Abstract
The mechanisms by which hyperthermophilic Archaea, such as “Pyrococcus abyssi” and Pyrococcus furiosus, survive high doses of ionizing gamma irradiation are not thoroughly elucidated. Following gamma-ray irradiation at 2,500 Gy, the restoration of “P. abyssi” chromosomes took place within chromosome fragmentation. DNA synthesis in irradiated “P. abyssi” cells during the DNA repair phase was inhibited in comparison to nonirradiated control cultures, suggesting that DNA damage causes a replication block in this organism. We also found evidence for transient export of damaged DNA out of irradiated “P. abyssi” cells prior to a restart of chromosomal DNA synthesis. Our cell fractionation assays further suggest that “P. abyssi” contains a highly efficient DNA repair system which is continuously ready to repair the DNA damage caused by high temperature and/or ionizing radiation.
The hyperthermophilic Archaea thrive in geothermal environments with optimal growth temperatures approaching the boiling point of water (25). Under these conditions, DNA is expected to be unstable due to the accumulation of many different types of damage (e.g., hydrolytic depurination, deamination of cytosine and adenine, and single- or double-strand breakage [DSB]; note that ionizing radiation and/or genotoxic chemicals can also induce similar damage [13, 18]). During the last few years, it has been shown that some hyperthermophilic archaea, such as Desulfurococcus amylolyticus and Thermococcus stetteri, are resistant to ionizing radiation (17). More recently, DiRuggiero and collaborators have reported that a hyperthermophilic archaeon, Pyrococcus furiosus, did not show any loss of viability at a dose of 2,000 Gy (gamma radiation), while the viability of Escherichia coli was already drastically decreased at a dose of 100 Gy (9). We also showed earlier that no specific DNA protection mechanisms exist in “Pyrococcus abyssi” (12), suggesting that an efficient mechanism(s) for repair of radiation-induced DNA damage must exist in Pyrococcus species. Presumably, these yet to be elucidated mechanisms, and their possible coordination with DNA replication, are critical for the accurate maintenance of genetic information in the hyperthermophilic Archaea.
We were previously unable to find any evidence for radiation-induced gene expression in Pyrococcus species by use of two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12), and two recA/rad51-like genes, radA and radB, of Pyrococcus sp. are constitutively expressed in P. furiosus (15). In this work we have further investigated physiological responses of “P. abyssi” to ionizing radiation by studying the kinetics of DSB repair and by investigating other possible responses to massive DNA damage in “P. abyssi” cells. Altogether, our results indicate that, despite the fact that the bacterium Deinococcus radiodurans and the archaeon “P. abyssi” grow under very different environments and contain nonhomologous repair and replication proteins, these two organisms respond in a similar fashion to DNA damage caused by ionizing radiation (1, 2).
Radiation-induced growth delay and DNA damage.
Previous experiments indicated that “P. abyssi” cells were able to fully recover growth within 2 h after gamma-ray irradiation at 2,500 Gy under optimal growth conditions in YPS medium (11) (data not shown), suggesting that irradiated cells efficiently repaired massive DNA damage within this repair period (12). To investigate the fate of damaged chromosomes during this period, DSB analysis was performed by the pulsed-field gel electrophoresis technique as described earlier (12) to visualize the DNA restoration of fragmented chromosomes immediately after irradiation of “P. abyssi” cells.
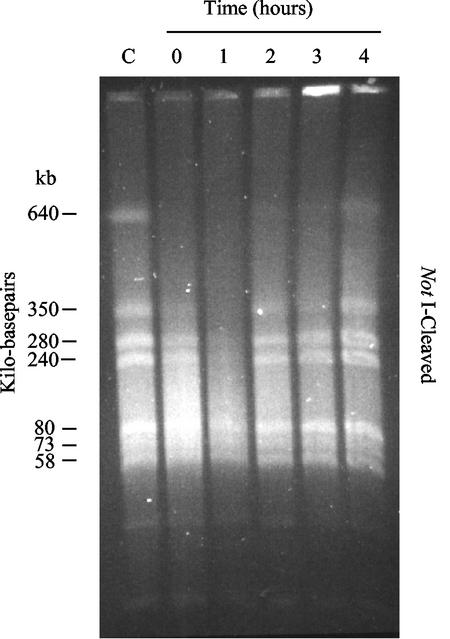
Our results showed that the chromosomal DNA of “P. abyssi” was fragmented following irradiation (Fig. 1). Interestingly, chromosomal DNA degradation appeared reproducibly more extensive at 1 h postirradiation under optimal growth conditions (Fig. 1; 1 h) than directly after irradiation (Fig. 1; 0 h). This observation could indicate the presence of active nuclease activities in “P. abyssi” cells that function in DNA repair (4, 14, 23, 24). Alternatively, this postirradiation DNA degradation could result from cleavage by heat of the topologically relaxed fragmented chromosomal DNA. Our analyses also showed that the fragmented chromosomal DNA was fully restored within 2 h after gamma-ray irradiation at 2,500 Gy under optimal growth conditions, which is clearly visible by the reappearance of the largest genomic band of 640 kbp. Note also that the pattern of NotI-digested chromosomal DNA was identical before and after radiation and was in good agreement with earlier data (9, 10). Thus, “P. abyssi” is able to efficiently repair DSBs, similar to what has been observed for P. furiosus (9) and Deinococcus radiodurans (2, 5, 6, 7, 20).
FIG. 1.
Pulsed-field gel electrophoresis analysis of “P. abyssi” chromosomal DNA following gamma irradiation of 2,500 Gy. Irradiation was performed on ice with a 137Cs gamma-ray source at a rate of 60 Gy/min (Institut Curie, Orsay, France). Each lane corresponds to DNA extracted from 107 cells/ml that was visualized by ethidium bromide staining (5 μg/ml). Lane C, control culture (without gamma irradiation); lanes 0 to 4, cells were subjected to 0, 1, 2, 3, and 4 h, respectively, of incubation at 96°C after irradiation. Chromosomal DNA was digested with NotI, and the sizes of the different NotI macrorestriction fragments are indicated in kilobase pairs (left side).
DNA repair and DNA synthesis are uncoupled during the repair period.
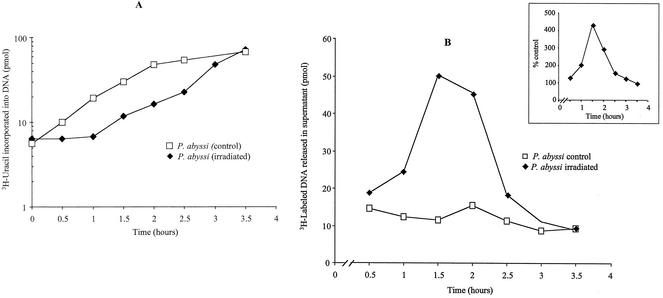
In Deinococcus radiodurans, DNA replication is known to cease following irradiation (8, 21), thus prompting us to investigate whether DNA repair and replication could also be coupled in our model organism. “P. abyssi” cells efficiently incorporate [3H]uracil (1μCi/ml [Amersham]; specific activity, 16.2 Ci/mmol) into DNA, a property which has been used to monitor DNA replication in this organism (22). To investigate whether ionizing irradiation inhibits DNA synthesis in “P. abyssi,” cells were irradiated at 2,500 Gy during stationary phase and were allowed to recover growth under optimal conditions. DNA synthesis in “P. abyssi” cells was monitored at different time points by measuring the incorporation of 3H-labeled uracil into alkaline-resistant trichloroacetic acid (TCA) precipitates (5% ice-cold TCA solution and 20 mM sodium pyrophosphate mixture) in both irradiated and nonirradiated cells (22). As shown in Fig. 2A, irradiated cultures showed a delay in DNA synthesis of approximately 60 to 90 min when cells were irradiated with 2,500 Gy. The nonirradiated culture did not show a similar lag phase, indicating that the observed delay is somehow related to DNA repair. The notion of repair-coupled DNA synthesis arrest is also supported by the observation that the measured end point of the replication delay corresponds well with the time of growth recovery and chromosome restoration. In the stationary-phase control culture (Fig. 2A) and exponential-phase cells (22), the incorporation into DNA of 3H-labeled uracil starts after 15 to 20 min (a similar delay is also observed for exponential-phase cells [Hannu Myllykallio, unpublished observations]) when cells are incubated at 96°C. These results suggest that DNA replication proteins, which are required for uracil incorporation into DNA, exist as functional forms in “P. abyssi” cells in exponential- and stationary-phase cultures.
FIG. 2.
Effect of gamma irradiation on DNA synthesis and export. (A) DNA synthesis restarts after gamma irradiation. The curves represent the restart of DNA replication in irradiated and nonirradiated cultures of “P. abyssi.” Cells were irradiated during stationary phase with 2,500 Gy of radiation. Fresh medium containing 3H-labeled uracil was inoculated and incubated at 95°C for 210 min. DNA replication was monitored by using scintillation counting to measure the incorporation of labeled uracil into DNA after TCA precipitation of whole cells. The curves show DNA replication in irradiated cells after irradiation at 2,500 Gy (⧫) and DNA replication in unirradiated cultures (□). These results are the means of two independent experiments. (B) Export of damaged and undamaged DNA from the “P. abyssi” cells following gamma irradiation. This experiment shows the release of radioactivity in the supernatant from “P. abyssi” cells in early stationary phase after gamma irradiation at 2,000 Gy. The “P. abyssi” culture was previously labeled overnight with [3H]uracil at a final concentration of 1 μCi per ml before irradiation. After irradiation, the measurement of radioactivity in the supernatant was performed every 30 min until 210 min at 96°C. The inset shows the percentages of labeled DNA material from the irradiated cells versus the nonirradiated cells. The radioactivity released in the supernatant of irradiated cells (♦) was monitored by using scintillation counting to measure the labeled uracil in DNA after TCA precipitation of whole cells. Percentages were determined from control cultures treated under the same conditions but without gamma irradiation. These values are averages of two independent experiments.
Export from the cells of damaged and undamaged DNA.
To investigate the fate of the damaged DNA in irradiated “P. abyssi” cells, cultures in the stationary growth phase were labeled with [3H]uracil as described above, washed two times with artificial seawater, and then suspended in fresh growth medium. Irradiation was performed at a sublethal dose of 2,000 Gy to avoid detection of the DNA in the supernatant fraction due to lysis by cell death. Following irradiation, cultures were allowed to recover under optimal growth conditions. One-milliliter aliquots of cultures were removed at intervals corresponding to each incubation time point. The amount of radioactivity in alkaline-resistant form in the culture supernatants was measured using TCA precipitation and scintillation counting.
In the case of irradiated cultures, the radioactivity released in the supernatant increased after the first 30 min postirradiation and reached its maximal level after 90 min of incubation (Fig. 2B). The amount of radioactivity in TCA precipitates started to decrease 2 h after irradiation, and it became close to the control values at 2.5 and 3 h postirradiation. “P. abyssi” cells thus actively expulsed damaged DNA (note that free nucleotides are not precipitated under these experimental conditions) into the growth medium just before a restart of DNA synthesis. As this material can be precipitated using TCA, the expulsed DNA corresponds to DNA fragments longer than 20 nucleotides, and our preliminary estimation reveals that 0.3 to 0.4% of labeled genomic DNA is released from the irradiated “P. abyssi” cells. The increase in supernatant radioactivity appears higher than has been observed for radiation-resistant “Chroococcidiopsis” cells (4). The mechanisms generating and transporting these DNA fragments out of the cell are currently unclear (3, 26). They could possibly correspond to DNA molecules generated during the repair processes during enzymatic processing of DSBs. A priori, the transport of damaged DNA should prevent the accumulation of genetic mistakes in irradiated Pyrococcus cells, similar to what was found earlier for Deinococcus radiodurans (1, 2) and “Chroococcidiopsis” cells (3).
A subset of “P. abyssi” DNA replication and repair proteins is chromatin bound before and after irradiation at 2,500 Gy.
A possibility for repair of DSBs in Pyrococcus sp. is homologous recombination (10, 15) by chromatin-bound DNA repair complexes. As constitutively expressed P. furiosus RadA presumably functions in homologous recombination and physically interacts with several replication proteins (e.g., replication protein A [RPA] and replication factor C [RFC]) (16), we have tested whether these proteins (RadA, RPA, and RFC) dissociate from the chromatin in stationary-phase and irradiated cells, thus explaining the observed block in DNA synthesis (Fig. 2A).
We examined by simple cell fractionation and immunodetection assays the level of chromatin association of “P. abyssi” RadA, RPA, and RFC in irradiated cells (early stationary phase). After irradiation, “P. abyssi” cells from irradiated and nonirradiated control cultures were incubated for 120 min. Culture aliquots were removed at the indicated intervals. Soluble proteins (supernatant) and the chromosomal DNA-enriched insoluble fraction (pellet) were separated by centrifugation through a sorbitol cushion, as described previously (19). The insoluble pellet fraction, containing DNA and chromatin-associated proteins, was washed once and dissolved in an equal volume of extraction buffer by brief sonication. This treatment yielded DNA fragments of 500 to 1,000 bp. Immunodetection of chromatin-associated proteins was performed as described previously (19).
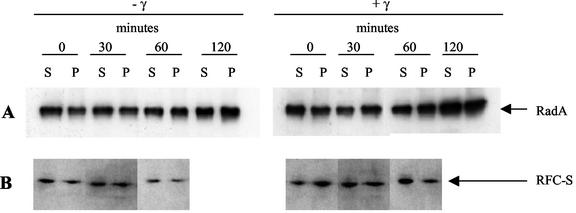
In accordance with earlier results (15), our data indicated that a steady-state expression level of RadA protein (expected molecular mass of 38.4 kDa) was not massively induced after gamma irradiation (Fig. 3A). Moreover, our results indicate that the amount of RadA protein in soluble and chromatin-associated forms remained similar for 120 min postirradiation. Similar results were obtained for RPA (data not shown) and RFC (Fig. 3B), which are implicated in DNA replication. Therefore, the delay of DNA synthesis (replication) observed after irradiation does not result from dissociation of RadA, RPA, and RFC proteins from the chromatin.
FIG. 3.
Western blotting analysis of RadA and a subunit of the RFC complex. Experiments were performed with early-stationary-phase “P. abyssi” cells after the cells were irradiated at 2,500 Gy (+γ) or not irradiated (−γ). Culture samples were removed every 30 min (up to 2 h at 96°C) and kept on ice. The soluble proteins (supernatant fraction [S]) and the DNA-bound proteins (pellet fraction [P]) were extracted from the cells as described earlier (19). Proteins were subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and were analyzed by Western blotting. The RadA (38.4 kDa) and RFC-S (37.4 kDa) proteins were immunodetected by specific rabbit antibodies by using the ECL Western blotting kit (Amersham Pharmacia Biotech). The protein band recognized by specific antibodies was visualized by exposure of the membranes to film.
Conclusion.
In this study we have shown that “P. abyssi” cultures in stationary phase are able to completely repair their fully fragmented chromosomes following gamma irradiation within 2 h. On the basis of our experimental data, “P. abyssi” utilizes an efficient strategy for responding to DNA damage that includes (i) an uncoupling of DNA repair and DNA synthesis and (ii) a prevention of accumulation of genetic mistakes by exporting damaged DNA. In the light of our finding that RadA, RPA, and RFC stay chromatin bound before and after irradiation, it is tempting to speculate that active chromatin-bound repair and replication complexes are continuously ready to counteract DNA damage in “P. abyssi.” Identification of possible novel functions in adaptive responses of hyperthermophilic Archaea to radiation-induced DNA damage will now require efforts in functional genomics and/or genetics. In particular, it will be of interest to investigate whether the radiation-induced DNA synthesis block could result either from a physical blockage of replication forks or, alternatively, from the presence of a regulated mechanism (a check point) that would uncouple DNA replication and repair processes.
Acknowledgments
We thank Vincent Favaudon for the use of a 137Cs gamma-ray source (Institut Curie, Orsay, France).
This work was supported by Electricité de France (EDF). Fujihiko Matsunaga thanks the Japan Society for the Promotion of Science for financial support.
REFERENCES
- 1.Battista, J. R., A. M. Earl, and M. J. Park. 1999. Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol. 7:362-365. [DOI] [PubMed] [Google Scholar]
- 2.Battista, J. R. 1997. Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51:203-224. [DOI] [PubMed] [Google Scholar]
- 3.Billi, D., E. I. Friedmann, K. G. Hofer, M. G. Caiola, and R. O. Friedmann. 2000. Ionizing radiation in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl. Environ. Microbiol. 66:1489-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constantinesco, F., P. Forterre, and C. Elie. 2002. nurA, a novel 5′-3′ nuclease gene linked to rad50 and mre11 homologs of thermophilic Archaea. EMBO Rep. 3:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly, M. J., and K. W. Minton. 1996. An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 178:4461-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daly, M. J., and K. W. Minton. 1995. Interchromosomal recombination in the extremely radioresistant bacterium Deinococus radiodurans. J. Bacteriol. 177:5495-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daly, M. J., L. Ouyang, P. Fuchs, and K. W. Minton. 1994. In vivo damage and recA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans. J. Bacteriol. 176:3508-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean, C. J., P. Feldshreiber, and J. T. Lett. 1966. Repair of X-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans. Nature 209:49-52. [DOI] [PubMed] [Google Scholar]
- 9.DiRuggiero, J., N. Santngelo, Z. Nackerdien, J. Ravel, and F. T. Robb. 1997. Repair of extensive ionizing-radiation DNA damage at 95°C in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 179:4643-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiRuggiero, J., J. R. Brown, A. P. Boggert, and F. T. Robb. 1999. DNA repair system in archaea: mementos from the last universal common ancestor? J. Mol. Evol. 49:474-484. [DOI] [PubMed] [Google Scholar]
- 11.Erauso, G., A. L. Reysenbach, A. Godfroy, J. R. Meunier, B. Crump, F. Partensky, J. A. Baross, V. T. Marteinsson, G. Barbier, N. R. Pace, and D. Prieur. 1993. Pyrococcus abyssi sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Arch. Microbiol. 160:338-349. [Google Scholar]
- 12.Gérard, E., E. Jolivet, D. Prieur, and P. Forterre. 2001. DNA protection is not involved in the radioresistance of the hyperthermophilic archaea Pyrococcus abyssi and Pyrococcus furiosus. Mol. Gen. Genet. 266:72-78. [DOI] [PubMed] [Google Scholar]
- 13.Grogan, D. W. 1998. Hyperthermophiles and the problem of DNA instability. Mol. Microbiol. 28:1043-1049. [DOI] [PubMed] [Google Scholar]
- 14.Hopfner, K. P., A. Karcher, D. Shin, C. Fairley, J. A. Tainer, and J. P. Carney. 2000. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol. 182:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komori, K., T. Miyata, J. DiRuggiero, R. Holley-Shanks, I. Hayashi, I. K. O. Can, K. Mayanagi, H. Shinagawa, and Y. Ishino. 2000. Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J. Biol. Chem. 275:33782-33790. [DOI] [PubMed] [Google Scholar]
- 16.Komori, K., and Y. Ishino. 2001. Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J. Biol. Chem. 276:25654-25660. [DOI] [PubMed] [Google Scholar]
- 17.Kopylov, V. M., E. A. Bonch-Osmolovskaya, V. A. Svetlichnyi, M. L. Miroshnicheko, and V. S. Skobin. 1993. Gamma-irradiation resistance and UV sensitivity of extremely thermophilic archaebacteria and eubacteria. Mikrobiologiya 62:90-95. [Google Scholar]
- 18.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 19.Matsunaga, F., P. Forterre, Y. Ishino, and H. Myllykallio. 2001. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl. Acad. Sci. USA 98:11152-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minton, K. W. 1996. Repair of ionizing-radiation resistant bacterium Deinococcus radiodurans. Mutat. Res. 363:1-7. [DOI] [PubMed] [Google Scholar]
- 21.Moseley, B. E. B., and H. J. R. Copland. 1976. The rate of recombination repair and its relationship to the radiation-induced delay in DNA synthesis in Micrococcus radiodurans. J. Gen. Microbiol. 93:251-258. [DOI] [PubMed] [Google Scholar]
- 22.Myllykallio, H., P. Lopez-Garcia, R. Heilig, W. Saurin, Y. Zivanovic, H. Philippe, and P. Forterre. 2000. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science 288:4119-4123. [DOI] [PubMed] [Google Scholar]
- 23.Petukhova, G., E. Y.-H. P. Lee, and P. Sung. 2001. DNA end-processing and heteroduplex DNA formation during recombinational repair of DNA double-strand breaks, p. 125-146. In J. A. Nickoloff and M. F. Hoestra (ed.), DNA damage repair. Advances from phage to humans. Humana Press, Inc., Totowa, N.J.
- 24.Sharples, G. J., and D. R. Leach. 1995. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17:1215-1217. [DOI] [PubMed] [Google Scholar]
- 25.Stetter, K. O. 1996. Hyperthermophilic procaryotes. FEMS Microbiol. Rev. 18:149-158. [Google Scholar]
- 26.Vukovik-Nagy, B., B. W. Fox, and M. Fox. 1974. The release of a deoxyribonucleic acid fragment after X-irradiation of Micrococcus radiodurans. Int. J. Radiat. Biol. 25:329-337. [DOI] [PubMed] [Google Scholar]