 smart minimizer). The poses were selected based on the scoring functions calculated in Cerius2 and visual inspection ensuring that both termini of paionin-1 were protruding from the surface of PAI-1. Remaining strain introduced during the molecular docking was relieved by partial minimization using NAMD [44] and the CHARMM27 force field [45]. Protein residues within 3.0 Å of docked paionin-1 in the structure of latent PAI-1 as well as paionin-1 itself were allowed to move during the final minimization.
Determination of the CMC (critical micelle concentration)
The conductivity at each peptide concentration was measured on a standard conductivity meter by titrating it into pure water adjusted to pH 7.4 with NaOH. As a reference, a profile for titrating the same volumes of buffer without peptide was determined and subtracted from the peptide profile. The change in hydrodynamic size as a determinant of the CMC was determined by photon correlation spectroscopy using a Zetasizer Nano ZS (Malvern Instruments). The analysis was performed at 25 °C in HBS while titrating in biotin–paionin-1. Sampling time and analysis was set to automatic.
Analysis of paionin-1 binding by ELISA
In the four ELISA formats described below, all incubations were performed with 100 μl per well in MaxiSorp 96-well plates (Nunc) coated with 250 ng of antibody in 0.1 M Na2CO3, pH 9.6 (formats 1, 2 and 4), or in Ni-NTA (Ni2+-nitrilotriacetate) HisSorb Strips (Qiagen) (format 3). Unless stated otherwise, PBS supplemented with 2% (w/v) BSA was used for blocking the wells after coating, PBS supplemented with 0.2% (w/v) BSA was used as the standard buffer for all incubations, and PBS supplemented with 0.05% (v/v) Tween 20 was used for washing. Phage was detected with a HRP (horseradish peroxidase)-conjugated monoclonal anti-M13 antibody (Amersham Biosciences) diluted 5000-fold. Wells were developed with 0.5 mg/ml OPD (o-phenylenediamine; Kem-En-Tech Diagnostics) in 50 mM citric acid (pH 5) supplemented with 0.03% H2O2 and quenched with 1 vol. 1 M H2SO4 before measuring the absorbance at 492 nm in a microplate reader.
In ELISA format 1, PBS was replaced with MBS and BSA was omitted from the reaction buffer in experiments involving XR-5118 or bis-ANS (4,4′-dianilino-1,1′-binaphthyl-5,5-disulfonic acid). PAI-1 from HT-1080 cells at the indicated concentrations was captured on monoclonal anti-PAI-1 antibody Mab-5. Phage (∼109 cfu/ml) was allowed to bind in the presence of various additions as indicated for each experiment and detected.
In ELISA format 2, wells were coated with a polyclonal rabbit-anti PAI-1 antibody. Samples of mostly active PAI-1 variants (100 nM) were prepared and split into three aliquots which were incubated for 16 h at 0 °C to preserve activity, incubated 16 h at 37 °C to obtain latent material or reacted with a 2-fold excess of uPA for 30 min at room temperature (23 °C) and then stored at 0 °C for 16 h. Three-fold dilution series of the different forms of PAI-1 variants were incubated in the wells and, after washing, paionin-1 phage clone B (∼109 cfu/ml) was allowed to bind PAI-1. The wells were washed and bound phage was detected.
In ELISA format 3, 50 nM His6-tagged PAI-1 variants were incubated for 16 h at 37 °C to obtain latent material. These samples and samples of active PAI-1 were immobilized in HisSorb wells. After washing, paionin-1 phage clone B (109 cfu/ml) was allowed to bind PAI-1. The wells were washed and bound phage was detected as described above. To ensure that equal amounts of the individual PAI-1 variants were bound to the wells, a parallel ELISA was performed with 1 μg/ml polyclonal rabbit anti-PAI-1 antibody instead of phage and with HRP-conjugated pig anti-rabbit serum (DAKO) diluted 2000-fold instead of anti-M13 antibody.
In ELISA format 4, wells were coated with a polyclonal rabbit anti PAI-1 antibody followed by incubation with recombinant PAI-1 variants (5 nM) and 0.3 μM biotin–paionin-1 peptide. After washing the wells, captured PAI-1–peptide complexes were detected with HRP-conjugated avidin (DAKO) diluted 10000-fold and a peroxidase reaction as described above. Equal loading of PAI-1 was assured by a parallel ELISA with 1 μg/ml monoclonal anti-PAI-1 Mab-1 antibody instead of peptide and with HRP-conjugated rabbit anti-mouse serum (DAKO) diluted 2000-fold instead of HRP-conjugated avidin.
Analysis of paionin-1 binding by SPR (surface plasmon resonance)
The peptide biotin–DVPCFGWCQDAGAKK-CONH2 was coupled to a CM5-chip (Biacore), according to the manufacturer's amine-coupling protocol, at pH 5.0 to a final immobilized ligand response of approx. 3000 RU (response units) (1.6 pmol). Detection of binding was carried out at 25 °C on a Biacore X instrument at a flow rate of 10–20 μl/min, using 10 mM Hepes, 140 mM NaCl, 3 mM EDTA and 0.005% (v/v) Tween 20, degassed and filtered (0.22 μm) as running buffer. The concentration of the applied proteins was 0.7 μM. Signals were corrected for those obtained from a serially connected ethanolamine-blocked flow cell on the same CM5 chip.
Solid-phase receptor-binding assays
Purified receptors were used to coat microtitre wells overnight in 50 mM NaHCO3, pH 9.6, with a receptor density adjusted to obtain 5–10% binding of 125I-uPA–PAI-1 complex, corresponding to receptor concentrations in the coating buffer of approx. 0.5 μg/ml. The wells were blocked with 10% (w/v) BSA in binding buffer (10 mM Hepes, pH 7.8, 140 mM NaCl, 2 mM CaCl2 and 1 mM MgCl2) for 2 h, washed, and incubated overnight at 4 °C with 20–40 pM 125I-ligand. Washing and incubation with ligand were carried out in binding buffer with 1% (w/v) BSA. After the incubation, the supernantants were removed for γ-counting and the wells were washed. Bound ligand was released from the wells with 10% (w/v) SDS and taken for γ-counting. Ligand binding was expressed as bound divided by total ligand and corrected for non-specific binding, i.e. the radioactivity recovered from wells without receptor. These data were then corrected for small variations in the actual relative amount of complex, by the use of the binding of complex to parallel wells coated with 20 μg/ml anti-PAI-1 Mab-2 antibody.
Receptor-mediated ligand degradation
U937 cells were washed three times with serum-free medium supplemented with 0.5% (w/v) BSA and incubated for 45 min at 37 °C in 0.5 ml of the same buffer with 10 pM 125I-labelled ligand and a cell density of 107 cells/ml. The cultures were then made 7% with respect to trichloroacetic acid and centrifuged at 9300 g for 10 min. Supernatant and pellets were γ-counted separately. Ligand degradation was defined as non-trichloroacetic-acid-precipitable radioactivity divided by the total amount of added radioactivity. The degradation in incubations without cells were scored as non-specific degradation and subtracted from all other data [31,46]. Under the conditions used, degradation proceeded linearly with time.
RESULTS AND DISCUSSION
Selection of PAI-1-binding peptides from phage display peptide libraries
A mixture of four different phage display random peptide repertoires was panned against PAI-1 in order to isolate specific PAI-1-binders. As bait, we used human glycosylated PAI-1 from HT-1080 fibrosarcoma cells. To avoid denaturation of PAI-1 by its immobilization on plastic [25,47], phage was allowed to bind PAI-1 in solution before capture of phage–PAI-1 complexes on immobilized monoclonal anti-PAI-1 antibody. Enrichment of antibody-binding peptides was avoided by using alternating antibodies, i.e. Mab-7, -5 or -3, for capture. The output/input ratio of phage increased 100-fold from round 1 to round 5 (results not shown). Individual phage from selection rounds 2–5 were tested for PAI-1-binding in a sandwich ELISA, and sequencing of positives identified a short disulfide-constrained loop CFG(W/Y/F)C as a well-defined consensus motif (Table 1). To compare the isolated phages, their EC50 values for PAI-1 binding were determined and revealed no gross variation among the different peptides, advocating that the consensus residues are the major determinants of binding (Table 1). Importantly, these EC50 values do not reflect the actual affinity between the displayed peptide and PAI-1, since each phage carries more than one copy of the peptide, thus causing avidity effects, i.e. an apparently tighter phage–PAI-1 binding than warranted by the affinity in the 1:1 peptide–PAI-1 complex. In the following, a peptide with the sequence DVPCFGWCQDA will be referred to as paionin-1. This sequence is a chimaera of clones A and B devoid of the flanking cysteine residues (Table 1). That this second disulfide was dispensable for binding could be inferred from the isolation of clone G (ILCFGMC; see Table 1).
Table 1. Selected PAI-1-binding peptides.
Sequence alignment and EC50 for the PAI-1-binding peptides isolated by phage display. Consensus motifs are underlined. The frequency indicates the occurrence of each clone among the sequenced binders. The EC50 values were determined as the amount of PAI-1 required for half-maximal binding in ELISA format 1 (see the Experimental section).
| Clone | Sequence | Frequency | EC50 (nM) |
|---|---|---|---|
| A | CDPPCFGWCQDAC | 2 | 10 |
| B | CDVPCFGWCVAWC | 12 | 8.2 |
| C | CQGGCRVQCFGFC | 1 | 30 |
| F | CTGACPVVCFGFC | 1 | 17 |
| G | ILCFGMC | 1 | 58 |
| H | CEGVCRVSCFGWC | 3 | 6.0 |
| I | CDVPCFGWCVSWC | 1 | 13 |
| J | CVCFGYCRYEC | 1 | 9.9 |
| N | CHILCFGYCGDLC | 1 | 4.2 |
| O | CEGKCVVSCFGFC | 1 | 9.8 |
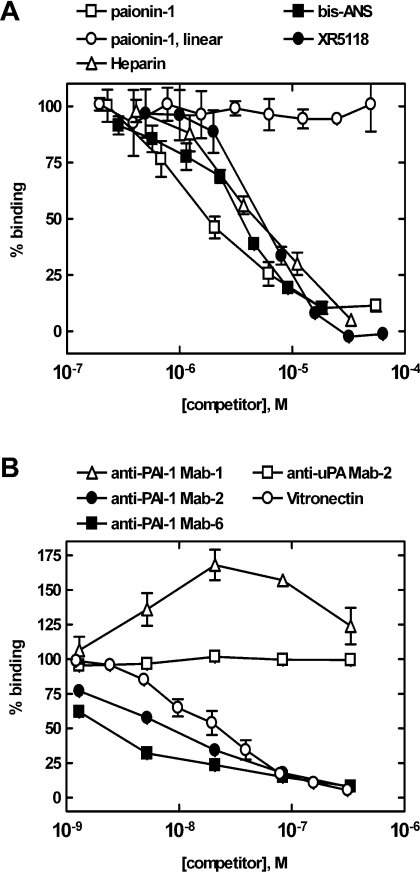
The paionin-1 peptide inhibited the PAI-1 binding of phage clone B with an IC50 of 1.8±0.8 μM (n=3), while a linear version with the two cysteine residues replaced by serines had no effect on binding (Figure 2A). This advocates that the constraint imposed by the disulfide locks the CFGWC loop in a conformation required for binding and/or that the cysteine contributes to PAI-1 binding by engaging in protein contacts. The binding of paionin-1 phage was evaluated further through competition experiments with monoclonal anti-PAI-1 antibodies with known epitopes and with vitronectin: Mab-1 binds the hI/s5A-loop and hC [26] and both Mab-2 and Mab-6 binds hF and the hF/s3A-loop [25]. The vitronectin-binding site is located between s1A, s2A, hE and hF [15,16]. Mab-2, Mab-6 and vitronectin competed with paionin-1 phage for binding, Mab-1 slightly augmented binding, and an irrelevant antibody (anti-uPA Mab-2) had no effect (Figure 2B). Considering the epitopes for the employed antibodies and vitronectin, these data are consistent with a binding site for paionin-1 close to hF of PAI-1. PAI-1 binding of phage was also inhibited by the organochemical PAI-1 neutralizers XR-5118 and bis-ANS, which are known to bind hydrophobic surfaces in the flexible joint region of PAI-1 [13,48,49], and by heparin which binds basic residues in and around hD [50] (Figure 2A). In summary, these competition experiments suggested a hydrophobic binding site for the disulfide-constrained CFGWC loop of paionin-1 in the flexible joint region of PAI-1.
Figure 2. Competition of paionin-1 phage binding in ELISAs.
Binding of paionin-1 phage clone B to 10 nM PAI-1 from HT-1080 cells was detected in ELISA format 1 (see the Experimental section) in the presence of additions as indicated for each experiment. (A) Paionin-1: NH2-DVPCFGWCQDA-CO2H. Paionin-1, linear: NH2-DVPSFGWSQDA-CO2H. Bis-ANS and XR-5118: see [13]. (B) Effects from monoclonal anti-PAI-1 antibodies Mab-1, Mab-2 and Mab-6 and vitronectin on paionin-1 phage binding. Monoclonal anti-uPA Mab-2 was included as a control.
Solution structure of paionin-1
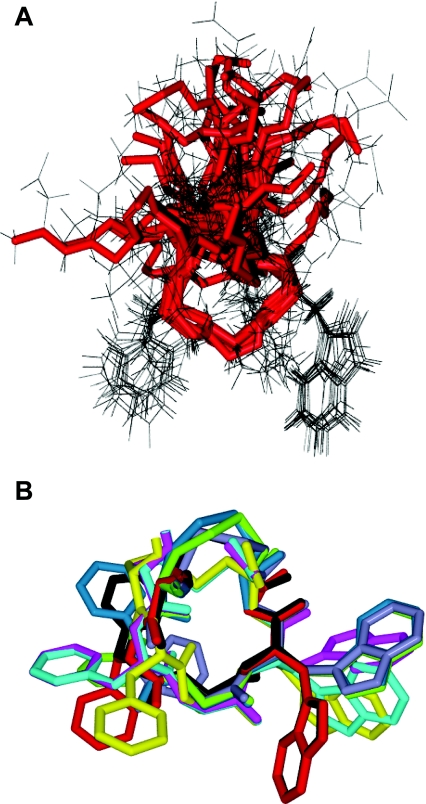
Expecting that the main chain and, to some extent, the side chains of a five-residue ring such as the CFGWC motif of paionin-1 would be stabilized by inherent strain, we analysed paionin-1 by liquid-state NMR spectroscopy. The peptide proton signals were well resolved in the resulting spectra, and all except for those of the N-terminal amide were assigned (see Supplementary Table S1). A total of 51 NOEs could be identified and used in the structure calculations (see Supplementary Table S2). In the 100 calculated structures with the lowest energy, the main chain of the CFGWC ring was well conserved with a low RMSD (root mean square deviation) (1.08±0.51 Å). The structure of the residues flanking the CFGWC ring is less well-defined as the only forces directing their conformation seem to be steric repulsion by other residues which leads to the predominantly orthogonal orientation of both termini relative to the planar CFGWC ring (Figure 3A). As expected from the primary structure, paionin-1 is polarized with respect to hydrophobicity, since the two hydrophobic side chains of the Phe5 and Trp7 points in the opposite direction of the more polar segments flanking the ring.
Figure 3. Structure of paionin-1 determined by NMR spectroscopy.
(A) Overlay of the 15 low-energy structures in conformational group number 1, aligned according to the backbone of the CFGWC ring, shown in red. (B) The CFGWC ring of one representative structure from each of the nine conformational groups, each with a distinct colour, aligned according to the backbone.
By visual inspection, we found that, in 88 of the 100 structures, the Phe5 and Trp7 side chains adapted one of nine different conformations relative to the CFGWC ring; one example is depicted in Figure 3(A). Accordingly, these 88 structures were divided into nine conformational groups based on the orientation of these two side chains (Table 2 and Figure 3B). Since there was no correlation between the calculated energy and the grouping of the 88 structures, we conclude that, from an energetic perspective, each of the conformations is equally likely to occur.
Table 2. NMR structure characteristics.
Among a total of 100 calculated low-energy structures of paionin-1 (NH2-DVPCFGWCQDA-CO2H), 88 could be grouped based on the orientation of the Phe5 and Trp7 side chains. The average RMSDs for the CFGWC ring is given for backbone and for all atoms for each group and for all 100 structures.
| RMSD (Å) | |||
|---|---|---|---|
| Group | Number of structures | Backbone | All atoms |
| 1 | 15 | 0.47±0.27 | 1.08±0.58 |
| 2 | 7 | 0.31±0.16 | 0.78±0.27 |
| 3 | 16 | 0.50±0.24 | 1.23±0.40 |
| 4 | 19 | 0.56±0.22 | 1.19±0.37 |
| 5 | 9 | 0.58±0.26 | 1.39±0.53 |
| 6 | 10 | 0.64±0.36 | 1.47±0.59 |
| 7 | 4 | 0.28±0.08 | 0.80±0.29 |
| 8 | 4 | 0.33±0.15 | 0.70±0.25 |
| 9 | 4 | 0.15±0.08 | 0.53±0.22 |
| – | 100 | 1.08±0.51 | 2.34±0.65 |
Molecular modelling of the paionin-1 interaction with PAI-1
The X-ray crystal structures of highest resolution of active PAI-1 carrying the stabilizing mutations N152H/K156T/Q321L/M356I and latent PAI-1 were searched for cavities capable of accommodating paionin-1. In both structures, the calculations revealed only one cavity with a total volume compatible with paionin-1 binding. The cavity was positioned in the flexible joint region between hD, hE and s2A, thus overlapping with that identified by Elliot et al. [14] (cavity 2). The total volume of the cavity was almost conserved between active PAI-114–1B (∼350 Å3) and latent PAI-1 (∼290 Å3), but the cavity in active PAI-114–1B consists of numerous narrow extensions from the centre of the cavity, whereas the cavity in the latent form consists almost entirely of one single moiety.
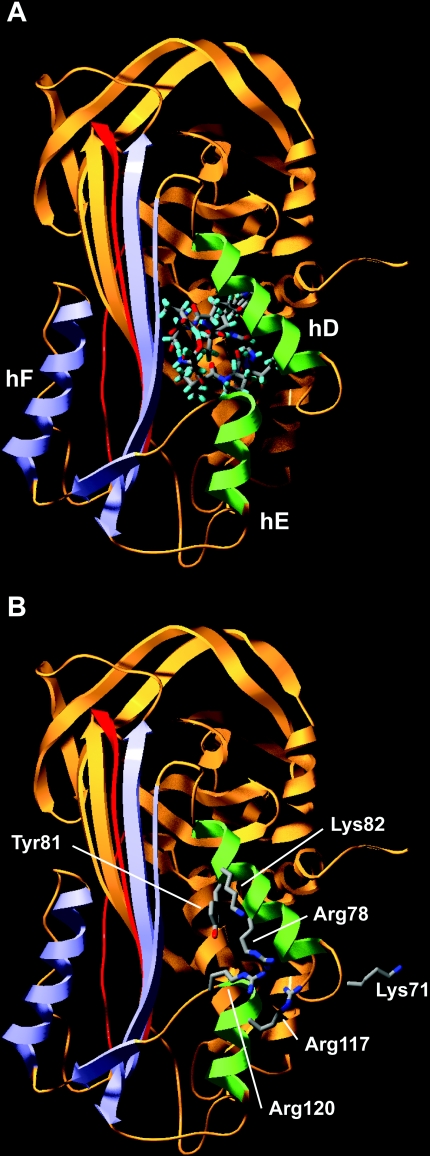
We next used in silico docking procedures to evaluate whether paionin-1 would fit into the identified cavities in active PAI-114–1B and latent PAI-1. The lowest-energy structure of paionin-1 from each of the nine conformational groups was used as the ligand for docking, and three converged solutions were obtained from each of the peptide structures. That the peptides containing the CFGWC motif listed in Table 1 can bind PAI-1 in the context of a phage, in which the displayed peptide sequence is preceded by Ala-Asp-Gly-Ala and followed by the phage coat protein [34], allowed us to disqualify docking solutions in which the termini of paionin-1 do not protrude from the surface of PAI-1. Accordingly, only three solutions were acceptable (one solution shown in Figure 4A), all with the entire CFGWC ring of paioinin-1 buried deep into the cavity of latent PAI-1. Importantly, since the ligand-docking algorithm mimics structural flexibility through the use of soft potentials, the hereby introduced strain was removed by a partial minimization yielding a final structure that could accommodate paionin-1 inside the cavity. Nonetheless, the cavity search and subsequent docking routines unequivocally pointed to the flexible joint region as the binding site for paionin-1 and suggested a preferential binding to the latent conformation of PAI-1.
Figure 4. Docking of paionin-1 into the structure of latent PAI-1 and residues important for paionin-1 binding in the flexible joint region.
Latent PAI-1 shown as a ribbon diagram with the inserted RCL in red, the large serpin fragment in orange, the small serpin fragment including hF in blue and hD and hE from the flexible joint region in green. (A) The docked structure of paionin-1 from conformational group number 1 is shown as a stick model. (B) Residues which based on binding assays with mutated PAI-1 variants are implicated in paionin-1 binding are indicated.
Solubility properties of biotin–paionin-1
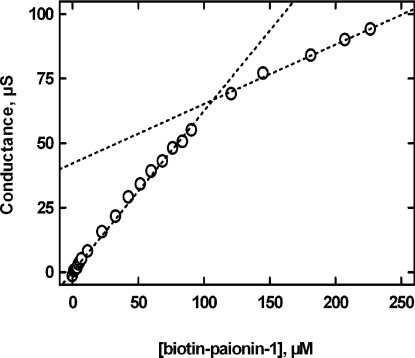
Handling of the paionin-1 peptide demanded the addition of DMSO to solubilize the peptide and 20% (v/v) DMSO was needed to keep the peptide in solution for NMR. For direct binding studies, we wished to employ a biotin-labelled peptide. Expecting that removal of the charge at the C-terminus by amidation and introduction of the apolar biotin at the N-terminus would reduce the solubility even further and obtaining inconsistent results with peptide concentrations above 50 μM, we decided to test whether the biotinylated peptide forms aggregates at high concentrations. The conductivity profile obtained when titrating biotin–paionin-1 into pure water demonstrated a clear break in the linear response at a concentration of ∼100 μM, suggesting that micelles are formed in water with a CMC close to this value (Figure 5), and since the CMC decreases with increasing saline concentration, the CMC of biotin–paionin-1 will be lower in HBS. However, the high dielectric constant of HBS made it impossible to determine the CMC of biotin–paionin-1 in this buffer by conductivity measurements. As an alternative approach, we used photon correlation spectroscopy to estimate the CMC for biotin–paionin-1 in HBS and found it to be between 30 and 40 μM (results not shown). In the light of these observations, it seemed reasonable to assume that biotin–paionin-1 has a CMC near 40 μM at physiological ionic strength. Hence, any data obtained at biotin–paionin-1 concentrations above this threshold may be flawed by non-specific biochemical and cell-biological effects owing to the formation of aggregates, presumably micelles.
Figure 5. Conductometric determination of the CMC for biotin–paionin-1.
The electric conductance was monitored while titrating biotin–paionin-1 into water. The presented data have been corrected for the conductance resulting from adding equal volumes of buffer without peptide. The two broken lines represent linear regressions of data points below and above 100 μM biotin–paionin-1 respectively, crossing each other at 107 μM.
Mapping of the paionin-1-binding site by site-directed mutagenesis
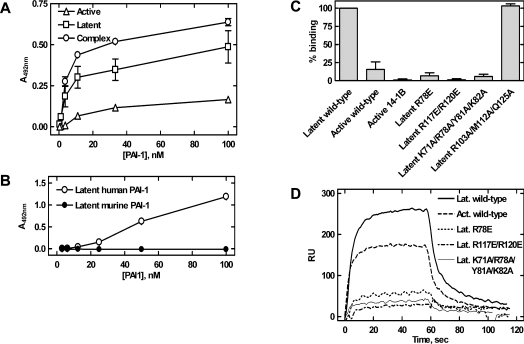
In order to map the paionin-1-binding site, we studied its binding to PAI-1 variants in ELISAs and by SPR. First, preferential binding of paionin-1 phage to latent PAI-1 and to the uPA–PAI-1 complex, as compared with active PAI-1, was shown (Figure 6A). Secondly, compared with the binding observed with latent human PAI-1, paionin-1 showed impaired binding to latent murine PAI-1 (Figure 6B). Thirdly, when the binding of the paionin-1 peptide with an N-terminal biotin and a C-terminal amide (biotin–paionin-1) to recombinant PAI-1 variants was tested, a clear preference for latent PAI-1 over active PAI-1 or PAI-114–1B was observed (Figure 6C). Furthermore, we found that biotin–paionin-1 showed decreased binding to latent forms of the variants PAI-1(R78E), PAI-1(R117E/R120E) and PAI-1(K71A/R78A/Y81A/K82A). In contrast, biotin–paionin-1 binding to latent PAI-1 was not affected by the triple mutation R103A/M112A/Q125A, which reduces the apparent affinity for vitronectin more than 3000-fold [51] (Figure 6C). The binding of paionin-1 to active and uPA-complexed PAI-1(R78E), PAI-1(R117E/R120E) and PAI-1(K71A/R78A/Y81A/K82A) was tested in an ELISA similar to the one depicted in Figure 6(A), and no binding was detected (results not shown).
Figure 6. Localization of the paionin-1-binding site.
(A) Binding between recombinant PAI-1, active, latent or in complex with uPA (complex), and paionin-1 phage in ELISA format 2 (see the Experimental section). Latent PAI-1 was prepared by incubating active PAI-1 at 37 °C for 16 h and uPA–PAI-1 complex was prepared by incubating active PAI-1 with a 2-fold molar excess of uPA at room temperature for 30 min. Results are means±S.D., representative of two independent experiments with similar results, each performed in triplicate. (B) Binding between recombinant human or murine PAI-1 and paionin-1 phage in ELISA format 3 (see the Experimental section). Results are means±S.D., representative of two independent experiments with similar results, each performed in triplicate. (C) Biotin–paionin-1 peptide binding to recombinant PAI-1 variants in ELISA format 4 (see the Experimental section). PAI-114–1B (active 14-1B) is a stable active PAI-1 variant carrying four mutations [42], other PAI-1 variants are indicated by their mutations. Except for active PAI-1 (active wild-type) and PAI-114–1B (active 14-1B), the tested variants were in their latent conformation. Signals were normalized to those obtained with latent PAI-1. Results are means±S.D. for three independent experiments. (D) SPR sensorgrams for association and dissociation of PAI-1 variants to immobilized biotin–paionin-1 with a C-terminal GAKK extension. Except for active (Act.), the variants were in their latent (Lat.) conformation. Protein (700 nM) was applied at a flow rate of 5 μl/min and the sensorgrams were corrected by subtraction of the signals from a reference cell.
Finally, we tested the recombinant variants in their latent form for biotin–paionin-1 binding by SPR analysis (Figure 6D). For these experiments, biotin–paionin-1 with a Gly-Ala-Lys-Lys extension on the C-terminus was immobilized on the sensor chip by amide coupling and used in SPR binding experiments. The obtained sensorgrams were in agreement with the ELISA data as an impaired binding of PAI-1(R78E), PAI-1(R117E/R120E) and PAI-1(K71A/R78A/Y81A/K82A) was evident (Figure 6D). Using the same SPR set-up, no binding between biotin–paionin-1 and the serpins antithrombin III and α1-proteinase inhibitor could be detected. In agreement with this, no binding site for paionin-1 in the flexible joint region of these serpins could be identified by an in silico cavity search (results not shown).
The charge-reversing mutations in PAI-1(R78E/R117E/R120E), located in hD (Arg78), hE (Arg117) and the hE/s1A loop (Arg120), were reported previously to impair binding to the PAI-1-neutralizing compounds AR-H029953XX and bis-ANS [12,13]. XR-5118 was found to have an overlapping, but not identical, binding site [13,49]. Thus the impaired binding of paionin-1 to variants carrying one or more of these mutations is consistent with paionin-1 binding to this hydrophobic surface area in the flexible joint and in agreement with the competition studies with bis-ANS and XR-5118 (Figure 2A). Likewise, the decreased binding of paionin-1 to PAI-1(K71A/R78A/Y81A/K82A) and the ability of heparin to compete with paionin-1 for binding (Figure 2A) is consistent with Lys71, Arg78 and Lys82 in hD being important for heparin binding [50]. The competition of paionin-1 binding by vitronectin (Figure 2B) is readily explainable by steric hindrance from the nearby vitronectin-binding site [15,16]. The impaired binding to murine PAI-1 may result from the replacement of Arg120 with glutamine in this orthologue.
In summary, these ELISA and SPR data consistently advocate a binding site for paionin-1 in the flexible joint region of human PAI-1 that becomes more accessible upon the S-to-R transition. As depicted in Figure 4(B), the residues found to be important for paionin-1 binding surround the entrance to the binding cavity proposed from the ligand-docking algorithms, thus lending support to this analysis.
Paionin-1 and the anti-proteolytic activity of PAI-1
We tested whether biotin–paionin-1 was able to affect the activity of PAI-1 towards uPA or to change the rate of latency transition of PAI-1. To this end, we employed a chromogenic assay which monitored the activity of uPA in the absence or presence of PAI-1 which had been treated with different concentrations of biotin–paionin-1 for various periods of time. Biotin–paionin-1 did not prevent formation of the uPA–PAI-1 complex, but did increase the rate of PAI-1 latency transition ∼2-fold at concentrations higher than 50 μM, i.e. above the determined CMC (results not shown). This effect is most likely to be unrelated to specific peptide binding, but rather a non-specific effect of biotin–paionin-1 aggregates or micelles, analogues to the acceleration of latency transition caused by >100 μM Triton X-100, which has a CMC of 200–300 μM [52]. That biotin–paionin-1 does not impair the activity of PAI-1, although it binds the same area as the PAI-1-neutralizers bis-ANS and XR-5118 exemplifies that the effect of targeting this area depends on the properties of the employed compound. For instance, negatively charged neutralizers including bis-ANS induce PAI-1 polymerization, whereas XR-5118, which carries a positive charge, does not [13,48,49].
Effects of paionin-1 on binding of the uPA–PAI-1 complex to VLDLR and LRP-1A
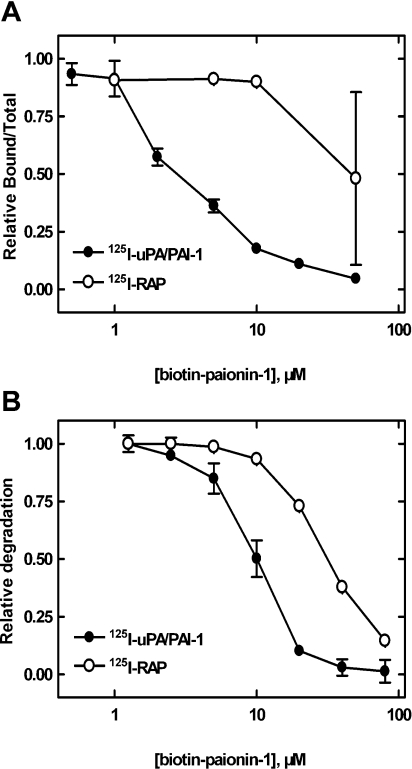
The four alanine substitutions in PAI-1(K71A/R78A/Y81A/K82A), located in the hC/hD-loop (Lys71) and hD (Arg78, Tyr81 and Lys82), reduces the affinity of the uPA–PAI-1 complex for endocytosis receptors of the LDLR family (S. Skeldal, J. V. Larsen, K. E. Pedersen, H. H. Petersen, R. Egelund, A. Christensen, J. K. Jensen, J. Gliemann, P. A. Andreasen, unpublished work). This is in agreement with a reported binding site for these receptors involving hD and hE of PAI-1 [18–20] and the impaired paionin-1 binding to PAI-1(K71A/R78A/Y81A/K82A) suggests an overlap between the binding sites for the peptide and the receptors. In the light of this, the effect of paionin-1 on uPA–PAI-1 complex binding to VLDLR and LRP-1A was tested. Receptor was immobilized on the solid phase and the binding of purified 125I-uPA–PAI-1 complex was determined in the presence of various concentrations of peptide. The results clearly showed that biotin–paionin-1 inhibited the binding of the complex to LRP-1A with an IC50 value of ∼2.5 μM (Figure 7A). Similar data were obtained with soluble VLDLR (IC50 ∼5 μM, results not shown). As a control, the peptide was shown not to affect the binding of 125I-RAP to the receptors, except when concentrations close to the CMC for biotin–paionin-1 were applied (Figure 7A, and results not shown). Noteworthily, a high variability of the effect on RAP binding was observed. Considering that the extent of micelle formation will be temperature-dependent close to the CMC, and that the experiments summarized in Figure 7(A) were performed at room temperature, this variability agrees with a non-specific effect of micelles. In contrast, paionin-1 without biotin on the N-terminus did not have an inhibiting effect even at high concentrations (50 μM). Biotin alone or attached to the linear version of paionin-1 was also found to not have an effect (results not shown). Our interpretation of these data is that the combination of the intact paionin-1 structure and the biotin are equally important for the function of the peptide, i.e. the structure dictates binding specificity and biotin provides the antagonizing function. The observed effect of the biotinylated paionin-1 peptide on uPA–PAI-1 complex binding to endocytosis receptors was confirmed in a cell culture system with the human histiocytic lymphoma-derived cell line U937. From this assay, it was evident that biotin–paionin-1 significantly reduced VLDLR-mediated endocytosis and degradation of 125I-uPA–PAI-1 complex with an IC50 value of ∼10 μM (Figure 7B). Again, a concentration in the area up to 30–40 μM was required to observe an effect of biotin–paionin-1 on 125I-RAP endocytosis, an effect probably explainable by the formation of micelles.
Figure 7. Effects of paionin-1 on the binding of the uPA–PAI-1 complex to endocytosis receptors.
(A) Effects of biotin–paionin-1 on the binding of 125I-uPA–PAI-1 (●) and 125I-RAP (○) to immobilized LRP-1A. Results are means±S.D. for at least two independent experiments. (B) Effects of biotin–paionin-1 on degradation of 125I-uPA–PAI-1 (●) and 125I-RAP (○) by U937 cells. results are means±S.D. for one experiment performed in duplicate, representative of three independent experiments.
In conclusion, we have isolated a PAI-1-binding peptide sequence, paionin-1, from a phage display peptide library. We have determined the solution structure of the peptide by NMR and used it for in silico docking of the peptide into X-ray crystal structures of PAI-1. These analyses suggested a binding site for paionin-1 in the flexible joint region of PAI-1. The predicted binding site was supported by the ability of other molecules targeting this region to compete with paionin-1 for binding and by site-directed mutagenesis. We demonstrated that the peptide with an N-terminal biotin extension is able to inhibit the binding of the uPA–PAI-1 complex to endocytosis receptors. However, the peptide needs to be developed further before being valuable as a tool for investigations of the molecular interactions of PAI-1. In particular, an improvement of affinity and solubility is needed in order to increase the presently small window between specific and non-specific effects. By being able to inhibit uPA–PAI-1 endocytosis receptor binding, biotin–paionin-1 exemplifies compounds which may prevent the recycling of free uPAR after internalization of the uPAR–uPA–PAI-1 complex. This would disrupt the signalling function of uPA–PAI-1 endocytosis receptor binding and down-regulate the pericellular proteolytic capacity.
Online data
Acknowledgments
We acknowledge Erkki Koivunen (Division of Biochemistry, University of Helsinki, Helsinki, Finland), Ann Gills (Faculty of Pharmaceutical Sciences, Katholieke Universiteit Leuven, Leuven, Belgium) and Jørgen Gliemann (Institute of Medical Biochemistry, University of Aarhus, Aarhus, Denmark) for materials. Axel Meissner (the Carlsberg Laboratory, Copenhagen, Denmark) is acknowledged for help with NMR experimental setup and for use of the Bruker 600 MHz NMR spectrometer, and Ken Howard [Interdisciplinary Nanoscience Center (iNANO), University of Aarhus, Aarhus, Denmark] for help with the dynamic light-scattering experiments. The Danish Cancer Research Foundation, iNANO, the Danish Cancer Society, the Danish National Research Foundation, Carlsbergfondet, the Danish Natural Science Research Council and the Danish Biotechnology Instrument Centre (DABIC) are thanked for financial support. T.W. is the recipient of a fellowship from Carlsbergfondet (ANS-1289/20).
References
- 1.Vaughan D. E. PAI-1 and atherothrombosis. J. Thromb. Haemostasis. 2005;3:1879–1883. doi: 10.1111/j.1538-7836.2005.01420.x. [DOI] [PubMed] [Google Scholar]
- 2.Durand M. K., Bødker J. S., Christensen A., Dupont D. M., Hansen M., Jensen J. K., Kjelgaard S., Mathiasen L., Pedersen K. E., Skeldal S., et al. Plasminogen activator inhibitor-I and tumour growth, invasion, and metastasis. Thromb. Haemostasis. 2004;91:438–449. doi: 10.1160/TH03-12-0784. [DOI] [PubMed] [Google Scholar]
- 3.Behrendt N. The urokinase receptor (uPAR) and the uPAR-associated protein (uPARAP/Endo180): membrane proteins engaged in matrix turnover during tissue remodeling. Biol. Chem. 2004;385:103–136. doi: 10.1515/BC.2004.031. [DOI] [PubMed] [Google Scholar]
- 4.Gettins P. G. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 5.Egelund R., Rodenburg K. W., Andreasen P. A., Rasmussen M. S., Guldberg R. E., Petersen T. E. An ester bond linking a fragment of a serine proteinase to its serpin inhibitor. Biochemistry. 1998;37:6375–6379. doi: 10.1021/bi973043+. [DOI] [PubMed] [Google Scholar]
- 6.Dementiev A., Dobo J., Gettins P. G. Active site distortion is sufficient for proteinase inhibition by serpins: structure of the covalent complex of α1-proteinase inhibitor with porcine pancreatic elastase. J. Biol. Chem. 2006;281:3452–3457. doi: 10.1074/jbc.M510564200. [DOI] [PubMed] [Google Scholar]
- 7.Huntington J. A., Read R. J., Carrell R. W. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 8.Huber R., Carrell R. W. Implications of the three-dimensional structure of α1-antitrypsin for structure and function of serpins. Biochemistry. 1989;28:8951–8966. doi: 10.1021/bi00449a001. [DOI] [PubMed] [Google Scholar]
- 9.Gettins P. G. The F-helix of serpins plays an essential, active role in the proteinase inhibition mechanism. FEBS Lett. 2002;523:2–6. doi: 10.1016/s0014-5793(02)02924-1. [DOI] [PubMed] [Google Scholar]
- 10.Mottonen J., Strand A., Symersky J., Sweet R. M., Danley D. E., Geoghegan K. F., Gerard R. D., Goldsmith E. J. Structural basis of latency in plasminogen activator inhibitor-1. Nature. 1992;355:270–273. doi: 10.1038/355270a0. [DOI] [PubMed] [Google Scholar]
- 11.Stein P., Chothia C. Serpin tertiary structure transformation. J. Mol. Biol. 1991;221:615–621. doi: 10.1016/0022-2836(91)80076-7. [DOI] [PubMed] [Google Scholar]
- 12.Björquist P., Ehnebom J., Inghardt T., Hansson L., Lindberg M., Linschoten M., Strömqvist M., Deinum J. Identification of the binding site for a low-molecular-weight inhibitor of plasminogen activator inhibitor type 1 by site-directed mutagenesis. Biochemistry. 1998;37:1227–1234. doi: 10.1021/bi971554q. [DOI] [PubMed] [Google Scholar]
- 13.Egelund R., Einholm A. P., Pedersen K. E., Nielsen R. W., Christensen A., Deinum J., Andreasen P. A. A regulatory hydrophobic area in the flexible joint region of plasminogen activator inhibitor-1, defined with fluorescent activity- neutralizing ligands: ligand-induced serpin polymerization. J. Biol. Chem. 2001;276:13077–13086. doi: 10.1074/jbc.M009024200. [DOI] [PubMed] [Google Scholar]
- 14.Elliott P. R., Pei X. Y., Dafforn T. R., Lomas D. A. Topography of a 2.0 Å structure of α1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci. 2000;9:1274–1281. doi: 10.1110/ps.9.7.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen J. K., Wind T., Andreasen P. A. The vitronectin binding area of plasminogen activator inhibitor-1, mapped by mutagenesis and protection against an inactivating organochemical ligand. FEBS Lett. 2002;521:91–94. doi: 10.1016/s0014-5793(02)02830-2. [DOI] [PubMed] [Google Scholar]
- 16.Zhou A., Huntington J. A., Pannu N. S., Carrell R. W., Read R. J. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat. Struct. Biol. 2003;10:541–544. doi: 10.1038/nsb943. [DOI] [PubMed] [Google Scholar]
- 17.Nykjær A., Willnow T. E. The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol. 2002;12:273–280. doi: 10.1016/s0962-8924(02)02282-1. [DOI] [PubMed] [Google Scholar]
- 18.Horn I. R., van den Berg B. M., Moestrup S. K., Pannekoek H., van Zonneveld A. J. Plasminogen activator inhibitor 1 contains a cryptic high affinity receptor binding site that is exposed upon complex formation with tissue-type plasminogen activator. Thromb. Haemostasis. 1998;80:822–828. [PubMed] [Google Scholar]
- 19.Rodenburg K. W., KjØller L., Petersen H. H., Andreasen P. A. Binding of urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex to the endocytosis receptors α2-macroglobulin receptor/low-density lipoprotein receptor-related protein and very-low-density lipoprotein receptor involves basic residues in the inhibitor. Biochem. J. 1998;329:55–63. doi: 10.1042/bj3290055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefansson S., Muhammad S., Cheng X. F., Battey F. D., Strickland D. K., Lawrence D. A. Plasminogen activator inhibitor-1 contains a cryptic high affinity binding site for the low density lipoprotein receptor-related protein. J. Biol. Chem. 1998;273:6358–6366. doi: 10.1074/jbc.273.11.6358. [DOI] [PubMed] [Google Scholar]
- 21.Munch M., Heegaard C. W., Andreasen P. A. Interconversions between active, inert and substrate forms of denatured/refolded type-1 plasminogen activator inhibitor. Biochim. Biophys. Acta. 1993;1202:29–37. doi: 10.1016/0167-4838(93)90059-z. [DOI] [PubMed] [Google Scholar]
- 22.Wind T., Jensen J. K., Dupont D. M., Kulig P., Andreasen P. A. Mutational analysis of plasminogen activator inhibitor-1. Eur. J. Biochem. 2003;270:1680–1688. doi: 10.1046/j.1432-1033.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 23.Nykjær A., Petersen C. M., MØller B., Jensen P. H., Moestrup S. K., Holtet T. L., Etzerodt M., ThØgersen H. C., Munch M., Andreasen P. A. Purified α2-macroglobulin receptor/LDL receptor-related protein binds urokinase·plasminogen activator inhibitor type-1 complex: evidence that the α2-macroglobulin receptor mediates cellular degradation of urokinase receptor-bound complexes. J. Biol. Chem. 1992;267:14543–14546. [PubMed] [Google Scholar]
- 24.Nielsen L. S., Andreasen P. A., GrØndahl-Hansen J., Huang J. Y., Kristensen P., DanØ K. Monoclonal antibodies to human 54,000 molecular weight plasminogen activator inhibitor from fibrosarcoma cells: inhibitor neutralization and one-step affinity purification. Thromb. Haemostasis. 1986;55:206–212. [PubMed] [Google Scholar]
- 25.Wind T., Jensen M. A., Andreasen P. A. Epitope mapping for four monoclonal antibodies against human plasminogen activator inhibitor type-1: implications for antibody-mediated PAI-1-neutralization and vitronectin-binding. Eur. J. Biochem. 2001;268:1095–1106. doi: 10.1046/j.1432-1327.2001.2680041095.x. [DOI] [PubMed] [Google Scholar]
- 26.Bødker J. S., Wind T., Jensen J. K., Hansen M., Pedersen K. E., Andreasen P. A. Mapping of the epitope of a monoclonal antibody protecting plasminogen activator inhibitor-1 against inactivating agents. Eur. J. Biochem. 2003;270:1672–1679. doi: 10.1046/j.1432-1033.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- 27.Offersen B. V., Nielsen B. S., HØyer-Hansen G., Rank F., Hamilton-Dutoit S., Overgaard J., Andreasen P. A. The myofibroblast is the predominant plasminogen activator inhibitor-1-expressing cell type in human breast carcinomas. Am. J. Pathol. 2003;163:1887–1899. doi: 10.1016/S0002-9440(10)63547-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.GrØndahl-Hansen J., Ralfkiaer E., Nielsen L. S., Kristensen P., Frentz G., DanØ K. Immunohistochemical localization of urokinase- and tissue-type plasminogen activators in psoriatic skin. J. Invest. Dermatol. 1987;88:28–32. doi: 10.1111/1523-1747.ep12464827. [DOI] [PubMed] [Google Scholar]
- 29.Rettenberger P. M., Oka K., Ellgaard L., Petersen H. H., Christensen A., Martensen P. M., Monard D., Etzerodt M., Chan L., Andreasen P. A. Ligand binding properties of the very low density lipoprotein receptor: absence of the third complement-type repeat encoded by exon 4 is associated with reduced binding of Mr 40,000 receptor-associated protein. J. Biol. Chem. 1999;274:8973–8980. doi: 10.1074/jbc.274.13.8973. [DOI] [PubMed] [Google Scholar]
- 30.Hansen M., Busse M. N., Andreasen P. A. Importance of the amino-acid composition of the shutter region of plasminogen activator inhibitor-1 for its transitions to latent and substrate forms. Eur. J. Biochem. 2001;268:6274–6283. doi: 10.1046/j.0014-2956.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- 31.KjØller L., Simonsen A. C., Ellgaard L., Andreasen P. A. Differential regulation of urokinase-type-1 inhibitor complex endocytosis by phorbol esters in different cell lines is associated with differential regulation of α2-macroglobulin receptor and urokinase receptor expression. Mol. Cell. Endocrinol. 1995;109:209–217. doi: 10.1016/0303-7207(95)03504-z. [DOI] [PubMed] [Google Scholar]
- 32.Moestrup S. K., Gliemann J. Analysis of ligand recognition by the purified α2-macroglobulin receptor (low density lipoprotein receptor-related protein). Evidence that high affinity of α2-macroglobulin–proteinase complex is achieved by binding to adjacent receptors. J. Biol. Chem. 1991;266:14011–14017. [PubMed] [Google Scholar]
- 33.Koivunen E., Wang B., Dickinson C. D., Ruoslahti E. Peptides in cell adhesion research. Methods Enzymol. 1994;245:346–369. doi: 10.1016/0076-6879(94)45019-6. [DOI] [PubMed] [Google Scholar]
- 34.Smith G. P., Scott J. K. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 35.Aue W. P., Batholdi E., Ernst R. R. Two-dimensional spectroscopy: application to nuclear magnetic resonance. J. Chem. Phys. 1976;64:2229–2246. [Google Scholar]
- 36.Macura S., Ernst R. R. Elucidation of cross relaxation in liquids by two-dimensional N.M.R. spectroscopy. Mol. Phys. 1980;41:95–117. [Google Scholar]
- 37.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Wüthrich K. New York: Wiley; 1986. NMR of Proteins and Nucleic Acids. [Google Scholar]
- 39.Johnson B. A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 40.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 42.Berkenpas M. B., Lawrence D. A., Ginsburg D. Molecular evolution of plasminogen activator inhibitor-1 functional stability. EMBO J. 1995;14:2969–2977. doi: 10.1002/j.1460-2075.1995.tb07299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stout T. J., Graham H., Buckley D. I., Matthews D. J. Structures of active and latent PAI-1: a possible stabilizing role for chloride ions. Biochemistry. 2000;39:8460–8469. doi: 10.1021/bi000290w. [DOI] [PubMed] [Google Scholar]
- 44.Kalé L., Skeel R., Bhandarkar M., Brunner R., Gursoy A., Krawetz N., Phillips J., Shinozaki A., Varadarajan K., Schulten K. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- 45.MacKerell A. D., Jr, Bashford D., Bellott R. L., Dunbrack R. L., Jr, Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 46.Jensen P. H., Christensen E. I., Ebbesen P., Gliemann J., Andreasen P. A. Lysosomal degradation of receptor-bound urokinase-type plasminogen activator is enhanced by its inhibitors in human trophoblastic choriocarcinoma cells. Cell Regul. 1990;1:1043–1056. doi: 10.1091/mbc.1.13.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng G., Royle G., Seiffert D., Loskutoff D. J. The PAI-1/vitronectin interaction: two cats in a bag? Thromb. Haemostasis. 1995;74:66–70. [PubMed] [Google Scholar]
- 48.Pedersen K. E., Einholm A. P., Christensen A., Schack L., Wind T., Kenney J. M., Andreasen P. A. Plasminogen activator inhibitor-1 polymers, induced by inactivating amphipathic organochemical ligands. Biochem. J. 2003;372:747–755. doi: 10.1042/BJ20021868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Einholm A. P., Pedersen K. E., Wind T., Kulig P., Overgaard M. T., Jensen J. K., Bødker J. S., Christensen A., Charlton P., Andreasen P. A. Biochemical mechanism of action of a diketopiperazine inactivator of plasminogen activator inhibitor-1. Biochem. J. 2003;373:723–732. doi: 10.1042/BJ20021880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ehrlich H. J., Gebbink R. K., Keijer J., Pannekoek H. Elucidation of structural requirements on plasminogen activator inhibitor 1 for binding to heparin. J. Biol. Chem. 1992;267:11606–11611. [PubMed] [Google Scholar]
- 51.Jensen J. K., Durand M. K., Skeldal S., Dupont D. M., Bødker J. S., Wind T., Andreasen P. A. Construction of a plasminogen activator inhibitor-1 variant without measurable affinity to vitronectin but otherwise normal. FEBS Lett. 2004;556:175–179. doi: 10.1016/s0014-5793(03)01405-4. [DOI] [PubMed] [Google Scholar]
- 52.Gils A., Declerck P. J. Modulation of plasminogen activator inhibitor 1 by Triton X-100: identification of two consecutive conformational transitions. Thromb. Haemostasis. 1998;80:286–291. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.