Abstract
The wild type Copia Gag precursor protein of Drosophila melanogaster expressed in Escherichia coli was shown to be processed autocatalytically to generate two daughter proteins with molecular masses of 33 and 23 kDa on SDS/PAGE. The active-site motif of aspartic proteinases, Asp-Ser-Gly, was present in the 23 kDa protein corresponding to the C-terminal half of the precursor protein. The coding region of this daughter protein (152 residues) in the copia gag gene was expressed in E. coli to produce the recombinant enzyme protein as inclusion bodies, which was then purified and refolded to create the active enzyme. Using the peptide substrate His-Gly-Ile-Ala-Phe-Met-Val-Lys-Glu-Val-Asn (cleavage site: Phe–Met) designed on the basis of the sequence of the cleavage-site region of the precursor protein, the enzymatic properties of the proteinase were investigated. The optimum pH and temperature of the proteinase toward the synthetic peptide were 4.0 and 70 °C respectively. The proteolytic activity was increased with increasing NaCl concentration in the reaction mixture, the optimum concentration being 2 M. Pepstatin A strongly inhibited the enzyme, with a Ki value of 15 nM at pH 4.0. On the other hand, the active-site residue mutant, in which the putative catalytic aspartic acid residue was mutated to an alanine residue, had no activity. These results show that the Copia proteinase belongs to the family of aspartic proteinases including HIV proteinase. The B-chain of oxidized bovine insulin was hydrolysed at the Leu15−–Tyr16 bond fairly selectively. Thus the recombinant Copia proteinase partially resembles HIV proteinase, but is significantly different from it in certain aspects.
Keywords: aspartic proteinase, autocatalytic processing, Copia proteinase, Drosophila melanogaster, recombinant proteinase, retrotransposon
Abbreviations: DAN, diazoacetyl-D,L-norleucine methyl ester; DTT, dithiothreitol; ORF, open reading frame; VLP, virus-like particles
INTRODUCTION
The retroviral proteinases (retropepsins), including HIV proteinase, share the common conserved active-site motif Asp-Ser/Thr-Gly with the ordinary aspartic proteinases [1]. The same amino acid sequence, Asp-Ser-Gly, was also reported to be present in several retrovirus-like transposon elements, such as the Drosophila Copia-like element and yeast transposon Ty1 [2]. Similarities in sequence organization were also recognized between the Copia-like element and the aspartic proteinase precursors of vertebrate retroviruses [3–6]. VLP (virus-like particles) were identified from the Drosophila melanogaster cell culture [7]. On the other hand, a 2 kb Copia RNA was reported to contain sufficient information to make the VLP, probably through autoprocessing of the Copia Gag precursor, in Drosophila cultured cells [8]. Furthermore, the precursor protein expressed in Escherichia coli was reported to be correctly processed to generate a unique laminate structure, indicating involvement of a proteinase in cleaving the Gag precursor itself [9]. So far, however, the proteinase responsible for the processing and release of the VLP protein has not been purified and characterized.
In the present study, the 50 kDa Copia Gag precursor protein expressed in E. coli was shown to be processed autocatalytically to give a proteinase-like protein identified on SDS/PAGE, and the processing site in the protein was determined by N-terminal amino acid sequencing. In order to isolate and characterize the proteinase-like protein, its coding region was expressed in E. coli, and the recombinant protein was purified from the inclusion bodies and refolded to create the active enzyme [called Copia proteinase in the present study; MEROPS (a peptidase database; http://merops.sanger.ac.uk/) name: Copia transposon (Drosophila type) peptidase; MEROPS classification: peptidase A11.001]. The enzymatic properties of the recombinant proteinase were partially similar to those of retroviral aspartic proteinases, but significantly different from them in certain aspects. To our knowledge, this is the first time that a retrotransposon proteinase has been expressed, purified and characterized.
EXPERIMENTAL
Materials
Restriction nucleases, T4 polynucleotide kinase and T4 DNA ligase were purchased from Toyobo Co. (Osaka, Japan). The Nco1 linker (5′-CAGCCATGGCTG-3′), the expression vector pKK233-2, E. coli JM 109 and Sephacryl S-200 were obtained from Pharmacia. DEAE cellulose (DE-52) was obtained from Whatman, and PVDF membrane was from Millipore. A synthetic peptide, His-Gly-Ile-Ala-Phe-Met-Val-Lys-Glu-Val-Asn, containing the sequence at the putative processing site (Met274–Val275) of the 50 kDa precursor protein was synthesized by using a model 431 A peptide synthesizer (Applied Biosystems) and purified by HPLC. Its purity and sequence was confirmed by amino acid sequencing.
Plasmid construction and expression
To express the entire ORF (open reading frame) of the 2 kb RNA (termed ORF2) in E. coli, the plasmid pEC1 [9] was used as a starting material. To express the enzyme protein, the dinucleotide sequence (TpT) near the first methionine codon in the processing site (Met274–Val275) of ORF2 was converted into CpC using oligonucleotide GCATTGCGTCCATGGTAAAA in order to create a NcoI site. The HpaI site located in the downstream of ORF2 was changed using the NcoI linker. The resulting 0.57 kb NcoI fragment (which covers the proteinase region of ORF2) was inserted into the NcoI site of the expression vector pKK233-2 to obtain the plasmid pEC3. Plasmid pEC16 was constructed in the same manner as pEC3, except for the site that harbours the GAT-GCT mutation responsible for the Asp→Ala mutation at the putative active-site motif (Asp-Ser-Gly). E. coli containing each expression plasmid was grown in Luria–Bertani broth at 37 °C until mid-exponential phase (D600 0.5–0.7). Gene expression was then induced by the addition of isopropyl β-D-thiogalactoside to a final concentration of 50 μg/ml. Bacteria were further grown at 37 °C for 6–8 h unless otherwise specified and the cells were collected and stored at −80 °C.
Production of antibodies against the enzyme protein
The recombinant enzyme protein was eluted from the SDS/PAGE gels of the extract of E. coli cells with pEC1 plasmid by electroelution. The purified protein was emulsified with Freund's complete adjuvant and injected intradermally into two rabbits. After 4 weeks, rabbits were boosted with the protein several times until a satisfactory antibody titre, judged by Western blot analysis, was obtained.
Western blotting
Samples were submitted to SDS/12%-(w/v)-PAGE [10] and electroblotted on to PVDF membrane. Protein bands were stained using a VECTASTAIN® ABC immunochemical staining kit (Vector Laboratories, Burlingame, CA, U.S.A.).
Amino acid sequence analysis
Proteins in the SDS/PAGE gel were electroblotted on to PVDF membrane with 10 mM Caps [3-(cyclohexylamino)propane-1-sulfonic acid] buffer, pH 11, and the membrane was stained with Coomassie Brilliant Blue R-250. Protein bands corresponding to 50, 33 and 23 kDa were cut out and submitted to sequencing using Applied Biosystems pulse-liquid protein sequencer model 477/120A.
Isolation and purification of the enzyme protein
Bacterial cells from a 4-litre culture were suspended in 280 ml of 50 mM Tris/HCl, pH 8.0, containing 1 mM EDTA, 1 mM PMSF and 1 mg/ml lysozyme. It was incubated for 15 min at 25 °C, and placed on ice for 10 min. Cells were disrupted by sonication with three 20 s bursts at 4 °C, and the resulting cell lysate was centrifuged at 3000 g. The pellet containing inclusion bodies was suspended in 560 ml of 50 mM Tris/HCl, pH 8.0, containing 1 mM EDTA, 1 mM PMSF, 100 mM NaCl and 0.5% Triton X-100. The resulting suspension was centrifuged at 30000 g for 30 min, and the pellet was resuspended in 280 ml of 0.1 M Tris/HCl buffer, pH 8.5, containing 1 mM EDTA, 1mM PMSF and 2 M urea. The suspension was centrifuged at 30000 g for 30 min to collect the pellet. The pellet was dissolved in 200 ml of 0.5 M Tris/HCl, pH 8.0, 1 mM EDTA, 1 mM PMSF and 6 M urea by homogenization. The resulting homogenate was centrifuged at 30 000 g for 30 min and the clear supernatant was dialysed against 5 litres of buffer A (20 mM Mops, pH 8.0, 1 mM EDTA, 1 mM PMSF, 30 mM 2-mercaptoethanol and 6 M urea).
The dialysed supernatant was applied to a DEAE-cellulose column (2.8 cm diameter×32 cm long) equilibrated with buffer A, and the column was washed with 2 litres of the same buffer. Protein was eluted with a linear gradient of 0–0.75 M NaCl in a total volume of 1 litre of buffer A. Fractions with absorbance at 280 nm were subjected to SDS/PAGE, and those with the 23 kDa protein were identified by Western blotting followed by immunochemical staining. The fractions containing the 23 kDa protein were pooled, dialysed against 4 litres of buffer A and concentrated by using a small DEAE-cellulose (DE-52) column (1.0 cm diameter×1.0 cm long). A 10 ml aliquot of the concentrated sample was gel-filtered on a Sephacryl S-200 column (3.1 cm diameter×145 cm long) equilibrated with buffer A containing 0.2 M NaCl. Fractions were analysed by SDS/PAGE, followed by Western blot analysis, and the major fractions containing the 23 kDa protein were pooled and stored at 4 °C.
A 10 ml aliquot of the above pooled Sephacryl S-200 fraction was diluted with buffer A to a final protein concentration of 0.08 mg/ml and dialysed against 1 litre of 50 mM sodium acetate buffer, pH 5.5, containing 10% (v/v) glycerol, 5% (v/v) ethylene glycol, 0.02% Nonidet P40, 1 mM EDTA and 30 mM 2-mercaptoethanol at 4 °C for 24 h. The dialysed sample was centrifuged at 100000 g and 4 °C for 60 min. The resulting supernatant was further dialysed successively against 1 litre each of 50 mM sodium acetate buffer, pH 5.5, containing 30, 20 and 10 mM 2-mercaptoethanol and centrifuged at 100000 g for 120 min. The dialysed sample was gel-filtered on a Tosoh TSKgel G3000 SW column (0.75 cm diameter×60 cm long) equilibrated with 50 mM sodium acetate buffer, pH 5.5. Each fraction was analysed by SDS/PAGE and immunoblotting using the antiserum against the 23 kDa protein.
Determination of proteolytic activity
Unless otherwise specified, 50 μl of 1 mM synthetic peptide in 0.05 M sodium acetate buffer, pH 5.5, containing 1 mM DTT (dithiothreitol) and 0.2 M NaCl was mixed with 10 μl of the refolded-proteinase solution and incubated at 37 °C for 30 min. The reaction was terminated by the addition of 240 μl of 5% (v/v) trifluoroacetic acid and freezing. The reaction products were analysed by HPLC using an ODS-032 (ABI) column with a linear gradient of 0–52.5% acetonitrile in 20 min at a flow rate of 1 ml/min. Absorbance of the effluent was monitored at 215 nm and the cleavage products were identified by amino acid analysis and sequencing.
The pH-dependence of activity was analysed as follows. A 120 μl volume of 1 mM synthetic peptide in 20 mM formate, 20 mM acetate, 10 mM methanesulfonate or 40 mM Tris buffer at different pH values containing 1 mM DTT was incubated with 0.4 μg of the refolded proteinase at 37 °C for 30 min. The reaction was stopped by the addition of 480 μl of 5% trifluoroacetic acid and the extent of peptide cleavage was analysed by HPLC as described above. The temperature dependence of activity was analysed in the same manner except that the assay was performed in 50 mM sodium acetate buffer, pH 4.0, containing 1 mM DTT at various temperatures. The effect of NaCl concentration on activity was analysed in the same manner except that the assay was performed in 200 mM sodium acetate buffer, pH 4.0, containing 1 mM DTT, at 37 °C at various concentrations of NaCl (0–5 M). The effects of various inhibitors on activity were analysed in the same manner, except that the assay was performed in 100 mM sodium acetate buffer, pH 4.0, containing 1 mM DTT and 0.2 M NaCl, at 37 °C in the presence of each inhibitor after pre-incubation at 37 °C for 30 min. The activity towards haemoglobin was determined as described in [11].
Measurements of the stability of the proteinase
To measure the thermal stability, 1 μg of the recombinant proteinase in 50 μl of 0.1 M sodium acetate buffer, pH 4.0, containing 1 mM DTT, was incubated at different temperatures for 1 h and the remaining activity was determined under the standard assay conditions. To measure the stability at pH 7.4, about 15 μg of the proteinase in 1.5 ml of 0.05 M Tris/HCl, pH 7.4, and 1 mM DTT was incubated at 37 °C. Aliquots were withdrawn at different time intervals and the remaining activity was determined under the standard assay conditions.
Reaction of DAN (diazoacetyl-D,L-norleucine methyl ester) with the proteinase
The proteinase (8 μg) was incubated with 50 μg of DAN in 0.8 ml of 0.05 M sodium phosphate buffer, pH 6.0, at 25 °C in the presence and absence of 1 mM cupric sulfate. Aliquots were removed at appropriate intervals and the remaining activity was determined.
Analysis of the cleavage specificity of the proteinase toward the B-chain of oxidized insulin
The B-chain of oxidized insulin (100 nmol) was digested with 8 μg of the proteinase in 300 μl of 0.05 M sodium acetate buffer, pH 4.0, containing 1 mM DTT and 0.2 M NaCl. Aliquots of 100 μl were removed at appropriate time intervals and submitted to HPLC using a Hitachi 655A-11 system on a column (0.46 cm long×25 cm diameter) of TSKgel ODS-120T (Tosoh Corp., Tokyo, Japan). The peptides were eluted with a linear gradient of acetonitrile (0–50%) in 30 min in 0.1% trifluoroacetic acid at a flow rate of 0.8 ml/min. The effluent was monitored by measuring the absorbance at 215 nm and the peptide peak fractions were collected and freeze-dried. An aliquot of each peptide fraction dissolved in water was submitted to amino acid analysis using an Applied Biosystems automated derivatizer/analyser (420A/130A).
RESULTS
Identification of the proteolytic processing site in the Copia Gag precursor protein
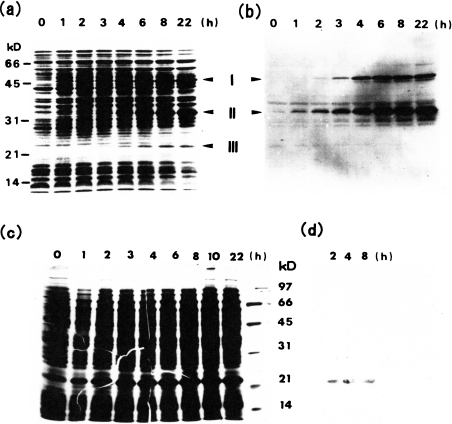
During the cultivation of E. coli containing the pEC1 plasmid, the intensities of three protein bands (bands I, II and III) were shown, when analysed by SDS/PAGE, to increase (Figure 1a). The bands corresponded to apparent molecular masses of 50, 33 and 23 kDa respectively. Band I and band II proteins were stained with an anti-VLP antiserum, whereas the band III protein was not (Figure 1b). This indicated that the 50 kDa Gag precursor protein was expressed and then processed into the 33 and 23 kDa daughter proteins.
Figure 1. Time course of processing of the wild-type Copia Gag precursor expressed in E. coli with pEC1 (a and b) and that of expression of the 23 kDa protein (Copia proteinase) in E. coli with pEC3 (c and d) as analysed by SDS/PAGE (a and c) and Western-blot analysis (b and d).
At the indicated times after addition of isopropyl β-D-thiogalactoside, the cells (2×107) were lysed in 10 μl of the lysis buffer, and the resulting lysates were submitted to SDS/PAGE (a, c) and immunoblot analysis using an anti-VLP serum (b) or an anti-serum against the 23-kDa protein obtained from E. coli with pEC1 (d). Approximate molecular masses are also shown for the standard marker proteins phosphorylase b (97.4 kDa), BSA (66.2 kDa), ovalbumin (45.0 kDa), carbonic anhydrase (31.0 kDa), soybean trypsin inhibitor (21.5 kDa) and lysozyme (14.4 kDa). kD=kDa.
The N-terminal amino acid sequences of these proteins were determined as shown below:
 |
The N-terminal sequences of band I and II proteins were the same and were identical with that deduced from the DNA sequence of the Gag precursor. On the other hand, the N-terminal 15-residue sequence of band III protein was identical with the sequence from Val275 to Phe289 of the deduced sequence of the precursor Gag protein. These results confirmed that band II and III proteins were produced from the expressed band I precursor Gag protein, through cleavage at the Met274–Val275 bond. The putative active-site motif of aspartic proteinases, namely Asp-Ser-Gly, was present in band III protein, and an antiserum raised against this protein reacted with band I precursor protein, but not with band II protein. These findings suggest that band III protein may be a proteinase and responsible for the observed processing of the Gag precursor protein. The soluble fraction of a crude extract of E. coli cells harbouring the pEC1 plasmid, however, showed no significant activity toward the synthetic peptide substrate.
Expression, isolation and purification of the enzyme protein
The region coding for band III protein was obtained from the Copia pZY2 plasmid, a mutant plasmid of the Copia sequence lacking the intron region, [8] and inserted into pKK233-2, and the resulting pEC3 plasmid was expressed in E. coli. The increase in intensity of the corresponding protein band was observed with isopropyl β-D-thiogalactoside induction up to 8 h (Figure 1c) and only this band reacted with the antiserum raised against band III protein (Figure 1d). The protein expressed in 1 litre of the E. coli cell culture was about 25 mg at 8 h after the induction as estimated by SDS/PAGE.
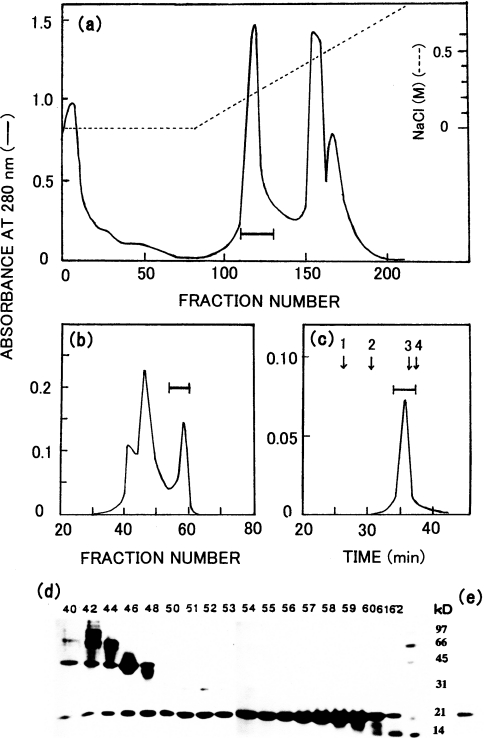
About 90% of the enzyme protein present in the purified inclusion-body pellet was solubilized in the buffer containing 6–8 M urea and 30 mM 2-mercaptoethanol. The enzyme protein was purified by successive steps of chromatography on DEAE-cellulose and Sephacryl S-200 gel filtration (Figures 2a and 2b). The purity of the fractions was analysed by SDS/PAGE. Fractions 54–60 on Sephacryl S-200 gel filtration were the major fractions containing the enzyme protein (Figure 2d). Thus about 25 mg of the purified protein was obtained from 100 mg of the expressed protein in 4 litres of the E. coli cell culture.
Figure 2. Isolation and purification of the 23 kDa protein (Copia proteinase) expressed in E. coli with pEC3.
(a) DEAE-cellulose chromatography. The solution of the solubilized inclusion body from E. coli with pEC3 was applied to a DEAE-cellulose column (2.8 cm diameter×32 cm long) at pH 7.9 and eluted with a linear gradient of 0–0.75 M NaCl in a total volume of 1 litre. The fraction size was 7.5 ml. (b) Sephacryl S-200 chromatography. The pooled fraction from the DEAE-cellulose chromatography was concentrated and applied to a Sephacryl S-200 column (3.1 cm diameter×145 cm long) at pH 7.9. The fraction size was 7.5 ml. (c) TSKgel G3000 SW chromatography. The pooled fraction from the Sephacryl S-200 column was submitted to the refolding procedure, and then applied to a TSKgel G3000 SW column at pH 5.5. Arrows indicate the elution positions of standard proteins: 1, BSA; 2, ovalbumin; 3, soybean trypsin inhibitor; 4, lysozyme. (d) SDS/PAGE of Sephacryl S-200 fractions. (e) SDS/PAGE of the pooled TSKgel G3000SW fraction. The same standard marker proteins were used in (d) and (e) as used in Figure 1. In each chromatographic run the fractions under the bar were pooled. The numbers above the gel in SDS/PAGE (d) indicate fraction numbers.
The enzyme protein was then refolded to the proper conformation with proteolytic activity by gradually removing urea (from 6 M to 0 M), followed by stepwise removal of 2-mercaptoethanol (from 30 to 20 mM and then from 20 to 10 mM). After refolding, the insoluble unfolded protein was removed by centrifugation at 100 000 g for 1 h, and the refolded proteinase was purified by gel filtration on a TSKgel G3000SW column (Figure 2c). The purified refolded enzyme gave a single band on SDS/PAGE (Figure 2e). Its molecular mass was estimated to be 23 kDa by SDS/PAGE under both reducing and non-reducing conditions and by gel filtration (results not shown).
Development of an assay procedure to characterize the Copia proteinase
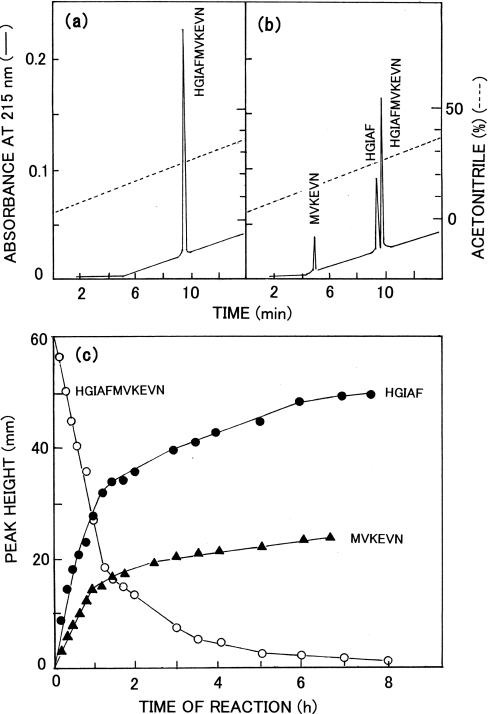
A synthetic 11-residue peptide, His-Gly-Ile-Ala-Phe-Met-Val-Lys-Glu-Val-Asn, corresponding to the sequence at the processing site and its vicinity of the Gag precursor protein was synthesized and used as the substrate. A typical HPLC analysis of the reaction mixture is shown in Figures 3(a) and 3(b); the peak height of the intact peptide was decreased with time, and two new peaks appeared. They were identified to be His-Gly-Ile-Ala-Phe and Met-Val-Lys-Glu-Val-Asn by amino acid sequencing. The increases in height of these peaks were linear up to 60 min (Figure 3c) at a saturating substrate concentration (around 1 mM) (Figure 4a). Therefore, a 30–60 min incubation period was used to determine the activity of the proteinase.
Figure 3. HPLC assay for Copia proteinase using the synthetic peptide His-Gly-Ile-Ala-Phe-Met-Val-Lys-Glu-Val-Asn as a substrate.
(a) HPLC of the intact peptide. (b) HPLC of a 1 h digest of the peptide. (c) Time course of the peptide hydrolysis. Peptides are shown using the single-letter notation.
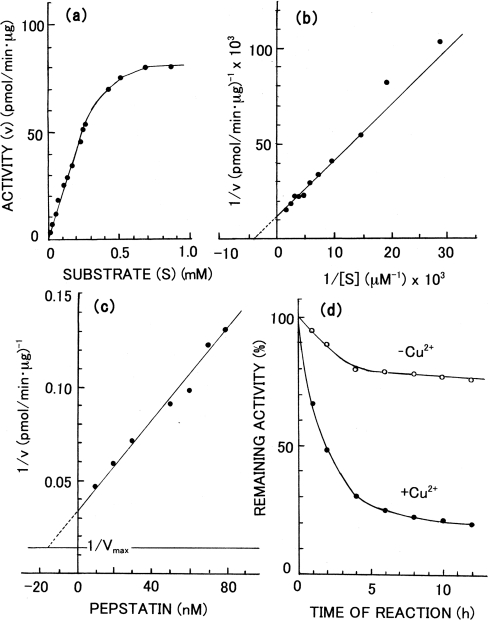
Figure 4. Effects of substrate concentration and inhibitors on the activity of Copia proteinase.
(a) The initial rates of hydrolysis were measured at various concentrations of substrate. (b) Lineweaver–Burk plot of the data in (a). (c) Dixon plot for pepstatin inhibition. (d) Time courses of changes in activity by reaction with DAN were measured in the presence (●) or absence (○) of 1 mM Cu2+.
Enzymatic properties of the proteinase
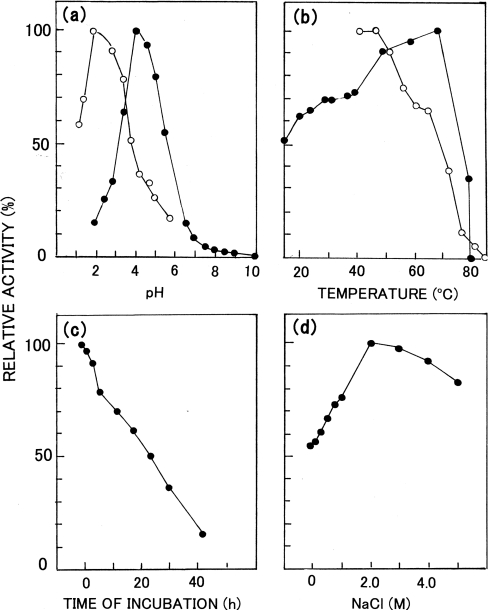
The Km and Vmax values of the proteinase toward the synthetic peptide at pH 5.5 and 37 °C were estimated to be 235 μM and 81.3 pmol·min−1·μg−1 respectively by using a Lineweaver–Burk plot (Figure 4b). The pH- and temperature-dependences of proteolytic activity of the enzyme are shown in Figures 5(a) and 5(b) respectively. The optimum pH of the activity toward the synthetic peptide was 4.0 (Figure 5a), and the specific activity at the optimum pH was determined to be 144 pmol·min−1·μg−1. Therefore the activity of the enzyme was assayed at pH 4.0 in the subsequent studies unless otherwise specified. On the other hand, the optimum activity toward acid-denatured haemoglobin was observed at pH 2.0 (Figure 5a). When the assay temperature was elevated, the activity of the enzyme increased up to 70 °C and a sharp decline was observed beyond 70 °C (Figure 5b). The activity at 37 °C was 69% of that observed at the optimum temperature, 70 °C.
Figure 5. Effects of various factors on the activity of Copia proteinase.
(a) Effect of pH on the activity toward the synthetic substrate (●) and haemoglobin (○). (b) Effects of temperature on the activity (●) and stability (○). To measure the stability, the enzyme was pre-incubated for 1 h at different temperatures, then assayed at 37 °C. (c) Effect of incubation at pH 7.4 on the activity. (d) Effect of NaCl concentration on the activity.
The stability of the enzyme at different temperatures and pH 4.0 and that at pH 7.4 and 37 °C are also shown in Figure 5. The enzyme was stable up to 50 °C at pH 4.0, and a gradual decrease of activity was observed above 50 °C; the enzyme was completely inactivated after a 1 h incubation at 85 °C (Figure 5b). The enzyme was not very stable at pH 7.4 and 37 °C, and a gradual reduction of activity was observed with incubation time. The half-life was 24 h, and nearly complete inactivation was observed after 48 h (Figure 5c).
When NaCl was added to the assay mixture, a gradual increase of activity was observed up to 2 M NaCl; the activity was decreased gradually above 2 M NaCl (Figure 5d). Pepstatin A strongly inhibited the enzyme; a Ki value of 15 nM was obtained at pH 4.0 by using Dixon plots (Figure 4c). DAN was shown to inactivate the enzyme to a significant extent in the presence of Cu2+ (Figure 4d). Under the reaction conditions used, in the presence of 1 mM Cu2+, 50% of the activity was lost in 2 h and the activity decreased further on longer incubation. In the absence of Cu2+, no significant inactivation took place. On the other hand, no inhibition was observed with any of the following inhibitors for serine peptidases, cysteine peptidases, metallopeptidases or aminopeptidases at the given concentrations: di-isopropyl fluorophosphate (0.1 mM), PMSF (1 mM), tosyl-l-phenylalanylchloromethane (‘TPCK’ 1 mM), E-64 [1-trans-epoxysuccinyl-L-leucylamido(4-guanidino)butane] (0.1 mM), o-phenanthroline (10 mM), EDTA (10 mM), soybean trypsin inhibitor (0.1 μg/ml), leupeptin (0.1 mM), antipain (0.1 mM), amastatin (0.1 mM) and diprotinin A (0.05 mM).
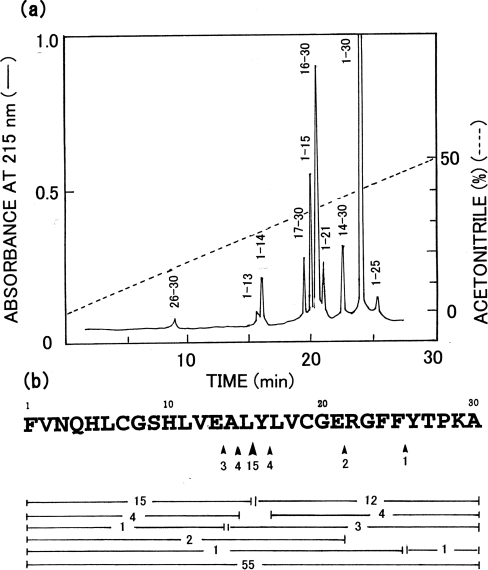
The cleavage profile of the enzyme toward the B-chain of oxidized insulin at pH 4.0 is shown in Figure 6. The Leu15–Tyr16 bond was cleaved fairly selectively, followed by slower cleavages at Ala14–Leu15, Tyr16–Leu17, Glu13–Ala14, Glu21–Arg22 and Phe25–Tyr26 bonds (in decreasing order).
Figure 6. Cleavage specificity of Copia proteinase toward oxidized insulin B-chain.
(a) HPLC pattern of a 3 h digest of oxidized insulin B-chain. The numbers against the peaks indicate the residue positions of the peptide in the substrate. (b) Yields of peptides and sites of cleavage. Arrowheads indicate the cleavage sites, and the number under each arrowhead shows estimated extent of cleavage as a percentage. Bars indicate peptide fragments produced, and the number in the middle of each bar shows the percentage yield of the peptide.
Loss of the proteolytic activity by active-site-residue mutation
The mutant enzyme in which the putative catalytic residue, Asp18, was changed to alanine was expressed in E. coli cells containing the pEC16 plasmid, purified and refolded by essentially the same procedures as used for the wild-type enzyme. The expression yield and the elution profiles on chromatography of the mutant protein were practically the same as those of the wild-type proteinase. The refolded mutant protein, however, showed no activity toward the synthetic peptide.
DISCUSSION
As described in the preceding section, when the 2 kb copia RNA was expressed in E. coli containing the plasmid pEC1, a protein with an apparent molecular mass of 23 kDa was shown to be produced, in addition to the 50 kDa Gag precursor protein and the 33 kDa VLP protein that had previously been found by Yoshioka et al. [9]. The N-terminal amino acid sequencing of these proteins clearly showed the processing of the 50 kDa precursor protein to produce the two daughter proteins, which represent the N- and C-terminal halves respectively of the 50 kDa protein.
Attempts to demonstrate the activity of the Copia proteinase in the lysate of E. coli cells with the plasmid pEC1 towards the synthetic peptide or towards the 50 kDa precursor protein obtained from E. coli containing the pEC2 plasmid were not successful, presumably due to a very low concentration of the active proteinase in the E. coli cell lysate. Thus direct purification of the enzyme protein from E. coli cells with the plasmid pEC1 was difficult; therefore the coding region of the enzyme protein in the copia gag gene was expressed in E. coli to obtain a high level of expression.
The simple purification and refolding procedures permitted us to obtain as much as 6.3 mg of the active proteinase from 1 litre of the E. coli culture. A selective cleavage of the synthetic peptide His-Gly-Ile-Ala-Phe-Met-Val-Lys-Glu-Val-Asn was observed at the Phe5–Met6 bond by the purified proteinase. On the other hand, the N-terminal amino acid sequence of the enzyme protein from E. coli cells containing the plasmid pEC1 started from Val275 in the sequence···-Phe273-Met274-Val275-···. These results suggest that the initial processing site in the 50 kDa Gag precursor protein is the Phe273–Met274 bond and that the resulting N-terminal Met274 residue was removed, presumably by the action of the methionyl aminopeptidase present in E. coli. Similarly, the removal of the N-terminal methionine residue from the enzyme protein obtained from E. coli cells containing the plasmid pEC3 was also observed. The methionine residues in the N-terminal Met-Asp-···sequence of the 50 and 33 kDa proteins were not removed. It is known that when the next residue has a side chain with a low radius of gyration (glycine, alanine, serine, threonine, proline, valine or cysteine), the N-terminal methionine residue is removed by methionyl aminopeptidase, but otherwise it is retained [12]. The above results are consistent with this specificity of methionyl aminopeptidase.
The ordinary aspartic proteinases are composed of two lobes that are highly homologous with respect to their tertiary structure [1]. Each lobe is composed of a single polypeptide chain of more than 150 residues with a conserved Asp-Ser/Thr-Gly motif constituting the active site of the enzyme. In contrast, retroviral proteinases are generally composed of two identical subunits, and each subunit is composed of less than 130 amino acids and contains only one Asp-Ser/Thr-Gly motif. In the case of HIV proteinase, its subunit was first demonstrated to dimerize to form the active enzyme [13]. Therefore, by analogy to HIV proteinase and other retroviral proteinases, dimerization of the present enzyme protein is thought to be a prerequisite for it to exhibit proteolytic activity. The actual molecular mass of the proteinase, based on its amino acid sequence of 152 residues, is 16.7 kDa. On the other hand, the active proteinase was eluted at the position of 23 kDa on gel filtration. This seems to indicate dimerization of the protein. However, the proteinase was also observed at the 23 kDa region on SDS/PAGE under both reducing and non-reducing conditions. This is presumably due to an anomalous behaviour of the proteinase subunit on SDS/PAGE such that it shows an apparently higher molecular mass than its theoretical one. Indeed, certain low-molecular-mass proteins are known to behave anomalously on SDS/PAGE [14]. For examples, ribonuclease A (13.7 kDa), ribonuclease T1 (11.1 kDa) and the heavy chain of aspergillopepsin II (18.3 kDa) were reported to give molecular masses of 18.5 kDa (35% overestimated) [14], 18.0 kDa (62% overestimated) [15] and 33.0 kDa (80% overestimated) [16] respectively on SDS/PAGE. The molecular mass of 23 kDa estimated by gel filtration is somewhat lower than the theoretical value of 33.4 kDa. The reason for this is not certain at present.
The present enzyme had a pH optimum at 4.0 toward the synthetic peptide. This is roughly similar to the result reported for HIV-1 proteinase, which, however, has a broader pH optimum, ranging from pH 3.5 to 5.5, toward a synthetic peptide [17]. The optimum pH values for other retroviral proteinases were reported to range from pH 5.5 to 7.2 [18]. Although the present enzyme was shown to act optimally at pH 2.0 toward haemoglobin, this is presumably due to more extensive denaturation of the substrate at lower pH values.
The optimum temperature (70 °C at pH 4.0) of the proteinase was remarkably higher than that of HIV-1 proteinase (37 °C at pH 6.0), which is inactive above 60 °C [17]. The present enzyme was fairly stable up to 50 °C at pH 4.0, and above this temperature it became less stable. On the other hand, human pepsin A was reported to be fairly stable up to 60 °C at pH 5.5, above which it is inactivated rapidly [11]. Taking these observations together, the present enzyme appears to be markedly more stable at higher temperatures than HIV-1 proteinase. The polypeptide chain of HIV-1 proteinase is composed of 99 amino acid residues, with the catalytic aspartic acid residue at position 25. On the other hand, that of the present enzyme is composed of 152 residues with the putative catalytic aspartic acid residue at position 18. Therefore, the present enzyme has a longer C-terminal tail than HIV-1 proteinase. This tail region might contribute to the stability of the present enzyme. The enzyme was not very stable at pH 7.4, but was relatively more stable than ordinary aspartic proteinases. For example, human pepsin A is rapidly inactivated at pH 7.0 and 37 °C; the half-life is about 1 min and it is almost completely inactivated within 10 min [11]. The structural basis for this stability of the present enzyme remains to be elucidated.
The enzyme was inhibited by the two aspartic-proteinase-specific inhibitors pepstatin A and DAN. Pepstatin A inhibited the enzyme with a Ki value of 15 nM. On the other hand, HIV-1 proteinase was reported to be inhibited by pepstatin A with a Ki value of 1.1 μM at pH 5.5 [19] or with IC50 values of 700 nM at pH 6.0 [20] and 360 nM at pH 5.5 [21]. The IC50 values for other retrovirus proteinases were reported to be in the range 100–500 nM [22]. Therefore the present enzyme is more sensitive to pepstatin A than retrovirus proteinases, although much less sensitive than ordinary aspartic proteinases, such as porcine pepsin (Ki 0.05 nM at pH 5.0 [23]). The enzyme was also inactivated by DAN in the presence of Cu2+, although the rate of inactivation appeared to be somewhat lower than those observed for ordinary aspartic proteinases. Moreover, the enzyme was not inhibited by any of the proteinase inhibitors tested, except for pepstatin A and DAN, and the mutant (Asp18→alanine) enzyme had no activity. These results are consistent with the notion that the enzyme is a member of the aspartic proteinase family.
The processing site of the Gag precursor protein was deduced to be the Phe273–Met274 bond. On the other hand, the enzyme cleaved most rapidly the Leu15–Tyr16 bond in the B-chain of oxidized insulin, with slower cleavages at five other peptide bonds, Ala14–Leu15, Tyr16–Leu17, Glu13–Ala14, Glu21–Arg22 and Phe25–Tyr26 in decreasing order. Thus the enzyme appears to show a preference for certain hydrophobic or aromatic or glutamic acid residues at the P1 position. These results indicate that the specificity of the enzyme is not restricted to the peptide bond at the processing site of the Copia Gag precursor, as in the cases of other retroviral proteinases [24]. The cleavage specificity toward oxidized insulin B-chain is fairly different from that of HIV-1 proteinase, which was reported to cleave the Glu13–Ala14 bond most rapidly, followed by the Tyr16–Leu17 bond, with slower cleavages at the Leu11–Val12 and Leu15–Tyr16 bonds [25]. Since the Leu15–Tyr16 bond is also one of the major sites cleaved by several ordinary aspartic proteinases [26], the cleavage specificity of the present enzyme toward oxidized insulin B-chain is similar to, though not identical with, those of ordinary aspartic proteinases.
To shed further light on the structure–function relationship of the present enzyme, it will be essential to elucidate its three-dimensional structure.
Acknowledgments
This study was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1.Rawlings N. D., Barrett A. J. Families of aspartic peptidases. Methods Enzymol. 1995;248:105–120. doi: 10.1016/0076-6879(95)48009-9. [DOI] [PubMed] [Google Scholar]
- 2.Rinckel L. A., Garfinkel D. J. Fungal, plant and animal transposon elements. In: Barrett A. J., Rawlings N. D., Woessner J. F., editors. Handbook of Proteolytic Enzymes, vol. 1. 2nd edn. London: Elsevier/Academic Press; 1998. pp. 190–195. [Google Scholar]
- 3.Shiba T., Saigo K. Retrovirus-like particles containing RNA homologous to the transposable element copia in Drosophila melongaster. Nature. 1983;302:119–124. doi: 10.1038/302119a0. [DOI] [PubMed] [Google Scholar]
- 4.Lewis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980;21:581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- 5.Bayev A. A., Krayev A. S., Lyubomirskaya N. V., Ilyin Y., Skryabin K. G., Georgiev G. P. The transposable element Mdg3 in Drosophila melanogaster is flanked with the perfect direct and mismatched inverted repeats. Nucleic Acids Res. 1980;8:3263–3273. doi: 10.1093/nar/8.15.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flavell A. J., Ruby S. W., Toole J. J., Roberts B. E., Rubin G. M. Translation and developmental regulation of RNA encoded by the eukaryotic transposable element copia. Proc. Natl. Acad. Sci. U.S.A. 1980;77:7107–7111. doi: 10.1073/pnas.77.12.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heine C. W., Kelly D. C., Avery R. J. The detection of intracellular retrovirus-like entities in Drosophila melanogaster cell cultures. J. Gen. Virol. 1980;49:385–395. doi: 10.1099/0022-1317-49-2-385. [DOI] [PubMed] [Google Scholar]
- 8.Yoshioka K., Homma H., Zushi M., Kondo S., Togashi S., Miyake T., Shiba T. Virus-like particle formation of Drosophila copia through autocatalytic processing. EMBO J. 1990;9:535–541. doi: 10.1002/j.1460-2075.1990.tb08140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshioka K., Kanda H., Kondo S., Togashi S., Miyake T., Shiba T. Autoprocessing of Drosophila copia gag precursor to generate a unique laminate structure in Escherichia coli. FEBS Lett. 1991;285:31–34. doi: 10.1016/0014-5793(91)80718-i. [DOI] [PubMed] [Google Scholar]
- 10.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Athauda S. B. P., Tanji M., Kageyama T., Takahashi K. A comparative study on the NH2-terminal amino acid sequences and some other properties of six isozymic forms of human pepsinogens and pepsins. J. Biochem. (Tokyo) 1989;106:920–927. doi: 10.1093/oxfordjournals.jbchem.a122952. [DOI] [PubMed] [Google Scholar]
- 12.Hirel P.-H., Schmitter J.-M., Dessen P., Fayat G., Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meek T. D., Dayton B. D., Metcalf B. W., Dreyer G. B., Strickler J. E., Gorniak J. G., Rosenberg M., Moore M. L., Magaard V. W., Debouck C. Human immunodeficiency virus 1 protease expressed in Escherichia coli behave as a dimeric aspartic protease. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1841–1845. doi: 10.1073/pnas.86.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunker A. K., Rueckert R. R. Observations on molecular weight determinations on polyacrylamide gel. J. Biol. Chem. 1969;244:5074–5080. [PubMed] [Google Scholar]
- 15.Fields R., Dixon H. B. F., Law G. R. Purification of ribonuclease T1 by diethylaminoethylcellulose chromatography. Biochem. J. 1971;121:591–596. doi: 10.1042/bj1210591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K. Proteinase A from Aspergillus niger. Methods Enzymol. 1995;248:146–155. doi: 10.1016/0076-6879(95)48012-9. [DOI] [PubMed] [Google Scholar]
- 17.Billich S., Knoop M. T., Hansen J., Štrop P., Sedláček J., Mertz R., Moelling K. Synthetic peptides as substrates and inhibitors of human immunodeficiency virus-1 protease. J. Biol. Chem. 1988;263:17905–17908. [PubMed] [Google Scholar]
- 18.Yoshinaka Y., Katoh I., Copeland T., Smythers G., Oroszlan S. Bovine leukemia virus protease: purification, chemical analysis, and in vitro processing of gag precursor polyproteins. J. Virol. 1986;57:826–832. doi: 10.1128/jvi.57.3.826-832.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darke P. L., Leu C.-T., Davis L. J., Heimbach J. C., Diehl R. E., Hill W. S., Dixon R. A., Sigal I. S. Human immunodeficiency virus protease: bacterial expression and characterization of the purified aspartic protease. J. Biol. Chem. 1989;264:2307–2312. [PubMed] [Google Scholar]
- 20.Kräusslich H. G., Ingraham R. H., Skoog M. T., Wimmer E., Pallai P. V., Carter C. A. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc. Natl. Acad. Sci. U.S.A. 1989;86:807–811. doi: 10.1073/pnas.86.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook N. D., Jessop R. A., Robinson R. S., Richards A. D., Kay J. Scintillation proximity assay: a rapid and novel assay technique applied to HIV proteinase. In: Dunn M. B., editor. Structure and Function of the Aspartic Proteinases: Genetics, Structures, and Mechanisms. New York: Plenum Press; 1991. pp. 525–528. [Google Scholar]
- 22.Katoh I., Yasunaga T., Ikawa Y., Yoshinaka Y. Inhibition of retroviral protease activity by an aspartyl proteinase inhibitor. Nature. 1987;329:654–656. doi: 10.1038/329654a0. [DOI] [PubMed] [Google Scholar]
- 23.Workman R. J., Burkitt D. W. Pepsin inhibition by a high specific activity radioiodinated derivative of pepstatin. Arch. Biochem. Biophys. 1979;194:157–164. doi: 10.1016/0003-9861(79)90605-2. [DOI] [PubMed] [Google Scholar]
- 24.Moore M. L., Bryan W. M., Fakhoury S. A., Maggard V. W., Huffman W. F., Dayton B. D., Meek T. D., Hyland L., Dreyer G. B., Metcalf B. W., et al. Peptide substrates and inhibitors of HIV-1 proteinase. Biochem. Biophys. Res. Commun. 1989;159:420–425. doi: 10.1016/0006-291x(89)90008-9. [DOI] [PubMed] [Google Scholar]
- 25.Tomasselli A. G., Hui J. O., Sawer T. K., Thaisrivongs S., Hester J. B., Heinrickson R. L. The evaluation of non-viral substrater of the HIV protease as leads in the design of inhibitors for aids therapy. In: Dunn M. B., editor. Structure and Function of the Aspartic Proteinases: Genetics, Structures, and Mechanisms. New York: Plenum Press; 1991. pp. 469–481. [DOI] [PubMed] [Google Scholar]
- 26.Athauda S. B. P., Takahashi K. Cleavage specificities of aspartic proteinases toward oxidized insulin B chain at different pH values. Protein Pept. Lett. 2002;9:289–294. doi: 10.2174/0929866023408698. [DOI] [PubMed] [Google Scholar]