Abstract
We describe the first virus-like particle of a hyperthermophilic euryarchaeote which was discovered in a strain of “Pyrococcus abyssi” previously characterized in our laboratory. This particle, named PAV1, is lemon-shaped (120 nm × 80 nm), with a short tail terminated by fibers, and resembles the virus SSV1, the type member of the Fuselloviridae, isolated from Sulfolobus shibatae. Sensitivity of the virus-like particle to organic solvents and detergents suggested that the envelope of PAV1 may contain lipids in addition to proteins. It contains a double-stranded circular DNA of 18 kb which is also present in high copy number in a free form in the host cytoplasm. No integrated form of the PAV1 genome could be detected in the host chromosome. Under standard growth conditions, the host cells continuously release PAV1 particles into the culture supernatant without spontaneous lysis, with a maximum reached in the late stationary phase. UV, gamma irradiation, treatment with mitomycin C, and various physiological stresses had no effect on PAV1 production. Screening of a large number of Thermococcales isolates did not permit to find a sensitive host. These results suggest that PAV1 persists in the host strain in a stable carrier state rather than a prophage.
The Archaea domain comprises two major phyla, namely, the Crenarchaeota, including the extremely thermophilic sulfur-metabolizing Archaea of the orders Sulfolobales and Thermoproteales, and the Euryarchaeota, containing mainly the methanogens, the extreme halophiles, and the hyperthermophilic order Thermococcales (29). Our knowledge about archaeal viruses is still rather limited, and among known archaeal viruses that have been reported, only few have been studied in detail at the molecular level.
All known crenarchaeotal viruses display unusual morphotypes and compose three novel families which were created to account for their unique features, namely, the filamentous Lipothrixviridae (2, 13, 32), the lemon-shaped Fuselloviridae (18, 25), and the rod-shaped Rudiviridae (20). The droplet-shaped Guttaviridae (3) have not yet been established as an acknowledged virus family. The best studied virus is the lemon-shaped SSV1, whose original host is the hyperthermophile Sulfolobus shibatae. SSV1 is temperate and forms stable lysogens by site specifically inserting its 15.5-kb circular genome into the host chromosome (31). Its complete nucleotide sequence has been determined (19). In contrast, all but two of the as-yet-described viruses of extreme halophiles and methanogens have the classical head-and-tail morphology typical of many bacterial phages and have therefore been assigned to the virus families Myoviridae or Siphoviridae (1). There are two known exceptions, both showing a lemon-shaped morphology resembling SSV1, the type member of the Fuselloviridae. The first one was described as a virus-like particle (VLP) isolated from M. voltae strain A3 containing a circular double-stranded DNA of 23 kb, of which an integrated copy was found in the host chromosome (30). The second exception is the lytic virus His1, which possesses a linear double-stranded DNA of 14.9 kb and infects the extreme halophile Haloarcula hispanica (4).
In the lowest branch of the Euryarchaeota, the Thermococcales, only a few plasmids have been reported from the genera Thermococcus and Pyrococcus; two of them have been studied in detail (11, 14, 27), but no virus is known so far.
Here, we report and discuss the isolation and characterization of a novel VLP, named PAV1, which was isolated from “Pyrococcus abyssi” strain GE23, a deep-sea isolate previously described in our laboratory (15).
MATERIALS AND METHODS
Thermococcales strains.
Reference strains, Pyrococcus furiosus DSM 3638, Pyrococcus woesei DSM 3773, Thermococcus stetteri DSM 5262, and Thermococcus celer DSM 2476 were obtained from the Deutsche Sammlung von Mikroorganismen Zellen (DSMZ), Braunschweig-Stokheim, Germany.
Strains GE5, GE9, and GE23 of “P. abyssi” used in this study have been previously described (9, 15). Strain GE5 is the type strain of “P. abyssi,” and strain GE23 is the host of the VLP PAV1.
A total of 180 novel Thermococcales organisms (Thermococcus and Pyrococcus spp.) isolated from deep-sea vents in the North Fiji Basin (site White Lady; 16°59′S, 173°55′W) or in the East Pacific Rise (13°N,104°W) were screened for genetic elements and used as potential hosts for PAV1.
Culture conditions and growth measurements.
Isolates and reference strains were cultured in the medium used by Ravot et al. (22) with slight modifications. The Ravot medium contained (per liter of distilled water) 1 g of NH4Cl, 0.2 g of MgCl2 · 6H2O, 0.1 g of CaCl2 · 2H2O, 0.1 g of KCl, 0.5 g of sodium acetate (anhydrous), 5 g of yeast extract, 5 g of bio-Trypcase, 20 g of NaCl, 3.45 g of piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), and 0.001 g of resazurin. The pH was adjusted to 7, and the medium was sterilized by autoclaving. After the medium cooled down, the following solutions, separately sterilized by autoclaving, were added: 5 ml of a 6% (wt/vol) KH2PO4 solution and 5 ml of 6% (wt/vol) K2HPO4 solution. The medium was dispensed (50 ml) into 100-ml sterile vials and completed by adding 1% (wt/vol) elemental sulfur previously sterilized by steaming at 100°C for 1 h on three successive days. Anaerobiosis was obtained by applying vacuum to the medium and saturating it with N2. Finally, the medium was reduced by adding a sterile solution of Na2S · 9H2O (final concentration, 0.05% [wt/vol]). The medium inoculated to a final concentration of 1% was incubated at 85°C with shaking (150 to 200 rpm).
Microbial growth was monitored by direct cell counting using a phase-contrast microscope and a Thoma counting chamber (0.01 mm depth; Weber, England).
Electron microscopy and counts of VLPs.
For negative staining, a droplet of the virus specimen was spotted onto a carbon-coated copper grid. The specimen was allowed to adsorb to the carbon layer for 2 min, and excess liquid was removed with a piece of filter paper (Whatman). A droplet of an uranyl acetate solution (2%) was then added to the carbon grid for 40 s, and excess liquid was removed again. After the specimen was air dried, it was examined using a Jeol JEM 100 CX II electron microscope. The approximate virus titer was determined by electron microscopy in a manner similar to that described for the virus TTV1 (13). The average number of VLPs, from a known volume of sample extrapolated to 1 ml, was counted on a measured area.
Attempts to establish a plaque assay.
Plaque assays were performed as previously described (10) with slight modifications (W. Zillig, personal communication). The soft layer, 1.5 ml, was prepared with 0.2% (wt/vol) phytagel (Gellan gum; Sigma) in Ravot medium and 75 μl of a suspension of colloidal sulfur (10 g per 100 ml of deionized water) (Riedel-de Haen, distributed by Sigma). Indicator cells (novel isolates, P. furiosus, P. woesei, T. stetteri, and T. celer) were cultured in Ravot medium to a density of 108cells/ml, and 150 μl of such culture was added to the soft layer. Finally, 300 μl of a suspension containing VLP, prepared by using host cell culture supernatants, by filtering host cells through a 0.2-μm-pore-size filter, or by diluting host cells (1/1,000), was also added to the mixture. This mixture was layered over 20 ml of 0.8% phytagel supporting gel. The plates were incubated in homemade anaerobic jars for 48 to 72 h at 85°C.
Attempts to infect a putative sensitive host were also made in liquid culture. Three milliliters of a culture of strain GE23 was mixed into 100 ml of cultures of indicator cells, both harvested in the exponential phase. The mixed cultures were incubated at 85°C for 15 to 20 h. The virus abundance was then checked by electron microscopy.
Purification of VLP from P. abyssi.
From a culture of strain GE23 in Ravot medium, 700 ml was harvested 12 h after inoculation with 108 cells, and cells were removed by low-speed centrifugation (6,000 × g, 20 min, 4°C). The VLP were precipitated from the supernatant in 1 M NaCl with 100 g of PEG 6000/liter overnight at 4°C with gentle stirring. The precipitate was collected by centrifugation (15,200 × g for 1 h 15 min at 4°C), well drained, and resuspended in TE buffer (10 mM Tris-HCl and 1 mM EDTA [pH 8]). A second precipitation with PEG 6000 (10%) and 1 M NaCl was performed for 1 h, and the precipitate was collected by centrifugation and resuspended in TE as described above. Cell debris were pelleted by centrifugation at 2,200 × g for 10 min at 4°C. The supernatant was kept, and the pellet was washed two more times under the same conditions with reduced volumes of TE. The VLP-containing supernatants were pooled and stored 1 night at 4°C. After a centrifugation (3,000 × g, 15 min, 4°C) to remove residual cell debris, the supernatant was concentrated by a centrifugation at 32,500 × g for 1 h 45 min, and the pellet was resuspended in TE. The VLP were purified from this suspension by centrifugation in a CsCl buoyant density gradient (1.298 g/ml) in a Beckman Optima LE-80 K SW55Ti rotor at 220,000 × g for 24 h. Fractions containing nucleic acids were detected at 254 nm and collected using a density gradient fractionator (model 185, ISCO). Fractions were then dialyzed against a large volume of TE buffer overnight at 4°C and concentrated at 18,800 × g for 3 h or directly concentrated after dilution in TE buffer in a Beckman ultracentrifuge at 150,000 × g for 3 h.
VLP sensitivity to an organic solvent, detergents, and proteolytic degradation.
A suspension of purified PAV1 was exposed to 25% (wt/vol) chloroform for 2.5 and 5 min at room temperature with constant agitation (4). PAV1 was also exposed to 0.3% (wt/vol) Triton X-100 for 1 and 3 min at room temperature (2) and to 0.1% (wt/vol) sodium dodecyl sulfate (SDS) for 3 min at 50°C (20). Finally, PAV1 was incubated with proteinase K (1 mg/ml) in buffer (10 mM Tris-HCl, 5 mM EDTA, 0.5% SDS) for 1 h at 56°C.
SDS-polyacrylamide gel electrophoresis.
PAV1 proteins were analyzed on discontinuous 16.5% SDS-polyacrylamide gels using tricine as the trailing ion for the separation of proteins in the range from 1 to 100 kDa, according the method of Schägger and Jagow (24). Gels were silver stained (0.1%).
Induction assays. (i) Mitomycin C.
Different concentrations of mitomycin C (Sigma) (0, 5, or 10 μg/ml) were added to a mid-exponential-phase culture of strain GE23. Incubation was continued, and aliquots (1.5 ml of culture) were collected at 0, 6, 18, and 30 h after addition of the mutagenic agent. A mitomycin C-free culture was used as control.
(ii) UV treatment.
All manipulations were essentially performed in an anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.) under a 93% N2-7% H2 atmosphere. A culture of strain GE23 in late exponential phase was diluted in sulfur-free medium to give a final concentration about 108 cells/ml and placed in plastic petri dishes. UV irradiation (254 nm) was performed with a Spectroline XX-15F/F lamp at a distance of 8 cm from the cell suspension at different exposure times, namely, 0, 5, 10 and 30 s, as previously described by Watrin and Prieur (28). After irradiation, the cell suspension was transferred in vials containing elemental sulfur at 1% (wt/vol). The irradiated cultures were then incubated at 85°C, and aliquots (1.5 ml) were collected at 2, 6, 9, 12, 19, and 24 h. A nonirradiated culture was used as control.
(iii) Oxidative stress.
One set of three vials containing a mid-exponential-phase culture of strain GE23 was opened and exposed to atmospheric air for 15 min and then closed and flushed with nitrogen. The control was an anaerobic culture flushed with nitrogen. After the treatment, each vial was reincubated at 85°C for 18 h. Aliquots were collected at 0, 4, 8, 12, and 18 h after the oxidative stress.
(iv) Hydrostatic pressure.
Cells were exposed to hydrostatic pressures (20 and 30 MPa) in sterile gas-tight glass syringes (Ultrafit; Heinke-Sass-Wolf, GMBH, Tutlingen, Germany) as previously described (16, 17). Syringes containing cultures were first incubated during 6 h at 85°C under atmospheric pressure; then they were transferred into the high-pressure, high-temperature reactor, set at 85°C for 2 h, decompressed, and reincubated at 85°C under atmospheric pressure. Aliquots were collected just before the incubation under hydrostatic pressure and 0, 3, 9, 16, and 28 h after that.
After each treatment, aliquots were collected to estimate the microbial growth and the production of VLPs by counting as described above.
Preparation and analysis of DNA.
Total DNA from the host strain GE23 was prepared essentially as previously described (8). Exponentially growing cells from 25-ml cultures were collected by centrifugation at 4°C at 6,200 × g for 20 min. Cell pellets were gently suspended in 800 μl of TNE buffer (100 mM Tris-HCl, 50 mM NaCl, 50 mM EDTA [pH 8]) and transferred to 1.5-ml microtubes kept on ice. For isolation of total DNA, 100 μl of 10% SDS, 100 μl of 10% lauryl-sarkosyl, and 10 μl of 10-mg/ml DNase-free RNase solution were added and mixed with the 800-μl cell suspension. After 30 min of incubation at room temperature, 50 μl of 20-mg/ml proteinase K was added to the lysate, and the mixture was incubated for 1 h at 56°C. The lysate was extracted with 1 volume of phenol-chloroform (1:1) mixture and then by 1 volume of chloroform. Total DNA was precipitated from the aqueous phase by adding 0.8 volume of isopropanol followed by centrifugation for 15 min at 18,000 × g in a benchtop centrifuge. The DNA pellet was washed in 70% ethanol and was again collected by centrifugation. The air-dried pellet was dissolved in 200 μl of TE buffer.
Viral DNA was also isolated from 50 μl of a purified and concentrated VLP suspension by the same procedure described above.
Covalently closed circular DNA (cccDNA) was isolated from exponentially growing strain GE23 and from VLP by the alkaline lysis method (23) followed by two extractions with 1 volume of phenol-chloroform-isoamylalcohol (25:24:1) and one extraction with 1 volume of chloroform. A physical map of the PAV1 genome was established after agarose gel electrophoresis of restriction fragments obtained with the following enzymes: BamHI, EcoRI, HindIII, PstI, and XbaI (New England Biolabs).
Total DNA and cccDNAs digested with HindIII or PstI and separated by electrophoresis on 0.8% agarose gel were transferred on a positively charged nylon membrane (Hybond N+, Amersham Pharmacia Biotech) by Southern blotting. cccDNA isolated from the strain GE23 was used as a specific probe: after complete digestion with HindIII, the preparation was purified using a QiaexII kit (Qiagen), and the resulting fragments were randomly prime labeled with fluorescein-11-dUTP with an ECF labeling and detection kit (Amersham Pharmacia Biotech). Labeling, hybridization, and detection procedures were performed in accordance with the manufacturer's instructions.
RESULTS
VLP shape and structure.
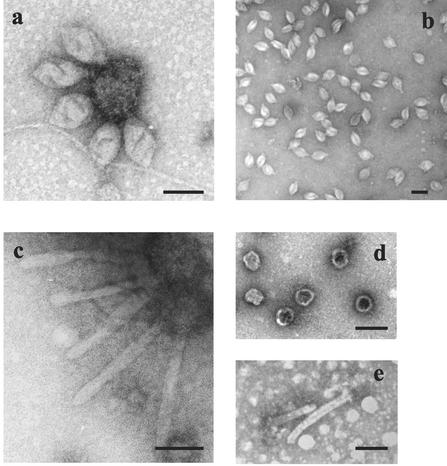
In the course of an extensive screening for extrachromosomal elements in deep-sea Thermococcales (reference 12; C. Geslin, M. Le Romancer, M. Gaillard, G. Erauso, and D. Prieur, unpublished data; and G. Erauso, A. Bideault, A.-C. Mattenet, C. Geslin, and M. Le Romancer, unpublished data), VLPs were discovered in the culture supernatant of the “P. abyssi” strain GE23. These particles, named PAV1 for “P. abyssi” virus 1, are regular lemon-shaped VLPs about 120 nm long and 80 nm wide with a tail approximately 15 nm in length terminated by fibers. Besides free VLP, clusters of VLPs adsorbed to cell-derived vesicles or cell fragments were also observed (Fig. 1a). PAV1 appeared to be similar in morphology to the S. shibatae virus SSV1 (18) of the Fuselloviridae family and also to the H. hispanica virus His1 (4), i.e., lemon-shaped with a short tail. PAV1 had a density of 1.28 g/ml, as determined by equilibrium centrifugation in the CsCl isopycnic gradient used in the process of VLP purification. This method of preparation yielded pure and intact VLP, as confirmed by transmission electron microscopy (TEM) (Fig. 1b). Nevertheless, under certain culture conditions, i.e., in media with an osmolarity higher than in the medium currently used for GE23 growth, VLP appeared more or less elongated and/or flattened, indicating plasticity of form (Fig. 1c).
FIG. 1.
Electron micrographs of PAV1 negatively stained with 2% uranyl acetate. (a) Virus particles attached to cellular material. (b) Purified free virus particles obtained after centrifugation in a CsCl buoyant-density gradient. (c) Elongated virus particles indicating plasticity of form. (d) Effect of exposure to chloroform on virus particle structure. (e) Effect of exposure to proteinase K on virus particle structure. Bars, 100 nm.
The physical stability of the PAV1 structure was assessed by exposing diluted particles to organic solvent, detergents, and proteolytic degradation and was checked by TEM observations. The PAV1 viability or infectivity could not be tested because a plaque assay was not available (see the next paragraph). PAV1 was shown to be sensitive to chloroform, because only spherical particles were observed in treated samples instead of the regular lemon-shaped ones (Fig. 1d). The VLPs were also partially disrupted by treatment with 0.3% Triton X-100 (data not shown), as well as after incubation for 1 h with proteinase K (1 mg/ml) (Fig. 1e); they were completely destroyed with 0.1% SDS (data not shown). These preliminary results suggested that the envelope of PAV1 was composed of proteins and lipids.
Genome of PAV1.
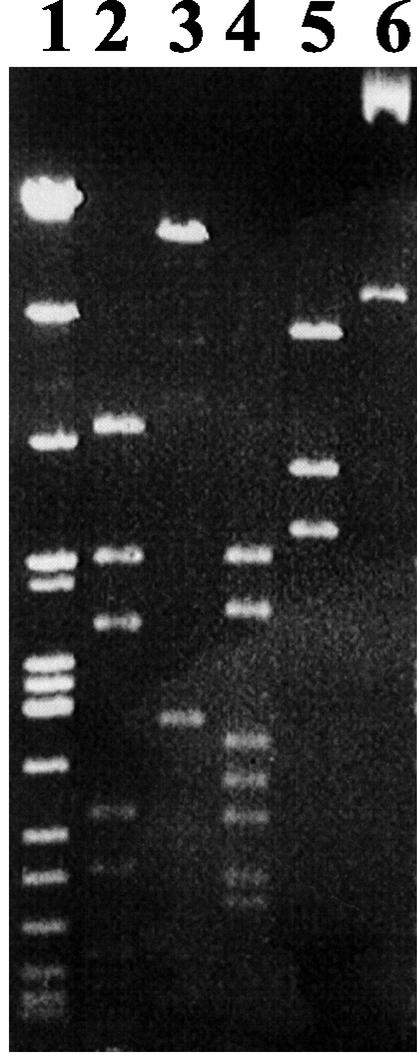
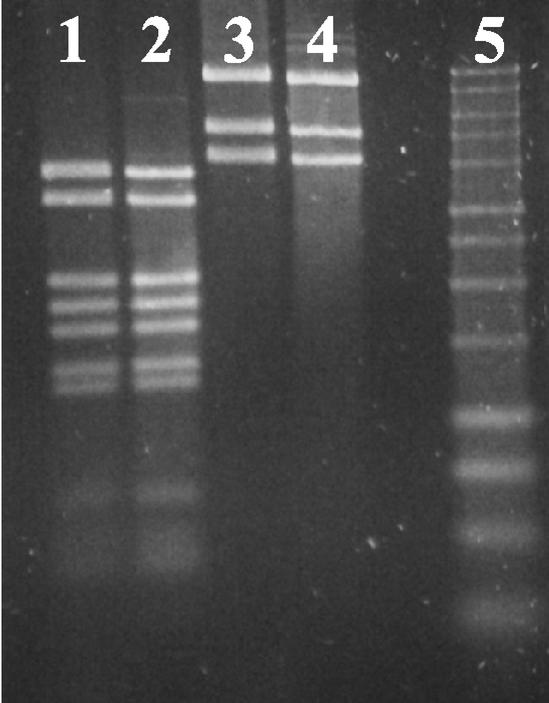
The cccDNA purified by the alkaline lysis method from strain GE23 was digested with restriction enzymes (BamHI, EcoRI, HindIII, and PstI), and its size was estimated to be 17.5 ± 0.5 kb (Fig. 2). We suspected this extrachromosomal DNA to be the free form of the PAV1 genome. To check this hypothesis, DNA prepared from purified virus particles was compared with plasmid DNA isolated from strain GE23. Agarose gel electrophoresis of viral DNA and plasmid DNA restricted with endonucleases HindIII or PstI gave the same pattern and proved the two to be identical. To investigate the possibility that viral DNA was in fact not circular in the virion, but rather linear with cohesive ends which cause circularization, samples of viral DNA but also plasmid DNA were heated to 70°C after digestion and chilled on ice before loading on gels. No change in the band pattern was observed (Fig. 3). We therefore conclude that the plasmid DNA is a free form of the PAV1 genome in strain GE23 and that PAV1 is composed with a double-strand circular DNA. Comparison of the intensity of the band corresponding to plasmid DNA with that of chromosomal DNA on total DNA preparation allow us to estimate the copy number of the plasmid form to be between 10 and 20 copies per chromosome (data not shown).
FIG. 2.
Agarose gel electrophoresis of the cccDNA from strain GE23 cut by BamHI (lane 2), EcoRI (lane 3), HindIII (lane 4) and PstI (lane 5) and uncut (lane 6). Lane 1, DNA size marker (kb) showing λDNA digested with HindIII, BamHI, and EcoRI.
FIG. 3.
Comparison of cccDNA from strain GE23 with DNA from purified PAV1 VLP. Lanes 1 and 3, plasmid DNA; lanes 2 and 4; VLP DNA. Lanes 1 and 2, digested with HindIII; lanes 3 and 4, digested with PstI. Lane 5, 1-kb DNA ladder.
VLP-associated proteins.
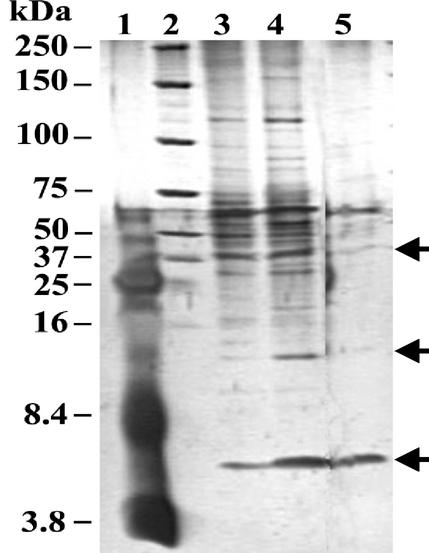
Proteins from CsCl-purified VLPs were analyzed by conventional SDS-polyacrylamide gel electrophoresis or by the method of Schägger and Jagow (24), which gives a better resolution of small proteins. Numerous bands of high molecular weight were often observed in such preparations, suggesting the presence of host proteins in addition to VLP proteins (Fig. 4, lanes 3 and 4). Indeed, TEM observations of these preparations showed they were contaminated by host cell flagella. In some cases, however, it was possible to obtain VLP preparations free of flagella (Fig. 4, lane 5), though in smaller amounts, making more difficult the visualization of the bands even with highly sensitive silver staining. Comparison of lane 5 with lanes 3 and 4 showed at least three VLP-specific proteins, the most abundant corresponding to the fat band of approximately 6 kDa and two others corresponding to fainter bands of about 13 and 36 kDa.
FIG. 4.
Silver-stained SDS-polyacrylamide gel electrophoresis of the PAV1 proteins. The results for three independent preparations of CsCl-gradient purified VLPs are shown. Lanes 3 and 4 are still contaminated with host proteins (presumably from host flagella); lane 5 contains mostly PAV1 proteins indicated by an arrow. Lanes 1 and 2, Kaleidoscope prestained standards and unstained Precision protein standards (Bio-Rad); the subunit sizes are listed in kDa at the left of the figure.
VLP-host relationship.
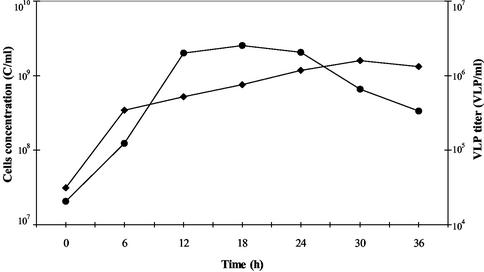
PAV1 was continuously and spontaneously released in the extracellular medium throughout growth of strain GE23, with the highest production during the stationary growth phase after about 30 h of incubation (1.6 × 106 VLP/ml) (Fig. 5). Virions were never observed by TEM inside cells or attached to intact cells. VLP production was never accompanied by the lysis of the culture nor by decrease of the cell density of the host. Nevertheless, it seems that PAV1 persistently infects the host cells, since attempts to obtain cured clones of GE23 by serial dilution in liquid medium and by plating failed. Moreover, the growth of strain GE23 is similar to that of strain GE9, a VLP-free “P. abyssi” strain, which indicates that strain GE23 tolerates the viral load very well.
FIG. 5.
Strain GE23 growth curve (cells/ml) (•) and time course of VLP production (⧫). The approximate titer of free VLP was estimated by a standardized electron microscopic count together with the corresponding host cell count.
Assuming that PAV1 could be a temperate virus, attempts were made to increase the VLP production by induction using chemical or physical treatments that damage DNA (mitomycin C, UV) and by exposing the host cells to physiological stresses like aerobiosis and hydrostatic pressure. Whatever the stress condition was, induction was never observed but rather a decrease of the viral production due to a slowdown of the host growth.
Southern hybridization experiments did not allow detection of any integrated prophage DNA in the host chromosome (Fig. 6). Indeed, in cross-hybridization reactions between labeled plasmid DNA (corresponding to a free form of the viral DNA) and total DNA of strain GE23, no additional hybridizing DNA fragments were observed in comparison to the restriction pattern of the plasmid DNA. Thus, the viral DNA did not integrate into the host chromosome.
FIG. 6.
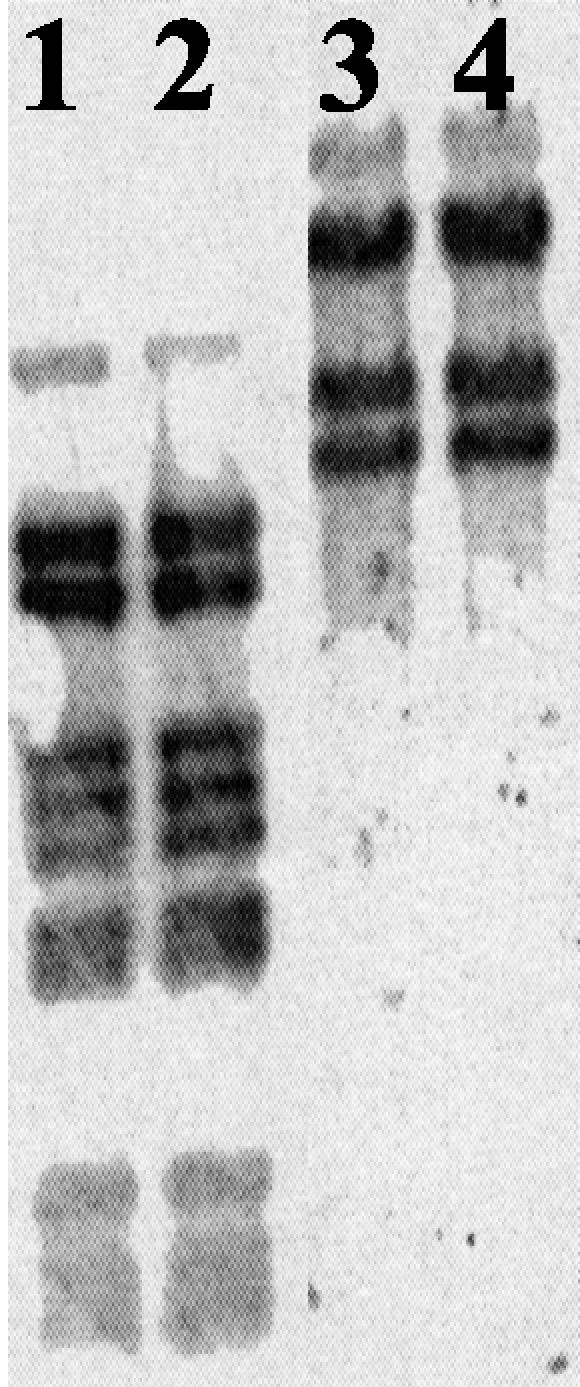
Southern hybridization analysis. Total DNA and cccDNA from the strain GE23 were digested with HindIII (lanes 1 and 2, respectively) or PstI (lanes 3 and 4, respectively) and separated on agarose gels (0.8%) before blotting. The pooled HindIII fragments of the cccDNA, labeled with fluorescein-11-dUTP, were used as a probe.
Lastly, to establish the possible infectivity of PAV1, we have tested almost 100 Thermococcales strains, including original isolates and reference species (“P. abyssi” strain GE5, P. furiosus, P. woesei, T. stetteri, and T. celer), for their abilities to act as a host, first by plaque assays and also in liquid culture. Despite numerous attempts, no strain was found to be a host.
DISCUSSION
Considering its lemon-shaped morphology, which is an unusual morphotype for euryarchaeotal viruses, PAV1 remarkably resembles the VLP A3 of M. voltae (30) and the viruses His1 of H. hispanica (4) and SSV1 of S. shibatae (18), the last one being the virus type of the Fuselloviridae family. Despite a plasticity of form, illustrated by the presence of elongated or flattened particles frequently observed by TEM, PAV1 particles were not altered by the purification procedures (ultracentrifugation in the presence of CsCl), indicating that they are osmotically resistant and can stand high gravity. The density of PAV1 in CsCl (1.28 g/ml) as deduced from the process of purification is very similar to those of VLP A3 (1.28 g/ml), His1 (1.30 g/ml), and SSV1 (1.24 g/ml), and such low density is characteristic of the enveloped viruses. This was confirmed by its sensitivity to organic solvent and detergents. Nevertheless, the sensitivity to lipid solvents as chloroform does not necessarily prove the presence of lipids. For example, despite the general absence of lipids, about 30% of tailed phages are readily inactivated by lipid solvents (acetone, chloroform, ether, toluene, etc.). Moreover, many viral proteins are susceptible to denaturation with chloroform and Triton X-100. Therefore, while these tests are commonly used for the characterization of new viruses, they cannot provide definite conclusions for PAV1 regarding the presence of an envelope containing lipids.
Several observations indicated that PAV1 persisted in its host in a stable carrier state. Very few particles are produced (estimated as 1.6 × 106 particles/ml for the best production). This could be compared with SSV1, for which 107 particles/ml of culture were released spontaneously in nonirradiated cultures (18). Virion structures were never observed inside host cells like in the case of SSV1. They were also not seen attached to intact cells, in contrast to SSV1, so it is unclear by which mechanism VLP enter, are assembled, and are released by the host cell. Attempts to induce VLP production or influence levels of production by subjecting GE23 culture to various chemical, physical, or physiological stresses failed. In this regard, PAV1 differs from SSV1, which is inducible by UV, but is comparable to the VLP A3, which is not. PAV1 is very stable in its host, as it has never been lost after many transfers in subcultures and was found in all colony clones checked.
Considering the host range of PAV1, we have been unable to demonstrate its infectivity. Possible reasons for this failure include biological inactivation of the particles during purification and/or lack of an appropriate uninfected host strain. To minimize the first possibility, infections were attempted by using simple filtration of GE23 supernatants, without any positive results. Regarding the second possibility, more than 100 strains, including novel isolates and type strains, were unsuccessfully tested. These results suggest that the host specificity for PAV1 is likely very restrictive or that PAV1 is defective for transmission. In the case of the bacteriophage P1, for example, some virions examined have smaller heads than usual and hence are defective. Most defective virions carry only 40% of the phage genome. However, because of the circularly permuted DNA, multiple infection of a single cell by defective viruses can result in progeny virus production as a consequence of complementation between defective phages (6). In the case of PAV1, the complementation between the defective phages may be not possible. The fact that we could not observe infection might also be due to our relative ignorance of the optimal conditions for phage infection and to the difficulties inherent to the cultivation of a strictly anaerobic sulfur-requiring hyperthermophile. In particular, the difficulty of plating potential hosts as a lawn hindered our investigation of the host-VLP interactions, and this point should be optimized for further studies.
The existence in strain GE23 of a large plasmid (pGN23) of 16.8 kb was previously reported (5). We estimated the size of this plasmid DNA to be 17.5 ± 0.5 kb, which is slightly larger than the first estimation (5). Our speculation that this element is in fact a free form of the viral genome has now been confirmed by showing that the DNA prepared from purified virus particles is identical to the plasmid DNA isolated from GE23 cells. In this regard, the DNA genome of PAV1 closely resembles those of the VLP A3 and SSV1 in being circular and double-stranded but differs from the genome of His1, which is linear. However, the genome of PAV1 is slightly larger than the genome of SSV1 (15.5 kb) and His1 (14.9 kb), in agreement with the significantly larger size of the PAV1 particles. An important difference between SSV1 and PAV1 is that no integrated prophage of PAV1 was detected in GE23 host chromosome. Indeed, most temperate bacteriophages integrate a copy of their genome into the host chromosome, as does SSV1 (18). However, others, for example, coliphages P1 and P4, can be stably maintained as plasmid prophages. Interestingly, P4 may lysogenize the host cell by integrating its genome into the bacterial chromosome (7), but it also can be maintained as a freely replicating multicopy plasmid (30 to 50 copies per Escherichia coli chromosome), a situation resembling that of PAV1 plasmid (about 20 copies per chromosome). To accomplish its lytic cycle, P4 exploits an unrelated helper phage of the P2 family for its viral morphogenesis. Such a system has also been described in Archaea for Sulfolobus, where the plasmid pSSVx requires the phage SSV2 to produce virion particles. PAV1 could be analogous to these systems by replicating its genome as a multicopy plasmid but differs because it seems defective for transmission, perhaps due to the absence of a helper virus.
The presence of lemon-shaped viruses seems to be more frequent in the Euryarchaeota lineage than previously expected. This morphotype has never been found among the Bacteria or Eucarya, but it is present in quite diverse extreme environments, such as acidic thermophilic continental solfatara (SSV1), salterns (His1), and deep-sea hydrothermal vents (PAV1), but also in subsurface anaerobic sediments (VLP A3). This strengthens the idea of their specificity to the archaeal domain and probably reflects a deep evolutionary history within this domain. It is interesting to speculate whether these different lemon-shaped viruses have evolved together with their respective hosts, which would imply that the common ancestor of Archaea was also infected by an ancestral lemon-shaped virus, or if such a virus originally present in one archaeal lineage has secondarily infected the other ones.
Based on its lemon-shaped morphology, the nature of its envelope, and its circular double-stranded DNA genome, PAV1 could be proposed as a new member of the recently described Fuselloviridae family (26). This family contains so far only one genus (Fusellovirus) represented by the best studied crenarchaeal virus, SSV1; therefore, the precise criteria for belonging to this family have not yet been clearly established. For these reasons, it is not clear whether the morphotype of His1 virus, for example, is sufficient to classify it in the Fuselloviridae family as previously proposed (4), although the linear type of its DNA suggests a different mode of replication than for SSV1. Usually the genomes of viruses belonging to the same genus have the same organization and mode of replication. This also seems to be valid for the few representatives of the novel archaeal viruses whose genome have been sequenced; e.g., the genomes of the Rudiviruses SIRV1 and SIRV2 are highly similar (20, 21). Sequencing of the complete genome of PAV1 is currently in progress and will be finished soon (unpublished data). Preliminary comparison of more than 80% of the total sequence with the genomes of SSVs viruses did not reveal any significant similarity, providing potential evidence that PAV1 is not closely related to known SSVs at least at the genus level. Unfortunately, the genome sequence of His1 and VLP A3 are not yet available for comparison studies. Obviously, more genome sequences of lemon-shaped viruses are needed to address the question of the PAV1 taxonomic status.
Acknowledgments
We thank Wolfram Zillig for his technical help and his critical reading of the manuscript. We thank also David Prangishvili for technical advice regarding the VLP purification procedure and Ken Stedman for the gift of the Sulfolobus shibatae strain B12 infected with SSV1. We thank Alain Hervé for generously allowing us to use his density gradient fractionator. We also thank Gérard Sinquin for assistance in electron microscopy.
This investigation was supported by a grant from Biomérieux.
REFERENCES
- 1.Ackerman, H.-W. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146:843-857. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, H. P., W. Zillig, U. Ziese, I. Holz, M. Crosby, T. Utterback, J. F. Weidmann, J. K. Kristjansson, H. P. Klenk, K. E. Nelson, and C. M. Fraser. 2000. A novel Lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267:252-266. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, H. P., U. Ziese, and W. Zillig. 2000. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272:409-416. [DOI] [PubMed] [Google Scholar]
- 4.Bath, C., and M. L. Dyall-Smith. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J. Virol. 72:9392-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benbouzid-Rollet, N., P. Lopez-Garcia, L. Watrin, G. Erauso, D. Prieur, and P. Forterre. 1997. Isolation of new plasmids from hyperthermophilic Archaea of the order Thermococcales. Res. Microbiol. 148:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Birge, E. A. 2000. Bacteriophage P1, p. 284-285. In E. A. Birge (ed.), Bacterial and bacteriophage genetics, 4th ed. Springer, New York, N.Y.
- 7.Briani, F., G. Deho, F. Forti, and D. Ghisotti. 2001. The plasmid status of satellite bacteriophage P4. Plasmid 45:1-17. [DOI] [PubMed] [Google Scholar]
- 8.Charbonnier, F., G. Erauso, T. Barbeyron, D. Prieur, and P. Forterre. 1992. Evidence that a plasmid from a hyperthermophilic archaebacterium is relaxed at physiological temperatures. J. Bacteriol. 174:6103-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erauso, G., A.-L. Reysenbach, A. Godfroy, J.-R. Meunier, B. Crump, F. Partensky, J. A. Baross, G. Barbier, N. R. Pace, and D. Prieur. 1993. Pyrococcus abyssi sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Arch. Microbiol. 160:338-349. [Google Scholar]
- 10.Erauso, G., A. Godfroy, G. Raguenes, and D. Prieur. 1995. Plate cultivation techniques for strictly anaerobic, thermophilic, sulfur-metabolizing archaea, p.25-29. In F. T. Robb (ed.), Thermophiles, archaea: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Erauso, G., S. Marsin, N. Benbouzid Rollet, M. F. Baucher, T. Barbeyron, Y. Zivanovic, D. Prieur, and P. Forterre. 1996. Sequence of plasmid pGT5 from the archaeon Pyrococcus abyssi: evidence for rolling-circle replication in a hyperthermophile. J. Bacteriol. 178:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geslin, C., M. Le Romancer, M. Gaillard, G. Erauso, and D. Prieur. 2003. Observation of virus-like particles in high temperature enrichment cultures from deep-sea hydrothermal vents. Res. Microbiol., in press. [DOI] [PubMed]
- 13.Janekovic, D., S. Wunderl, I. Holz, W. Zillig, A. Gierl, and H. Neumann. 1983. TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic, anaerobic, sulfur reducing archaebacterium Thermoproteus tenax. Mol. Gen. Genet. 192:39-45. [Google Scholar]
- 14.Lucas, S., L. Toffin, Y. Zivanovic, D. Charlier, H. Moussard, P. Forterre, D. Prieur, and G. Erauso. 2002. Construction of a shuttle vector and spheroplasts transformation of the hyperthermophilic archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 68:2258-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marteinsson, V. T., L. Watrin, D. Prieur, J. C. Caprais, G. Raguénès, and G. Erauso. 1995. Phenotypic characterization, DNA similarities, and protein profiles of twenty sulfur-metabolizing hyperthermophilic anaerobic Archaea isolated from hydrothermal vents in the south western Pacific ocean. Int. J. Syst. Bacteriol. 45:623-632. [Google Scholar]
- 16.Marteinsson, V. T., P. Moulin, J.-L. Birrien, A. Gambacorta, M. Vernet, and D. Prieur. 1997. Physiological responses to stress conditions and barophilic behavior of the hyperthermophilic vent archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 63:1230-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marteinsson, V. T., A.-L. Reysenbach, J.-L. Birrien, and D. Prieur. 1999. A stress protein is induced in the deep-sea barophilic hyperthermophile Thermococcus barophilus when grown under atmospheric pressure. Extremophiles 3:277-282. [DOI] [PubMed] [Google Scholar]
- 18.Martin, A., S. Yeats, D. Janekovic, W.-D. Reiter, W. Aicher, and W. Zillig. 1984. SAV1, a temperate U.V.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius. EMBO J. 3:2165-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palm, P., C. Schleper, B. Grampp, S. Yeats, P. McWilliam, W.-D. Reiter, and W. Zillig. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242-250. [DOI] [PubMed] [Google Scholar]
- 20.Prangishvili, D., H. P. Arnold, D. Götz, U. Ziese, I. Holz, J. K. Kristjansson, and W. Zillig. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the Sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prangishvili, D., K. Stedman, and W. Zillig. 2001. Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol. 9:39-43. [DOI] [PubMed] [Google Scholar]
- 22.Ravot, G., B. Ollivier, M. Magot, B. K. C. Patel, J.-L. Crolet, M.-L. Fardeau, and J.-L. Garcia. 1995. Thiosulfate reduction, an important physiological feature shared by members of the order Thermotogales. Appl. Environ. Microbiol. 61:2053-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Schägger, H., and G. V. Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 25.Schleper, C., K. Kubo, and W. Zillig. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. USA 89:7645-7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.). 2000. Virus taxonomy: seventh report of The International Committee on Virus Taxonomy, p. 131-136. Academic Press, San Diego, Calif.
- 27.Ward, D. E., I. M. Revet, R. Nandakumar, J. H. Tuttle, W. M. De Vos, J. Van Der Oost, and J. DiRuggiero. 2002. Characterization of plasmid pRT1 from Pyrococcus sp. strain JT1. J. Bacteriol. 184:2561-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watrin, L., and D. Prieur. 1996. UV and ethyl methanesulfonate effects in hyperthermophilic archaea and isolation of auxotrophic mutants of Pyrococcus strains. Curr. Microbiol. 33:377-382. [DOI] [PubMed] [Google Scholar]
- 29.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eukarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood, A. G., W. B. Whitman, and J. Konisky. 1989. Isolation and characterization of an archaebacterial virus-like particle from Methanococcus voltae A3. J. Bacteriol. 171:93-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zillig, W., W.-D. Reiter, P. Palm, F. Gropp, H. Neumann, and M. Rettenberger. 1988. Viruses of archaebacteria. p. 517-555. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y.
- 32.Zillig, W., A. Kletzin, C. Schleper, I. Holz, D. Janekovic, J. Hain, M. Lanzendörfer, and J. K. Kristjansson. 1994. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfatars. Syst. Appl. Microbiol. 16:609-628. [Google Scholar]