Abstract
Ubiquilin proteins have been shown to interact with a wide variety of other cellular proteins, often regulating the stability and degradation of the interacting protein. Ubiquilin contains a UBL (ubiquitin-like) domain at the N-terminus and a UBA (ubiquitin-associated) domain at the C-terminus, separated by a central region containing Sti1-like repeats. Little is known about regulation of the interaction of ubiquilin with other proteins. In the present study, we show that ubiquilin is capable of forming dimers, and that dimerization requires the central region of ubiquilin, but not its UBL or the UBA domains. Furthermore, we provide evidence suggesting that monomeric ubiquilin is likely to be the active form that is involved in binding presenilin proteins. Our results provide new insight into the regulatory mechanism underlying the interaction of ubiquilin with presenilins.
Keywords: dimerization, immunoprecipitation, oligomerization, presenilin, ubiquilin, yeast two-hydrid assay
Abbreviations: GFP, green fluorescent protein; GST, glutathione S-transferase; HA, haemagglutinin; Ni-NTA, Ni2+-nitrilotriacetate; ODC, ornithine decarboxylase; PS, presenilin(s); UBA, ubiquitin-associated; UBL, ubiquitin-like
INTRODUCTION
Ubiquilin is a conserved protein that has been found in all eukaryotes examined [1]. This novel protein is characterized by a UBL (ubiquitin-like) domain at its N-terminus, a central more variable domain containing several different short repeats, and a UBA (ubiquitin-associated) domain at its extreme C-terminus. There are three ubiquilin genes in humans: ubiquilin-1 is expressed ubiquitously, ubiquilin-2 is expressed with a more restricted tissue expression pattern than ubiquilin-1, and ubiquilin-3 is expressed only in the testis [1–3]. Human ubiquilin-1 encodes a protein of 589 amino acids, whereas ubiquilin-2 and -3 encode proteins of 624 and 655 amino acids respectively. The three proteins differ from each other primarily by the presence or absence of long insertions in the central region of the protein. The functional significance of these insertions is unknown.
Ubiquilin proteins have been reported to interact with numerous proteins that are apparently unrelated [1,3–16]. In most cases, overexpression of ubiquilin has been found to increase the stability of the interacting protein, although it is not yet clear how ubiquilin functions in this capacity [1,4,5,8,15,17,18]. This role is particularly puzzling, as ubiquilin, similar to Rad23 with which it shares similar structural organization [19–22], has also been proposed to function as a shuttle factor acting to deliver polyubiquitinated proteins to the proteasome for degradation [5,6,12,23–26]. In this scenario, the UBA domain of ubiquilin, which has been shown to bind polyubiquitinated chains, would bind polyubiquitinated proteins through interaction with their conjugated polyubiquitinated chains and bind the proteasome via UBL interaction with the S5a subunit of the 26 S proteasome [18,22,27,28].
Although ubiquilin has been shown to interact with a number of proteins, little is known of how the interaction of ubiquilin with its various binding partners is regulated. Ubiquilin may be subject to spatial or temporal regulation, post-translational modification or some other regulatory modification. One possibility is that oligomerization of ubiquilin proteins regulates its ability to bind its other interactors. To test this hypothesis we examined whether ubiquilin forms oligomers in vitro and in vivo, and whether oligomerization is required for interaction with PS2 (presenilin-2). We report that ubiquilin is capable of forming dimers, and that the monomer is likely the active form that binds PS2.
EXPERIMENTAL
Yeast two-hybrid liquid assay
In addition to constructs encoding the full-length ubiquilin-1 and -2 proteins, ubiquilin-1 constructs encoding the UBL domain, UBA domain, ubiquilin(ΔUBL), ubiquilin(ΔUBA) and ubiquilin(ΔUBL/ΔUBA) were prepared. All constructs were cloned into both bait and prey vectors. Yeast strain EGY48 was sequentially transformed with lacZ reporter plasmid pSH18-34, and various permutations of ubiquilin constructs cloned into bait and prey vectors pEG202 and pJG4-5 respectively. Yeast transformants were selected for by plating on the appropriate drop-out plates. Interaction between ubiquilin constructs was measured by assaying for β-galactosidase enzyme activity in liquid cultures using ONPG (O-nitrophenyl-β-D-galactopyranoside) as a substrate [29]. All interactions were normalized to a lamin bait in pEG202 as a negative control [30]. Assays were performed in triplicate.
Statistical analysis
For statistical analysis, the Microscoft Excel program was used to calculate S.D., and the significance was determined using the P value function.
HeLa cell culture, DNA transfection and immunoprecipitation
Ubiquilin-1 constructs described above were cloned into both pEGFP-N1 and pCMV-Myc vectors (ClonTech). HeLa cells were grown in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS (fetal bovine serum). Cells were co-transfected with pEGFP-N1-ubiquilin and pCMV-Myc-ubiquilin constructs using Lipofectamine™ 2000 reagent according to the manufacturer's instructions (Invitrogen). Each plasmid cDNA (2 μg) was used to transfect cells plated on 10 cm dishes. Cells were collected 20 h after transfection in standard RIPA buffer with 0.5% Nonidet P40 and protease inhibitors. Cell lysates were sheared using a 21-gauge needle and spun for 10 min at 13000 g prior to BCA assay. Total protein (500 μg; at a 1 μg/1 μl concentration) was used for each immunoprecipitation reaction. For immunoprecipitation, cells were first pre-incubated with Protein A–Sepharose beads for 30 min. The mixture was then spun at 1000 g for 1 min, and the resultant supernatant was added to fresh beads along with 10 μl rabbit anti-GFP (green fluorescent protein) antibody. Reactions were incubated at 4 °C for 2 h with gentle rotation. Beads were then washed 5 times with RIPA buffer prior to adding sample buffer and boiling for 5 min. Immunoprecipitation reactions and total lysates were separated by SDS/PAGE.
Western blot analysis
Proteins were transferred on to nitrocellulose membrane, which was blocked with non-fat dried milk prior to probing with antibody. Antibodies used were mouse anti-Myc (1:100; hybridoma supernatant 9E10), rabbit anti-HA (haemagglutinin) (1:1000; Sigma) and mouse anti-ODC (ornithine decarboxylase) (1:200; Sigma) and rabbit anti-GFP (1:1000; generated against recombinant GFP). Secondary antibodies conjugated to horseradish peroxidase were used at a concentration of 1:3000 (Amersham).
Cell staining and immunofluorescence microscopy
HeLa cells were plated on to glass coverslips in 10 cm dishes and transfected with 2 μg each of CMV (cytomegalovirus) expression plasmids encoding the following combinations of fusion proteins: Myc–ubiquilin-1 and ubiquilin-1–GFP, Myc–ubiquilin-1 and ubiquilin-2–GFP, Myc–ubiquilin-2 and ubiquilin-1–GFP, or Myc–ubiquilin-2 and ubiquilin-2–GFP. Cells were fixed and stained for immunofluorescence microcopy as described previously [1,31]. Primary antibodies used were mouse anti-Myc (hybridoma supernatant 9E10) antibody at a final concentration of 1:100, and rabbit anti-GFP antibody diluted (1:500). Fluorescence staining of cells was visualized using a ×100 objective under an inverted Leica DM IRB microscope and images were captured using a Photometrics SenSys camera and merged using IPLab Software.
Assay for ubiquilin-1 dimerization or oligomerization
Ubiquilin-1 was cloned into pGST-T1 vector or pET-21a(+) vector to generate GST (glutathione S-transferase)–ubiquilin-1 or ubiquilin-1–His6 respectively. For this assay, GST–ubiquilin-1 was purified as described previously [31]. Ubiquilin-1–His6 was purified using Ni-NTA (Ni2+-nitrilotriacetate)–agarose beads using the manufacturer's instructions (Qiagen). Proteins were mixed at 1:1 and 10:1 ratios (ubiquilin-1–His6/GST–ubiquilin-1). For 1:1 ratio, 250 μg of purified GST–ubiquilin-1 and 250 μg of purified ubiquilin-1–His6 were mixed together in a 1 ml volume with 1×PBS. The protein mixture was incubated for 20 min at 4 °C with gentle rotation. After 1 h, 1.5 ml of glutathione–agarose beads were added to the mixture, followed by rotation for 1 h at 4 °C. The slurry was then packed into a column, washed twice with GST wash buffer, and then the protein eluted with 10 mM glutathione in 50 mM Tris/HCl (pH 8.0). To this eluant, 2 ml Ni-NTA agarose beads were added and the slurry was again incubated with rotation for 1 h at 4 °C. The slurry was next packed into a column and washed twice with His wash buffers (Qiagen). Finally, the protein was eluted with 250 mM imidazole. Fractions were saved from each step outlined above and separated by SDS/PAGE (8.5% gels). Proteins were transferred on to nitrocellulose membrane, which was blocked in non-fat dried milk prior to probing with antibody. Antibodies used were rabbit anti-ubiquilin (1:1000) [1], rabbit anti-GST (1:1000) (raised against recombinant GST protein) and mouse anti-His5 (1:1000) (Qiagen). Secondary antibodies conjugated to horseradish peroxidase were used at a concentration of 1:3000. Relative band intensities were quantified using IPGel software.
Assay for PS interaction with ubiquilin-1 dimers or monomers
Ubiquilin-1–His6 and GST–ubiquilin-1 were purified as described above. [35S]Methionine-labelled PS1 or PS2 was synthesized in a coupled in vitro transcription–translation reaction [1]. For the PS-binding assay, 100 μg each of ubiquilin-1–His6 and GST–ubiquilin-1, and 100 μl of [35S]methionine-labelled PS1 in vitro translation product were mixed together in a 1 ml volume in 1×PBS and incubated for 1 h at 4 °C with gentle rotation. After 1 h, 1.5 ml glutathione–agarose beads were added to the mixture, followed by rotation for 1 h at 4 °C. The slurry was then added to a column, washed twice with GST wash buffer, and then the bound protein eluted with 10 mM glutathione in 50 mM Tris/HCl (pH 8.0). To this eluant, 1.5 ml Ni-NTA–agarose beads were added and the slurry was again incubated with rotation for 20 min at 4 °C. The slurry was next packed into a column and washed twice with His wash buffers. The protein mixture was then eluted with 250 mM imidazole. Fractions from each step were separated by SDS/PAGE (8.5% gels). The first gel was stained with Coomassie Blue for 30 min prior to destaining in methanol buffer. This gel was next dried and exposed to film by autoradiography. For the second gel, proteins were transferred on to nitrocellulose membrane and immunoblotted for ubiquilin. Rabbit anti-ubiquilin was used at a dilution of 1:1000, followed by incubation with horseradish peroxidase.
RESULTS
Ubiquilin-1 and -2 isoforms interact with each other in yeast
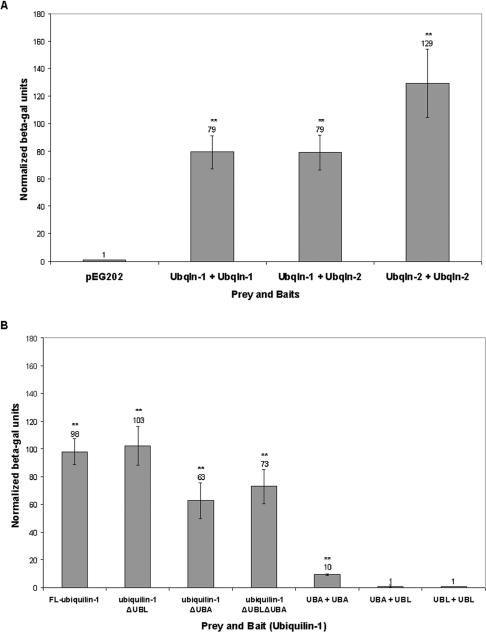
Ubiquilin was previously found to be self-activating in yeast two-hybrid screens [6]. To investigate whether ubiquilin proteins can interact with one another, we measured the interaction of the proteins using yeast two-hybrid β-galactosidase liquid assays. Ubiquilin-1 and -2 proteins were used as both bait and prey. We found that ubiquilin-1 and ubiquilin-2 isoforms interacted with one another in all their different combinations (Figure 1A). Ubiquilin-1 interacted with ubiquilin-1 and ubiquilin-2 to the same extent, whereas homophilic interaction between ubiquilin-2 was stronger than the homophilic interaction between ubiquilin-1 proteins. Similar results were obtained when the baits and prey were switched (results not shown). These data demonstrate that ubiquilin is capable of self-interaction in yeast.
Figure 1. Ubiquilin-1 and -2 interact with each other in yeast.
Interaction of ubiquilin (Ubqln)-1 and -2 isoforms in yeast: demonstration that the central region and not the UBL or UBA domains are responsible for the interaction. (A) Yeast co-transformed with ubiquilin-1 or -2 as prey and ubiquilin-1 or -2 as bait were assayed using a β-galactosidase liquid assay to test for interaction. All experimental values are normalized to yeast that were co-transformed with the preys together with lamin in bait vector (pEG202). Note that ubiquilin-1 interacts with ubiquilin-1 and -2 to the same extent, with a relative intensity of 79 units. Also note, ubiquilin-2 interacts more strongly with ubiquilin-2, with a relative intensity of 129 units. Each assay was performed in triplicate. The interactions marked with ** were found to be statistically significant (P<0.01). (B) Yeast were co-transformed with ubiquilin-1 constructs as indicated as both bait and prey. Ubiquilin-1 constructs include (from left to right): the full-length protein, deletion of the UBL domain, deletion of the UBA domain, deletion of both the UBL and UBA domains, the UBA domain alone, the UBA domain as prey and the UBL domain as bait, and the UBL domain alone. The interactions marked with ** were found to be statistically significant (P<0.01). Compared with the full-length interaction, deletion of the UBL domain does not significantly decrease interaction (P>0.05). Deletion of the UBA domain leads to a slight decrease in interaction, although it does not completely abolish interaction. The UBA domain alone cannot reconstitute the intensity of interaction seen with the full-length protein. Together, these results indicate that the central region of ubiquilin is critical for interaction.
Interaction between ubiquilin isoforms does not require the UBL or the UBA domain
We next investigated which regions of ubiquilin are critical for self-interaction. Ubiquilin has a UBL domain near the N-terminus and a UBA domain at the C-terminus. As the UBL domain is structurally similar to ubiquitin, and as UBA domains have been shown to bind ubiquitin moieties, we examined whether self-interaction of ubiquilin occurs through interaction of its UBL and UBA domains. Again using β-galactosidase liquid assays, we measured the interaction of ubiquilin-1 constructs lacking either the UBL domain or the UBA domain, or both the UBL and the UBA domains, when expressed as both bait and prey in yeast (Figure 1B). We found that deletion of the UBL domain did not decrease homophilic interaction of ubiquilin-1 relative to the full-length protein. Deletion of the UBA produced a slight decrease in self-interaction as compared with the full-length protein. However, neither deletion of the UBA domain or the UBL domain alone, nor simultaneous deletion of both the UBL and UBA domains, was sufficient to abrogate ubiquilin-1 self-interaction (Figure 1B).
To further investigate whether the UBL and UBA domains of ubiquilin-1 are capable of self-interaction, we used the domains alone as both bait and prey. We found that the UBL domain neither self-interacted nor did it interact with the UBA domain (Figure 1B). We did observe some slight interaction between the UBA domains; however, this interaction was only one tenth of the strength observed with the full-length protein (Figure 1B). We conclude that, although the UBA domain may contribute to self-interaction of ubiquilin, it is the central region of ubiquilin that is critical for this interaction.
Ubiquilin isoforms oligomerize in HeLa cells
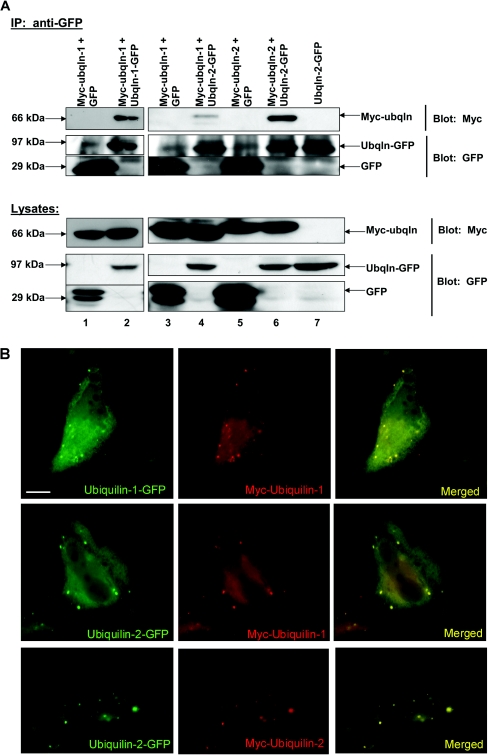
The yeast two-hybrid assays revealed that ubiquilin isoforms interact with each other in yeast. We next examined whether ubiquilin isoforms can bind one another in mammalian cells by co-immunoprecipitation assays. To do so, we co-transfected HeLa cells with Myc- and GFP-tagged ubiquilin constructs. Myc–ubiquilin was co-transfected with GFP alone as a negative control for these experiments. After transfection (20 h) the cells were lysed and rabbit anti-GFP antibody was added to the lysates to immunoprecipitate GFPs. Immunoblot analysis of the immunoprecipitates revealed that the Myc–ubiquilin proteins co-immunoprecipitate when co-expressed with ubiquilin–GFP proteins, but not when co-expressed with GFP alone (Figure 2A). Myc–ubiquilin-1 was co-immunoprecipitated with ubiquilin-1–GFP or with ubiquilin-2–GFP (Figure 2A). The co-immunoprecipitation of ubiquilin proteins is unlikely to be an artifact of tagging of the proteins with GFP, because we observed similar co-immunoprecipitation of HA- and Myc-tagged ubiquilin proteins (results not shown). Likewise Myc–ubiquilin-2 was co-immunoprecipitated with ubiquilin-1–GFP (results not shown) and with ubiquilin-2–GFP (Figure 2A). Interestingly, although the proteins were expressed at similar levels, more Myc–ubiquilin-2 was co-immunoprecipitated with ubiquilin-2–GFP than with ubiquilin-1–GFP. We speculate that this is because of stronger interactions of ubiquilin-2 proteins, as found in yeast two-hybrid assays. We next used immunofluorescence microscopy to determine whether ubiquilin-1 and ubiquilin-2 proteins co-localize in cells. As shown in Figure 2(B), cells co-transfected with Myc- and GFP-tagged ubiquilin constructs revealed excellent co-localization of the two ubiquilin-tagged proteins, in all possible combinations, strongly suggesting that ubiquilin-1 and ubiquilin-2 proteins interact with each other in cells. These results confirm the yeast two-hybrid data and demonstrate that ubiquilin can form oligomers in mammalian cells.
Figure 2. Ubiquilin-1 and -2 oligomerize in HeLa cells.
(A) HeLa cells were co-transfected with Myc–ubiquilin (Ubqln) and ubiquilin–GFP or Myc–ubiquilin and GFP. The GFP tag was immunoprecipitated from cell lysates using rabbit anti-GFP and Protein A–Sepharose beads. The immunoprecipitates and cell lysates were separated by SDS/PAGE. Immunoblotting for the Myc tag and the GFP tag reveals that Myc–ubiquilin and ubiquilin–GFP or GFP were expressed as expected (lysates). The immunoprecipitate (IP) lanes confirm that proteins containing the GFP tag were indeed immunoprecipitated (IP: anti-GFP, bottom two panels). There is a non-specific band that co-migrates around the same molecular mass as ubiquilin–GFP; however, the presence of ubiquilin–GFP in the appropriate lanes is easily distinguished by the much greater band intensity as compared with the negative control lanes. Lanes 1, 3 and 5: negative controls where Myc–ubiquilin does not co-immunoprecipitate with GFP as expected. Lanes 2, 4 and 6: ubiquilin isoforms interact and co-immunoprecipitate from HeLa cell lysates, confirming the yeast two-hybrid data. Lane 7: note that cells transfected with ubiquilin–GFP alone still show immunoprecipitation of ubiquilin–GFP from cell lysates, and this ubiquilin–GFP protein is not cross-reactive with the anti-Myc antibody. (B) HeLa cells grown on coverslips were co-transfected with plasmid DNA encoding Myc–ubiquilin-1 and ubiquilin-1–GFP, or with Myc–ubiquilin-1 and ubiquilin-2–GFP, or Myc–ubiquilin-2 and ubiquilin-2–GFP. The cells were stained for Myc (red) or GFP (green), and the result of merging the two images is shown on the right-hand side. Note the extensive co-localization of the two tagged ubiquilin proteins in the cells. Scale bar, 5 μm.
UBA domain of ubiquilin is not required for oligomerization in HeLa cells
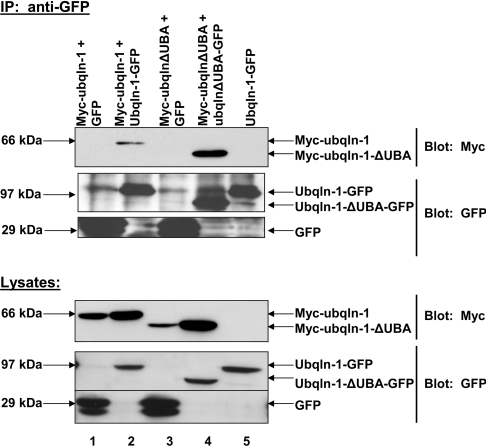
We next confirmed that deletion of the UBA domain did not abolish ubiquilin oligomerization in HeLa cells. HeLa cells were co-transfected with plasmids encoding Myc–ubiquilin-1(ΔUBA) and ubiquilin-1(ΔUBA)–GFP or Myc–ubiquilin-1(ΔUBA) and GFP alone. Immunoprecipitation from cell lysates was performed as described above. In agreement with yeast two-hybrid data, deletion of the UBA domain did not abolish ubiquilin oligomerization (Figure 3). It is interesting to note that oligomerization of the ubiquilin-1(ΔUBA) protein appears to be greater than oligomerization of the full-length ubiquilin protein. Similar experiments with the ubiquilin-1(ΔUBL) and ubiquilin-1(ΔUBL/ΔUBA) constructs also confirmed that deletion of these domains does not abolish ubiquilin oligomerization (results not shown).
Figure 3. Ubiquilin-1(ΔUBA) proteins oligomerize in HeLa cells.
HeLa cells were co-transfected with Myc–ubiquilin-1(ΔUBA) and ubiquilin-1(ΔUBA) –GFP or Myc–ubiquilin-1-(ΔUBA) and GFP. Myc–ubiquilin-1 and ubiquilin-1–GFP or GFP were included as a positive control. The GFP tag was immunoprecipitated as described in the Experimental section. Probing the lysates with anti-Myc and anti-GFP confirmed that the proteins were expressed as expected. The immunoprecipitate lanes (IP: anti-GFP section, lanes 2 and 4) show that the GFP tags were immunoprecipitated (lower two panels). The upper panel of the immunoprecipitation reactions probed with anti-GFP shows that Myc–ubiquilin-1 co-immunoprecipitates with ubiquilin-1–GFP, but not with GFP alone. Also, Myc–ubiquilin-1(ΔUBA) co-immunoprecipitates with ubiquilin-1(ΔUBA)–GFP, but not GFP alone, further supporting the data from the yeast two-hybrid analysis. Lane 5 confirms that ubiquilin-1–GFP does not cross-react with the anti-Myc antibody. Ubqln, ubiquilin.
Ubiquilin forms dimers in vitro
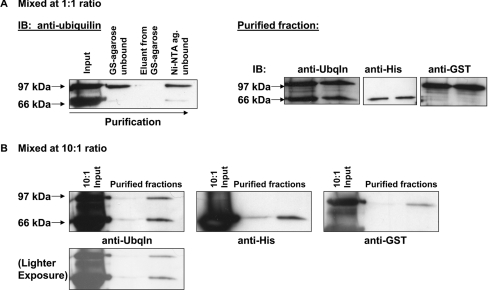
Having shown that ubiquilin proteins can self-interact in yeast and form oligomers in mammalian cells, we next investigated the stoichiometry of these oligomers. Specifically, we wanted to know whether ubiquilin forms dimers or a higher-order oligomer. To do this, we mixed purified GST–ubiquilin-1 and purified ubiquilin-1–His6 proteins together in various ratios (Figure 4). After an incubation period, the proteins were first passed over a column containing glutathione–agarose beads. The bound proteins were then eluted and next passed over a Ni-NTA column. The bound proteins were again eluted. This final protein fraction, which binds to both the glutathione–agarose and the Ni-NTA columns, should only contain complexes comprising both GST–ubiquilin-1 and ubiquilin-1–His6 proteins. This fraction was separated by SDS/PAGE and then transferred on to nitrocellulose. Probing with anti-ubiquilin antibody reveals that the bands corresponding to GST- and His-tagged ubiquilin are at a 1:1 ratio, regardless of the initial ratio (1:1 or 10:1) at which they were mixed. Separate Western blots confirm that the 97 kDa band corresponds to GST–ubiquilin-1 and the 66 kDa band corresponds to ubiquilin-1–His6 (Figure 4). These data demonstrate that ubiquilin forms dimers in vitro.
Figure 4. Ubiquilin-1 forms dimers in vitro.
(A) Purified proteins ubiquilin-1–His6 and GST–ubiquilin-1 were mixed together in vitro in a 1:1 ratio. The protein mixture was first added to a column with glutathione–agarose beads. The unbound fraction was collected, the beads were then washed, and finally the proteins were eluted using 10 mM GSH. This eluted protein mixture was then added to a column with Ni-NTA beads. Again, the unbound fraction was collected, the beads washed and the proteins eluted using 250 mM imidazole buffer. Fractions were separated by SDS/PAGE. Immunoblotting with anti-ubiquilin shows the proteins present in each fraction of the purification process. Ubiquilin-1–His6 has a molecular mass of 66 kDa and GST–ubiquilin-1 of 97 kDa. Note that the final ratio of ubiquilin-1–His6 to GST–ubiquilin-1 is 1:1 (two lanes containing the purified fractions are shown). Anti-His and anti-GST blotting confirmed that the 66 kDa and 97 kDa bands correspond to ubiquilin-1–His6 and GST–ubiquilin-1 respectively. (B) Purified proteins ubiquilin-1–His6 and GST–ubiquilin-1 were mixed together in vitro in a 10:1 ratio. The proteins were passed through glutathione–agarose and Ni-NTA columns as described for (A). Note that even though the two different tagged-proteins were mixed together in a 10:1 ratio, they were recovered after purification over the two affinity columns in a 1:1 ratio. Ubqln, ubiquilin.
Monomeric ubiquilin interacts with PS in vitro
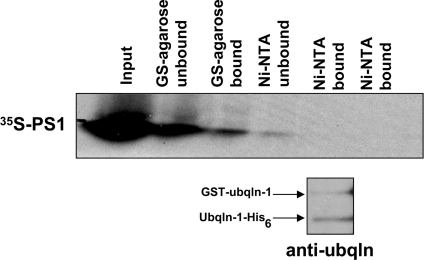
In order to determine whether it is the monomeric form of ubiquilin or the dimer that interacts with another protein, such as PS [1], we mixed the purified proteins described above together with [35S]methionine-labelled PS1. Similar to the in vitro dimerization experiment, we incubated the proteins together before passing over successive columns for each tag. The protein mixture was first passed over a column containing glutathione–agarose beads. The bound proteins were next eluted and passed over a column containing Ni-NTA beads. Finally, the bound proteins were eluted from the Ni-NTA column. Unbound fractions were collected at each step prior to eluting the bound fractions, which were passed over the subsequent column. Fractions from each step were separated by SDS/PAGE. Duplicate gels were run for each experiment, one of which was dried and used to obtain an autoradiogram, whereas the other was used to transfer proteins on to nitrocellulose prior to probing for ubiquilin.
If the dimeric form of ubiquilin is responsible for binding PS1, a known ubiquilin interactor, then we expected to observe binding of PS1 protein to both the glutathione–agarose and Ni-NTA columns. Instead, however, we found that PS1 was only present after binding the glutathione–agarose column, but not after passing over the subsequent Ni-NTA column (Figure 5, upper panel). Immunoblotting with anti-ubiquilin revealed that GST–ubiquilin-1 and ubiquilin-1–His6 are both present after passing over the two columns (Figure 5, lower panel). Reversing the order of the columns confirmed that PS1 is unable to bind simultaneously with both GST–ubiquilin-1 and ubiquilin-1–His6 (results not shown). This indicates that the ubiquilin dimer does not interact with PS1, but rather it is the monomeric form that interacts with this protein. Similar results were obtained using PS2 (results not shown). Note that [35S]methionine-labelled full-length PS1 protein is more easily distinguishable than PS2 (see [1]), and it is for this reason it is the only result shown. Interestingly, immunoblot analyses of proteins that were pulled down from the in vitro-translated PS1 or PS2 reactions using GST–ubiquilin-1 revealed strong anti-ubiquitin reactivity, suggesting that ubiquilin most likely binds poly-ubiquitinated PS proteins (results not shown). This result leads us to speculate that ubiquilin most likely binds polyubiquitinated PS proteins via its UBA domain, because the UBA domain can bind polyubiquitin chains [18] and because we found it was necessary and sufficient for binding PS proteins [1].
Figure 5. Monomeric ubiquilin-1 binds PS1 and PS2 in vitro.
Purified proteins GST–ubiquilin-1 and ubiquilin-1–His6 were mixed together in vitro along with [35S]methionine-labelled PS1 or PS2. The protein mixture was sequentially passed over glutathione–agarose and Ni-NTA columns. Unbound protein was collected from each column prior to washing with the appropriate column wash buffer. The eluted (bound) protein mixture from the glutathione–agarose column was subsequently added to the Ni-NTA column. The bound protein mixture was eluted from each column using 10 mM GSH or 250 mM imidazole buffer. Protein fractions were separated by SDS/PAGE. Autoradiography revealed that PS1 binds to the glutathione–agarose column, but not the subsequent Ni-NTA column. Immunoblotting the [35S]methionine-labelled PS1 blot for ubiquilin revealed that GST–ubiquilin-1 and ubiquilin-1–His6 both remain after passing through the two columns. Ubqln, ubiquilin.
Increased expression of PS2 disrupts ubiquilin dimerization in vivo
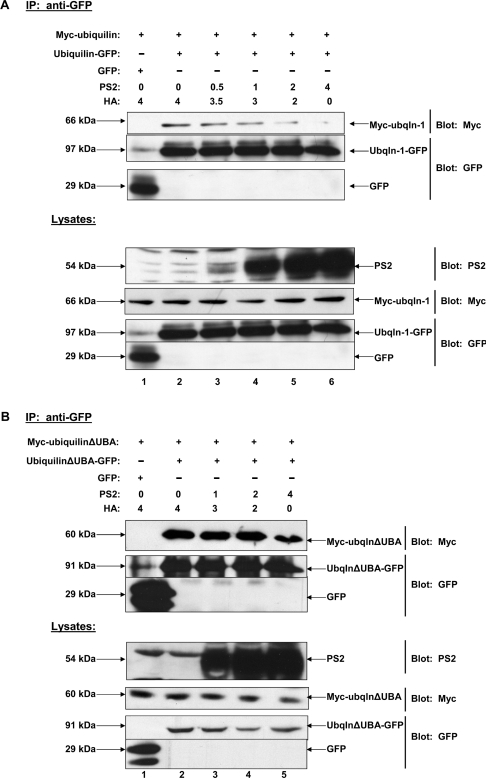
We next sought to determine whether it is the monomeric form of ubiquilin or the dimer that interacts with PS in vivo. We previously demonstrated that ubiquilin interacts strongly with PS2 both in vitro and in vivo [1]. We therefore examined the effects of increasing PS2 levels on ubiquilin dimerization. Therefore HEK-293 cells were co-transfected with equal amounts of cDNAs coding for Myc–ubiquilin-1, ubiquilin-1–GFP or GFP alone, and increasing amounts of PS2. GFPs were then immunoprecipitated from the transfected cell lysates and immunoblotted for the Myc tag and subsequently for the GFP tag. In parallel, the total cell lysates were probed for Myc, GFP and PS2. We found that, as increasing amounts of PS2 were co-transfected, there was a dose-dependent decrease in the amount of ubiquilin that dimerized (Figure 6A). This effect was not due to differences in expression of ubiquilin proteins, as approximately equal levels of Myc–ubiquilin and ubiquilin–GFP proteins were expressed regardless of the levels of PS2 expression. These findings suggest that it is the monomeric form of ubiquilin that interacts with PS2.
Figure 6. Over-expression of PS2 leads to dose-dependent decrease in dimerization of full-length ubiquilin-1 in HeLa cells but not of ubiquilin-1(ΔUBA).
(A) HeLa cells were co-transfected with plasmids encoding Myc–ubiquilin-1, ubiquilin-1–GFP or GFP alone, and increasing amounts of PS2 cDNA. Various amounts of pCMV-HA cDNA were co-transfected in order to keep the total amount of DNA transfected equal. Co-immunoprecipitations were performed as described in the text. Note that with increasing PS2 expression there is a dose-dependent decrease in ubiquilin dimerization. Immunoblots of the same lysates confirmed that Myc–ubiquilin-1 and ubiquilin-1–GFP expression levels were expressed at approximately the same level in each sample. (B) HeLa cells were co-transfected with plasmids encoding Myc–ubiquilin-1(ΔUBA), ubiquilin-1(ΔUBA)–GFP or GFP alone, and increasing amounts of PS2 cDNA. Immunoprecipitations were performed as described in (A). Note that deletion of the UBA domain of ubiquilin results in a failure of increased PS2 expression to disrupt ubiquilin dimerization. Ubqln, ubiquilin.
We next examined whether the PS2-induced disruption of ubiquilin dimerization would still occur if the UBA domain of ubiquilin was deleted. The reason we examined this possibility was that our yeast two-hybrid data suggested the UBA domain does not appear to be involved in ubiquilin dimerization, whereas according to our in vitro binding assays the domain was necessary and sufficient for binding PS [1]. Therefore we predicted that deletion of the PS-binding site in ubiquilin (i.e. deletion of its UBA domain) should result in a protein that might not dissociate into the monomer upon overexpression of PS2. Thus, we repeated the same experiment shown in Figure 6(A), but this time transfected the cells with Myc–ubiquilin-1(ΔUBA) and ubiquilin-1(ΔUBA)–GFP and increasing amounts of PS2. In agreement with our prediction, increased expression of PS2 had little, to no, effect on the disruption of ubiquilin-1(ΔUBA) dimerization (Figure 6B). This result suggests that interaction of the UBA domain of ubiquilin with PS may be necessary for the disruption of ubiquilin dimerization.
Overexpression of ODC does not disrupt ubiquilin dimerization in vivo
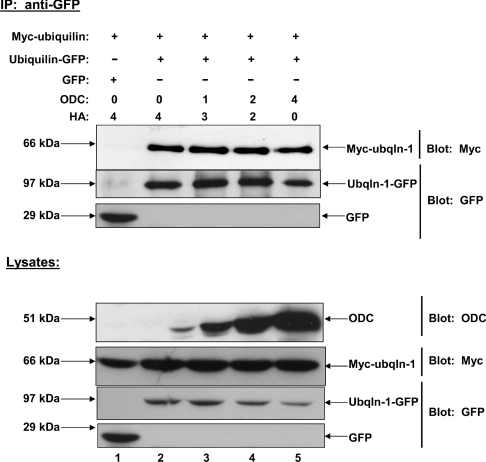
To establish that the dose-dependent decrease in ubiquilin dimerization induced by over-expression of PS2 was due to increased binding of PS2 to ubiquilin, we repeated the experiment this time using ODC instead of PS2. We hypothesized that ODC would not interact with ubiquilin, because, unlike PS2, and other ubiquilin interactors, ODC is degraded by the proteasome in a ubiquitin-independent manner [32–34]. For this experiment, HEK-293 cells were co-transfected with equal amounts of cDNAs coding for Myc–ubiquilin, ubiquilin–GFP or GFP, and increasing amounts of ODC. Due to the short half-life of ODC, cells were treated with MG132 for 8 h prior to collecting lysates. Co-immunoprecipitations were performed as described above. We found that increasing amounts of ODC had no effect on the amount of ubiquilin that dimerized (Figure 7). These findings further support the idea that the ubiquilin monomer is the active form of the protein.
Figure 7. Over-expression of the ubiquilin non-interactor ODC does not disrupt dimerization of ubiquilin-1 in HeLa cells.
HeLa cells were co-transfected with Myc–ubiquilin-1, ubiquilin-1–GFP or GFP, and increasing amounts of HA–ODC cDNA. Varying amounts of pCMV-HA cDNA were co-transfected in order to keep the total amount of DNA transfected equal. Co-immunoprecipitations were carried out as previously described. Increasing the amount of ODC does not induce a decrease in ubiquilin dimerization, in contrast with the dose-dependent decrease in ubiquilin dimerization induced by PS2. Immunoblots of lysates confirmed that Myc–ubiquilin-1 and ubiquilin-1–GFP expression levels were comparable between samples. Ubqln, ubiquilin.
DISCUSSION
In the present study we have shown that ubiquilin forms dimers, dependent upon the central region of the protein and independent of its UBL and UBA domains. Furthermore, our results strongly suggest that the ubiquilin monomer is the active form of the protein responsible for interacting with PS1 and PS2. These data provide new insight into the regulation of ubiquilin interaction with PS, and will likely be a common theme for ubiquilin interaction with its other binding partners.
Our findings are the first to conclusively and unambiguously demonstrate that ubiquilin forms dimers. Although two other groups have suggested that ubiquilin or the putative yeast ubiquilin homologue, Dsk2, form homodimers [5,35], their data did not reveal the stoichiometry of ubiquilin oligomerization. In our present study, we show conclusively that two different ubiquilin proteins bind to one another in a stoichiometry of 1:1, strongly indicating that ubiquilin forms dimers. However, although ubiquilin clearly forms dimers, this does not preclude the possibility that the dimers then further assemble to form higher-order oligomers.
We have also demonstrated that ubiquilin can form both homodimers and heterodimers, as shown by the interaction of ubiquilin-1 and -2 isoforms in all their possible combinations in cells. Although the physiological role of these combinations is unknown at this time, it is likely that dimerization of the different isoforms may add an additional layer of regulation or complexity with regard to their function. For example, our results suggest ubiquilin-2 homodimers have a stronger interaction than ubiquilin-1 homodimers. This may allow differential activation of ubiquilin-1 versus ubiquilin-2 to the active monomeric form. It is likely that differences in the strength of interaction of ubiquilin -1 and -2 proteins arise from differences in the primary sequence of the two proteins that facilitate their dimerization. One possibility is that the collagen-like domain, which is unique to the central region of ubiquilin-2 [1,5], may contribute to stronger homophilic interaction of ubiquilin-2 proteins. The differences in the strength of self-interaction may provide a mechanism utilized by cells to control the dynamics of interaction of a specific ubiquilin isoforms with a particular target.
It is interesting to note that the ubiquilin-1 construct devoid of the UBA domain dimerizes to a greater extent than the full-length ubiquilin-1 construct, despite being expressed at approximately equal levels. With many of ubiquilin's interactors, the UBA domain of ubiquilin is required and even sufficient to bind the interacting protein [1,8,11–14,18,24]. One possibility for the greater dimerization of the ubiquilin-1(ΔUBA) protein is that deletion of the UBA domain, which normally binds polyubiquitin chains conjugated on to proteins, will lead to a build-up of the unbound ubiquilin-1(ΔUBA) protein, increasing its chance for self-association. The UBA domain in the context of the full-length protein might also obscure the sites involved in ubiquilin self-interaction, suggesting that ubiquilin self-interaction may be both negatively and positively regulated by the structure of the protein.
It has been has reported that the putative yeast homologue of ubiquilin-1, Dsk2, is capable of forming homodimers [36,37]. In these studies, it was found that the UBA domain of Dsk2 is required and sufficient for homodimerization of Dsk2. This absolute dependency on the UBA domain for dimerization is contrary to what we observe in the present study. However, yeast Dsk2 and human ubiquilin-1 share weak homology, with only 19% sequence identity between the two proteins. Thus it is likely that human ubiquilin and yeast Dsk2 would have different biochemical properties, and possibly different functional properties. Furthermore, since the homology between Dsk2 and ubiquilin-1 is primarily confined to the UBL and the UBA domains, and we found that human ubiquilin-1 dimerization is dependent upon the central region of the protein, it is possible that dimerization of the two proteins occurs via different mechanisms. With regard to ubiquilin, it is interesting to note that variants within the central region of ubiquilin-1 have been found to be genetically associated with late-onset Alzheimer's disease [38,39], although others have failed to find such an association [40,41]. The putative genetic association of the ubiquilin-1 gene with Alzheimer's disease suggests that the central domain of the ubiquilin protein might play some function in human disease, and raise the question of whether the variants in this region affect ubiquilin function via alterations in the dimerization properties of the proteins.
Another group has reported an interaction between ubiquilin-1 and K7 protein, a small membrane protein encoded by Kaposi's sarcoma-associated herpes virus that is involved in protecting cells from apoptosis [14]. They found that an increase in K7 protein levels leads to a disruption of ubiquilin-1 dimerization, which is analogous to our results showing that increased PS2 levels induce a dose-dependent decrease in ubiquilin dimerization. The disruption of ubiquilin dimerization appears to be dependent on the interaction of ubiquilin with its targets, because we found that increased expression of ODC, a ubiquilin-non-interacting protein, does not affect ubiquilin dimerization. Coupled with our in vitro data directly showing that it is the monomeric form of ubiquilin that binds PS1 and PS2, we conclude that the monomeric form of ubiquilin is responsible for binding PS and that the ubiquilin dimer probably cannot bind PS. Together, these data suggest an emerging trend for the regulation of ubiquilin interaction with other proteins in which the monomeric form of ubiquilin is likely responsible for interacting with the protein. Further studies on additional ubiquilin interactors will be necessary to determine whether monomeric ubiquilin is the only form that is able to bind its target proteins, or if there are any circumstances where the dimeric ubiquilin form might also be able to interact with the proteins. At this point, the data suggest that the dimeric ubiquilin form may serve more in a regulatory role to prevent interaction with other proteins.
One question that inevitably arises from the present study is: what is the trigger for conversion of the ubiquilin dimer to the active monomeric form? Our in vitro studies, in which we mixed two differently tagged species of ubiquilin-1 together and then tested for dimerization, indicate that the proteins in vitro may be dynamic and able to dissociate from the homodimer form and re-associate into a heterodimer. Alternatively, it is possible that the in vitro protein consists of both monomers and homodimers, and upon mixing the differently tagged proteins, some of the monomers associate to form heterodimers. It is unclear at this point whether ubiquilin dimers in vivo can readily associate/dissociate, or if additional factors exist within the cell milieu to regulate the interconversion between monomer and dimer. One possibility is that a build-up of polyubiquitinated proteins in cells with which ubiquilin can interact could trigger the dissociation of the ubiquilin dimer to the monomer. In support of this idea, we found that deletion of the UBA domain of ubiquilin, which is required for binding PS2, results in the failure of PS to induce the dose-dependent conversion of ubiquilin from the dimer to monomer.
Another possibility is that ubiquilin might be subject to some transient modification that triggers the conversion between the monomer and the dimer. One such modification might be phosphorylation, as ubiquilin is a known phosphoprotein [3]. However, the role of phosphorylation of ubiquilin is not known. In our experiments, we used recombinant bacterially expressed ubiquilin proteins, which are unlikely to be phosphorylated, to demonstrate that ubiquilin proteins dimerize. Further experiments are needed to determine what enzyme(s) are responsible for phosphorylation of ubiquilin and how this modification affects ubiquilin function. Future studies will focus on the role of the ubiquilin dimer and monomer, and the dynamics of ubiquilin interaction with its targets.
Acknowledgments
We thank Dr Philip Coffino for kindly providing the ODC cDNA. We thank the anonymous referee for suggesting the experiment shown in Figure 6(B). This work was supported mainly by NIH (National Institutes of Health) grant (GM066287) and in part by NIH grant (AG016839) to M.J.M.D.L.F. is supported by a Training Grant in Integrative Membrane Biology (T32 GM08181; R. Bloch, Principal Investigator).
References
- 1.Mah A. L., Perry G., Smith M. A., Monteiro M. J. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J. Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conklin D., Holderman S., Whitmore T. E., Maurer M., Feldhaus A. L. Molecular cloning, chromosome mapping and characterization of UBQLN3 a testis-specific gene that contains an ubiquitin-like domain. Gene. 2000;249:91–98. doi: 10.1016/s0378-1119(00)00122-0. [DOI] [PubMed] [Google Scholar]
- 3.Wu S., Mikhailov A., Kallo-Hosein H., Hara K., Yonezawa K., Avruch J. Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim. Biophys. Acta. 2002;1542:41–56. doi: 10.1016/s0167-4889(01)00164-1. [DOI] [PubMed] [Google Scholar]
- 4.Funakoshi M., Geley S., Hunt T., Nishimoto T., Kobayashi H. Identification of XDRP1; a Xenopus protein related to yeast Dsk2p binds to the N-terminus of cyclin A and inhibits its degradation. EMBO J. 1999;18:5009–5018. doi: 10.1093/emboj/18.18.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleijnen M. F., Shih A. H., Zhou P., Kumar S., Soccio R. E., Kedersha N. L., Gill G., Howley P. M. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 6.Davidson J. D., Riley B., Burright E. N., Duvick L. A., Zoghbi H. Y., Orr H. T. Identification and characterization of an ataxin-1-interacting protein: A1Up, a ubiquitin-like nuclear protein. Hum. Mol. Genet. 2000;9:2305–2312. doi: 10.1093/oxfordjournals.hmg.a018922. [DOI] [PubMed] [Google Scholar]
- 7.Kaye F. J., Modi S., Ivanovska I., Koonin E. V., Thress K., Kubo A., Kornbluth S., Rose M. D. A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett. 2000;467:348–355. doi: 10.1016/s0014-5793(00)01135-2. [DOI] [PubMed] [Google Scholar]
- 8.Bedford F. K., Kittler J. T., Muller E., Thomas P., Uren J. M., Merlo D., Wisden W., Triller A., Smart T. G., Moss S. J. GABAA receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat. Neurosci. 2001;4:908–916. doi: 10.1038/nn0901-908. [DOI] [PubMed] [Google Scholar]
- 9.Ko H. S., Uehara T., Nomura Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J. Biol. Chem. 2002;277:35386–35392. doi: 10.1074/jbc.M203412200. [DOI] [PubMed] [Google Scholar]
- 10.N'Diaye E. N., Brown E. J. The ubiquitin-related protein PLIC-1 regulates heterotrimeric G protein function through association with Gβγ. J. Cell Biol. 2003;163:1157–1165. doi: 10.1083/jcb.200307155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao L., Tu H., Shi S. T., Lee K. J., Asanaka M., Hwang S. B., Lai M. M. Interaction with a ubiquitin-like protein enhances the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2003;77:4149–4159. doi: 10.1128/JVI.77.7.4149-4159.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko H. S., Uehara T., Tsuruma K., Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 13.Doi H., Mitsui K., Kurosawa M., Machida Y., Kuroiwa Y., Nukina N. Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett. 2004;571:171–176. doi: 10.1016/j.febslet.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 14.Feng P., Scott C. W., Cho N. H., Nakamura H., Chung Y. H., Monteiro M. J., Jung J. U. Kaposi's sarcoma-associated herpesvirus K7 protein targets a ubiquitin-like/ubiquitin-associated domain-containing protein to promote protein degradation. Mol. Cell. Biol. 2004;24:3938–3948. doi: 10.1128/MCB.24.9.3938-3948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ficklin M. B., Zhao S., Feng G. Ubiquilin-1 regulates nicotine-induced up-regulation of neuronal nicotinic acetylcholine receptors. J. Biol. Chem. 2005;280:34088–34095. doi: 10.1074/jbc.M506781200. [DOI] [PubMed] [Google Scholar]
- 16.Regan-Klapisz E., Sorokina I., Voortman J., de Keizer P., Roovers R. C., Verheesen P., Urbe S., Fallon L., Fon E. A., Verkleij A., et al. Ubiquilin recruits Eps15 into ubiquitin-rich cytoplasmic aggregates via a UIM–UBL interaction. J. Cell Sci. 2005;118:4437–4450. doi: 10.1242/jcs.02571. [DOI] [PubMed] [Google Scholar]
- 17.Riley B. E., Xu Y., Zoghbi H. Y., Orr H. T. The effects of the polyglutamine repeat protein ataxin-1 on the UbL-UBA protein A1Up. J. Biol. Chem. 2004;279:42290–42301. doi: 10.1074/jbc.M406284200. [DOI] [PubMed] [Google Scholar]
- 18.Massey L. K., Mah A. L., Ford D. L., Miller J., Liang J., Doong H., Monteiro M. J. Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J. Alzheimers Dis. 2004;6:79–92. doi: 10.3233/jad-2004-6109. [DOI] [PubMed] [Google Scholar]
- 19.Chen L., Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medicherla B., Kostova Z., Schaefer A., Wolf D. H. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raasi S., Pickart C. M. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 22.Ryu K. S., Lee K. J., Bae S. H., Kim B. K., Kim K. A., Choi B. S. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J. Biol. Chem. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- 23.Wang H., Lim P. J., Yin C., Rieckher M., Vogel B. E., Monteiro M. J. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum. Mol. Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 24.Kleijnen M. F., Alarcon R. M., Howley P. M. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol. Biol. Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma R., Oania R., Graumann J., Deshaies R. J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 28.Rao H., Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds A., Lundblad V. Current Protocols in Molecular Biology. In: Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. New York: John Wiley & Sons; 1989. pp. 13.6.1–13.6.4.. [Google Scholar]
- 30.Stabler S. M., Ostrowski L. L., Janicki S. M., Monteiro M. J. A myristoylated calcium-binding protein that preferentially interacts with the Alzheimer's disease presenilin 2 protein. J. Cell Biol. 1999;145:1277–1292. doi: 10.1083/jcb.145.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janicki S., Monteiro M. J. Increased apoptosis arising from increased expression of the Alzheimer's disease-associated presenilin-2 mutation (N141I) J. Cell Biol. 1997;139:485–495. doi: 10.1083/jcb.139.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol. Cell. Biol. 1992;12:3556–3562. doi: 10.1128/mcb.12.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoyt M. A., Zhang M., Coffino P. Ubiquitin-independent mechanisms of mouse ornithine decarboxylase degradation are conserved between mammalian and fungal cells. J. Biol. Chem. 2003;278:12135–12143. doi: 10.1074/jbc.M211802200. [DOI] [PubMed] [Google Scholar]
- 34.Elias S., Bercovich B., Kahana C., Coffino P., Fischer M., Hilt W., Wolf D. H., Ciechanover A. Degradation of ornithine decarboxylase by the mammalian and yeast 26 S proteasome complexes requires all the components of the protease. Eur. J. Biochem. 1995;229:276–283. [PubMed] [Google Scholar]
- 35.Wu A. L., Wang J., Zheleznyak A., Brown E. J. Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol. Cell. 1999;4:619–625. doi: 10.1016/s1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki T., Funakoshi M., Endicott J. A., Kobayashi H. Budding yeast Dsk2 protein forms a homodimer via its C-terminal UBA domain. Biochem. Biophys. Res. Commun. 2005;336:530–535. doi: 10.1016/j.bbrc.2005.08.126. [DOI] [PubMed] [Google Scholar]
- 37.Lowe E. D., Hasan N., Trempe J. F., Fonso L., Noble M. E., Endicott J. A., Johnson L. N., Brown N. R. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr. D Biol. Crystallogr. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- 38.Bertram L., Hiltunen M., Parkinson M., Ingelsson M., Lange C., Ramasamy K., Mullin K., Menon R., Sampson A. J., Hsiao M. Y., et al. Family-based association between Alzheimer's disease and variants in UBQLN1. N. Engl. J. Med. 2005;352:884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 39.Kamboh M. I., Minster R. L., Feingold E., DeKosky S. T. Genetic association of ubiquilin with Alzheimer's disease and related quantitative measures. Mol. Psychiatry. 2006;11:273–279. doi: 10.1038/sj.mp.4001775. [DOI] [PubMed] [Google Scholar]
- 40.Smemo S., Nowotny P., Hinrichs A. L., Kauwe J. S., Cherny S., Erickson K., Myers A. J., Kaleem M., Marlowe L., Gibson A. M., et al. Ubiquilin 1 polymorphisms are not associated with late-onset Alzheimer's disease. Ann. Neurol. 2006;59:21–26. doi: 10.1002/ana.20673. [DOI] [PubMed] [Google Scholar]
- 41.Slifer M. A., Martin E. R., Bronson P. G., Browning-Large C., Doraiswamy P. M., Welsh-Bohmer K. A., Gilbert J. R., Haines J. L., Pericak-Vance M. A. Lack of association between UBQLN1 and Alzheimer disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141:208–213. doi: 10.1002/ajmg.b.30298. [DOI] [PubMed] [Google Scholar]