Abstract
The two-component system, consisting of a histidine (His) protein kinase that senses a signal input and a response regulator that mediates the output, is an ancient and evolutionarily conserved signaling mechanism in prokaryotes and eukaryotes. The identification of 54 His protein kinases, His-containing phosphotransfer proteins, response regulators, and related proteins in Arabidopsis suggests an important role of two-component phosphorelay in plant signal transduction. Recent studies indicate that two-component elements are involved in plant hormone, stress, and light signaling. In this review, we present a genome analysis of the Arabidopsis two-component elements and summarize the major advances in our understanding of Arabidopsis two-component signaling.
Two-component systems are involved in various signal transduction pathways in many prokaryotes, fungi, slime molds, and plants (Stock et al., 2000). The prototypical two-component system is a major signaling mechanism that mediates the response to various environmental stimuli in bacteria. It typically consists of a membrane-localized His protein kinase that senses the input signal, and a response regulator, e.g. a transcription factor, that mediates the output (Fig. 1A). Signaling is initiated when the His protein kinase, modulated by the environmental stimulus, autophosphorylates its conserved His residue. The phosphoryl group is transferred to a conserved Asp residue on the response regulator that results in modulation of its activity. In bacteria, yeasts, slime molds, and plants, multistep phosphorelay has evolved with additional phosphotransfer steps involving His phosphotransfer and receiver domains or proteins that connect to final response regulators or other signaling outputs (Fig. 1B; Wurgler-Murphy and Saito, 1997; Stock et al., 2000; Thomason and Kay, 2000). For example, the SLN1/YPD1/SSK1 phosphorelay in yeast translates a change in osmolarity into the phosphorylation state of the response regulator SSK1, which then modulates the HOG1 MAPK cascade in the cytosol to control gene expression (Fig. 1B; Wurgler-Murphy and Saito, 1997). The completion of the Arabidopsis genome sequence has revealed 54 genes encoding putative His kinase (AHK), His phosphotransfer (AHP), response regulator (ARR), and related proteins (Fig. 2), suggesting that two-component signaling is likely involved in many facets of plant cell regulation (Imamura et al., 1999; D'Agostino et al., 2000; Sakakibara et al., 2000; Urao et al., 2000b; Lohrmann and Harter, 2002; Schaller et al., 2002). The identification of putative His protein kinases as the photoreceptor phytochromes, a putative osmosensor, and the cytokinin and ethylene receptors supports the importance of two-component proteins in plant signal transduction (Elich and Chory, 1997; Hughes and Lamparter; 1999; Urao et al., 1999; Bleecker and Kende, 2000; Inoue et al., 2001). Recent studies have also provided evidence that AHPs and ARRs play essential roles in cytokinin and light signaling (Hwang and Sheen, 2001; Sakai et al., 2001; Sweere et al., 2001; Haberer and Kieber, 2002). However, the functions of most plant two-component proteins have not been determined. This review presents a genome-wide analysis of the two-component proteins in Arabidopsis and provides a framework for future functional dissection of two-component systems in plant signal transduction.
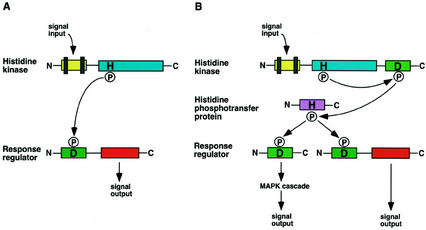
Figure 1.
Schematic representation of the two-component and the multistep phosphorelay signaling systems. A, The prototypical two-component pathway uses a single phosphoryl transfer event between a His protein kinase and its cognate response regulator. B, The multistep His-to-Asp phosphorelay system in which a His-containing phosphotransfer protein serves as a phosphoryl acceptor and donor between the hybrid protein kinase and the response regulator. In yeast (Saccharomyces cerevisiae) osmosensing, the phosphorelay connects to an MAPK cascade (Wurgler-Murphy and Saito, 1997). In Arabidopsis cytokinin signaling, a response regulator directly regulates its target gene expression (Hwang and Sheen, 2001). The vertical bars represent transmembrane domains. H, His; D, Asp; P, phosphoryl group.
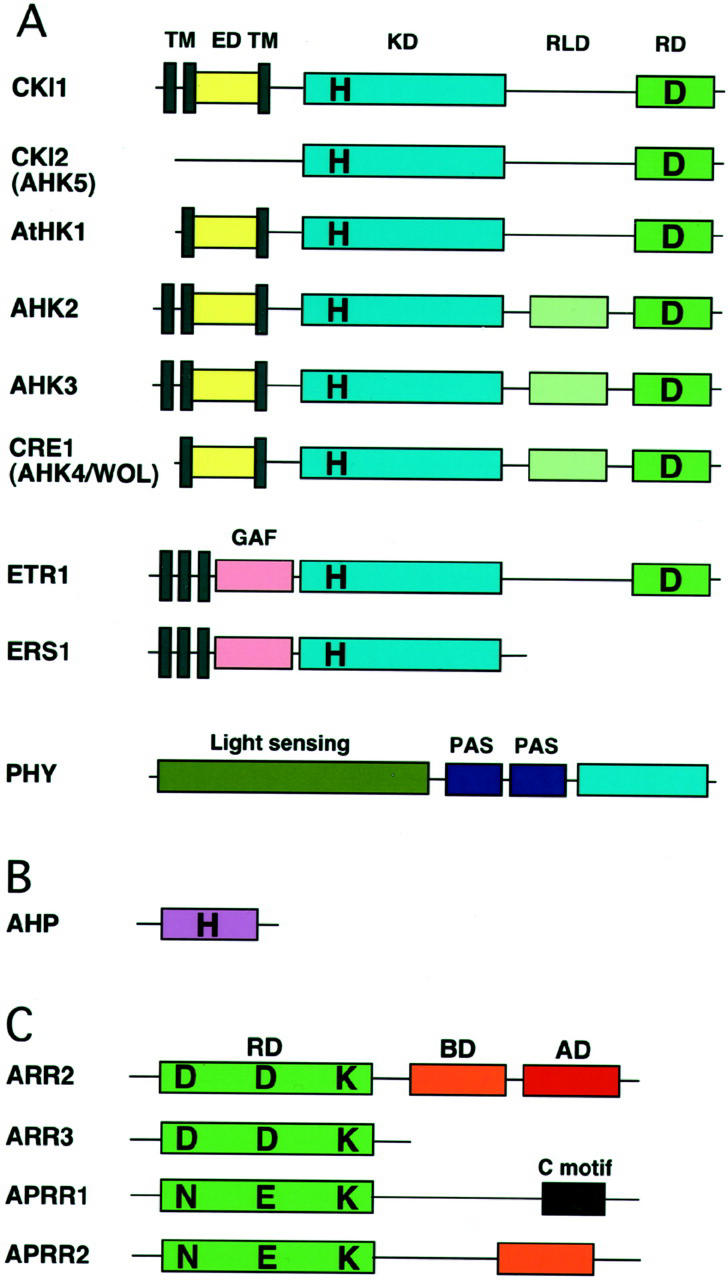
Figure 2.
Primary domain structure of representative two-component elements in Arabidopsis. A, CRE1/AHK4/WOL and CKI1 are similar in domain structure but quite diverged in amino acid sequence. ETR1 and ERS1 are representatives of the ethylene receptor family with/without a receiver domain. Phytochromes (PHY) are soluble proteins with similar overall structure and consisting of the light-sensing domain (the chromophore-binding domain), the PAS repeats, and a domain with His protein kinase homology. TM, Transmembrane domain; ED, extracellular putative input domain; KD, kinase domain; RD, receiver domain; RLD, receiver-like domain; H, His; D, Asp. B, His-containing phosphotransfer protein. C, Response regulators and response regulator-like proteins: ARR2, a B-type response regulator; ARR3, an A-type response regulator; and APRR1 and APRR2 (pseudo-type response regulators), response regulator-like proteins. ARR2 has a receiver domain followed by a DNA-binding domain (B motif) and a Pro-/Gln-rich transactivation domain, whereas ARR3 only carries a receiver domain. APRR1 and APRR2 have an atypical receiver domain similar to ARR2, but with N, E, and K motifs. In addition to a receiver-like domain, APRR1 and APRR2 have C and B motifs, respectively. Diagrams are not to scale. RD, Receiver domain; BD, DNA-binding B motif; AD, transactivation domain; D, Asp; N, Asn; E, Glu; K, Lys.
HIS PROTEIN KINASES AND RELATED PROTEINS
Sequence analysis of the entire Arabidopsis genome has revealed that there are at least 16 distinct Arabidopsis genes encoding putative His protein kinases and related proteins (Table I). Analyzed by the Clustal X program (Thompson et al., 1997), Arabidopsis histidine protein kinase homologs are represented by three distinct families: the ethylene receptors, the phytochrome photoreceptors, and the AHK family including a cytokinin receptor (CRE1/AHK4/WOL1) and a putative osmosensing receptor (AtHK1; Fig. 3; Table I). A typical His protein kinase has five conserved signature motifs, H, N, G1, F, and G2, with the conserved His as the central feature in the H motif. The other four motifs define the nucleotide-binding cleft (Fig. 4A; Stock et al., 2000). The five conserved signature motifs are known to be functionally important for His protein kinase activity (Stock et al., 2000). These motifs are not always conserved together (Chang and Stadler, 2001). Two of the ethylene receptors, ETR2 and ERS2, and all of the PHYs lack all five motifs and may not act as His protein kinases. EIN4 has the conserved His residue but is missing the other four signature motifs. Thus, only eight of the 16 genes likely possess His protein kinase activity (Fig. 4A). With the exception of ERS1, these Arabidopsis proteins are hybrid His kinases that carry both the His protein kinase domain and receiver and/or receiver-like domains at the C terminus (Figs. 2 and 4B; Chang et al., 1993; Urao et al., 2001; Schaller et al., 2002). Despite extensive analyses of the phytochrome photoreceptors and ethylene receptors, the in vivo functions of His protein kinase activities and/or His protein kinase-like domains remain unclear (Bleecker and Kende, 2000; Krall and Reed, 2000; Smith, 2000; Schaller et al., 2002). Currently, the importance of His protein kinase activities in plant signal transduction has only been demonstrated for the AHK family members (Urao et al., 1999; Hwang and Sheen, 2001; Inoue et al., 2001; Ueguchi et al., 2001).
Table I.
Putative his protein kinases and related proteins in Arabidopsis
| Namea | Arabidopsis Genome Initiative (AGI) IDb | BACc | Expressiond | AAe | Mutantf | Function | Refsg |
|---|---|---|---|---|---|---|---|
| CK11 | At2g47430 | AC002535 | Yes* | 1,122 | 1G | Cytokinin? | 1 |
| CK12 (AHK5) | At5g10720 | AL392144 | 1 Expressed sequence tag (EST) | 950 | 2G | Cytokinin? | 1 |
| AtHK1 (AHK1) | At2g17820 | AC003952 | Yes | 1,190 | 0 | Osmosensor? | 2 |
| AHK2 | At5g35750 | AB011485 | 1 ESTs | 1,173 | 2G/3S | Cytokinin | 3 |
| AHK3 | At1g27320 | AC004557 | 3 ESTs | 1,176 | 1G | Cytokinin | 3 |
| CRE1 (AHK4 and WOL1) | At2g01830 | AC007069 | 11 ESTs | 1,057 | 1S | Cytokinin | 4, 5, 6 |
| ETR1 | At1g66340 | AC020665 | Yes | 738 | 1S | Ethylene | 7 |
| ERS1 | At2g40940 | AC002409 | 6 ESTs | 613 | 1G/1S | Ethylene | 8 |
| ETR2 | At3g23150 | AB025608 | 2 ESTs | 773 | 4G/1S | Ethylene | 9 |
| EIN4 | At3g04580 | AC011437 | 4 ESTs | 766 | 1G | Ethylene | 10 |
| ERS2 | At1g04310 | AC000104 | 5 ESTs | 645 | 2G | Ethylene | 10 |
| PHYA | At1g09570 | AC003970 | 28 ESTs | 1,122 | 2G/1S | Photoreceptor | 11 |
| PHYB | At2g18790 | AC005724 | 3 ESTs | 1,172 | 1G | Photoreceptor | 11 |
| PHYC | At5g35840 | AB005236 | 2 ESTs | 1,111 | 1G/3S | Photoreceptor | 11 |
| PHYD | At4g16250 | AL161543 | 1 ESTs | 1,164 | 1G/2S | Photoreceptor | 12 |
| PHYE | At4g18130 | AL110123 | 5 ESTs | 1,112 | 3S | Photoreceptor | 12 |
Alternative names are given in parentheses.
Systematic names given to genes by Munich Information Center for Protein Sequence (MIPS) (http://mips.gsf.de/proj/thal/) from the AGI sequencing project.
Bacteria artificial chromose (BAC) accession nos. are for genomic DNA deposited in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/).
Gene expression based on cloned cDNA or the presence of EST. The no. of ESTs in the MIPS database is indicated. An asterisk indicates that there is no EST in the database.
The no. of predicted amino acids in the proteins.
The no. of Garlic (G) and SALK (S) T-DNA insertion lines available from Torrey Mesa Research Institute (TMRI)/Syngenta (http://www.tmri.org) and Salk Institute Genomic Analysis laboratory (http://signal.salk.edu), respectively.
References: 1, Kakimoto (1996); 2, Urao et al. (1999); 3, Ueguchi et al. (2001); 4, Inoue et al. (2001); 5, Mahonen et al. (2000); 6, Suzuki et al. (2001a); 7, Chang et al. (1993); 8, Hua et al. (1995); 9, Sakai et al. (1998b); 10, Hua et al. (1998); 11, Sharrock and Quail (1989); 12, Clack et al. (1994).
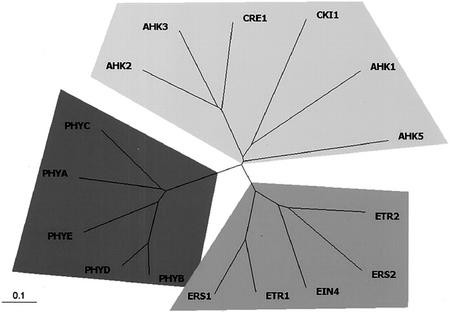
Figure 3.
Unrooted relationship tree of His protein kinases and related proteins in Arabidopsis. The tree is branched into three groups: the ethylene receptors, the photoreceptor phytochromes, and the AHK members. The entire amino acid sequences of His protein kinases were aligned by the Clustal X program (Thompson et al., 1997) and the relationship tree was produced by the TreeView program (Page, 1996). The AHK members, the ethylene receptors, and the phytochromes are in light-gray, gray, and dark-gray shade, respectively.
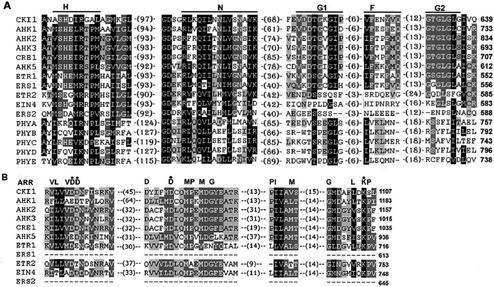
Figure 4.
Amino acid sequence alignment of His protein kinase transmitter (A) and receiver (B) domains of His protein kinases and related proteins in Arabidopsis. Sequences were aligned by the Clustal X program. Conserved amino acids are highlighted. Black and gray backgrounds indicate percentage of amino acid similarity: black, at least 75%; darker gray, 50%; and lighter gray, 25%. Amino acid similarity groups are: D, N; E, Q; S, T; K, R; F, Y, and W; and L, I, V, and M. Conserved motifs are indicated above the alignment. The numbers indicate the amino acid gaps between the motifs.
THE AHK FAMILY
The AHK family consists of six hybrid His protein kinases, AtHK1, AHK2, AHK3, AHK4 (CRE1/WOL), AHK5 (CKI2), and CKI1, with a receiver domain fused to the kinase domain, which contains the conserved His phosphorylation site (Figs. 2 and 4; T. Kakimoto, personal communication). These proteins share 38% to 69% similarity in the entire amino acid sequences. A subfamily contains three members (AHK2, 3, and AHK4/CRE1/WOL) sharing 52% to 54% amino acid sequence identity. The N terminus of AtHK1 and CKI1 consist of two or three transmembrane domains and an extracellular domain, followed by a C-terminal kinase domain and a receiver domain. AHK2, AHK3, and AHK4 (CRE1/WOL) exhibit a similar structure with an additional receiver-like domain. AHK5 (CKI2) lacks the N-terminal transmembrane domains, but instead contains two coiled domains, probably for interaction with other proteins in the cytoplasm (Fig. 2A).
Similar to the bacterial and yeast osmosensing pathways, a plant His protein kinase, AtHK1, has been implicated in osmosensing (Urao et al., 1999). Expression of AtHK1 complemented a yeast double mutant (sln1 sho1) lacking its two osmosensors and allowed the mutant to grow in high-salt media. Interestingly, the AtHK1 transcript is most abundant in roots and is up-regulated by external osmolarity changes (Urao et al., 1999). However, the precise function of AtHK1 in plant cells requires further investigation.
Recent genetic and molecular studies have provided strong evidence that cytokinins are perceived by AHK2, 3, and CRE1/AHK4/WOL (Kakimoto, 1996; Hwang and Sheen, 2001; Inoue et al., 2001; Suzuki et al., 2001a; Ueguchi et al., 2001). Explants of the cre1-1 (cytokinin response 1) mutant showed a lack of cytokinin responses such as cell proliferation, greening, and shoot formation (Inoue et al., 2001). The wol (wooden leg) mutants displayed defects in the root vascular system due to the impairment of proper asymmetric cell division in the early stages of embryogenesis (Mahonen et al., 2000). These phenotypes bring together the role of CRE1/WOL as a cytokinin receptor and demonstrate cytokinin functions in cell division. In heterologous yeast expression systems, CRE1/AHK4 complemented budding and fission yeast mutants that lacked the endogenous His protein kinase and restored the ability to grow in a cytokinin-dependent manner. The conserved His and Asp mutations abolished a cytokinin-dependent growth (Inoue et al., 2001; Suzuki et al., 2001a). Cytokinin-dependent His protein kinase activity of AHK4 was also shown to complement an Escherichia coli mutant (Suzuki et al., 2001a). In addition, binding assays in yeast revealed that AHK4 proteins directly bind active cytokinins such as isopentenyladenine, t-zeatin, and benzyladenine with a Km of 4.5 nm (Yamada et al., 2001). In Arabidopsis protoplasts, expression of CRE1/AHK4/WOL enhanced binding of t-zeatin on the cell surface (I. Hwang and J. Sheen, unpublished data). Interestingly, the wol mutation in the putative extracellular ligand-binding domain disrupted cytokinin binding to CRE1/AHK4/WOL (Mahonen et al., 2000; Yamada et al., 2001). These experiments provide compelling evidence that CRE1/AHK4/WOL is a cytokinin receptor and perceives extracellular cytokinins. In Arabidopsis protoplasts, CRE1/AHK4/WOL enhanced the promoter activity of a primary cytokinin-responsive gene ARR6 in a cytokinin-dependent manner (Hwang and Sheen, 2001). Furthermore, CRE1/AHK4/WOL with mutations in the conserved His and Asp residues exerted dominant negative effects and diminished the cytokinin response, suggesting that His protein kinase activity and phosphoryl transfer are required for CRE1/AHK4/WOL function in cytokinin signaling (Hwang and Sheen, 2001).
Interestingly, although cytokinin is important for leaf and shoot meristem development, wol and cre1 mutant plants lack obvious mutant leaf and shoot phenotypes. Because CRE1/AHK4/WOL is predominantly expressed in roots (Mahonen et al., 2000), cytokinin perception may be exerted by the closely related AHK2 and AHK3 in leaf and shoot development in wild-type plants. AHK3 is expressed in root, leaf, flower, and stem, and AHK2 is expressed in root, leaf, and flower (Ueguchi et al., 2001). Although AHK2 could not complement the yeast His protein kinase mutant (Ueguchi et al., 2001), AHK2 expression in Arabidopsis protoplast could activate the cytokinin-responsive ARR6 promoter in a cytokinin-dependent manner (Hwang and Sheen, 2001). In addition, AHK3 also enhanced promoter activity of the cytokinin-responsive ARR6 gene in the presence of cytokinin (Hwang and Sheen, 2001) even though it complemented the yeast His kinase mutant in the absence of cytokinin (Ueguchi et al., 2001). More recent progress has shown that AHK2 and AHK3 could complement the yeast sln1 His protein kinase mutant in a cytokinin-dependent manner (T. Kakimoto, personal communication). Based on the analysis using the PSORT program (Nakai and Horton, 1999), AHK2, AHK3, and CRE1/AHK4/WOL are probably localized to the plasma membrane (Inoue et al., 2001; Ueguchi et al., 2001).
CKI1, a hybrid His protein kinase with one conserved receiver domain, has also been implicated in cytokinin signaling. Overexpression of CKI1 confers cytokinin-independent cell division and shoot formation on transgenic callus (Kakimoto, 1996). CKI1 has low amino acid sequence similarity to CRE1 with only 25% identity and the putative extracellular domain (presumably the ligand-binding domain) of CKI1 is completely different from that of CRE1. This sequence divergence and the failure of cytokinin binding to CKI1 in yeast argue against the function of CKI1 as a cytokinin receptor (Yamada et al., 2001). In Arabidopsis protoplasts, however, the expression of CKI1 activated the cytokinin-responsive ARR6 promoter in the absence of exogenous cytokinin, indicating that CKI1 is a constitutively active His protein kinase connected to cytokinin signaling. Mutations in the conserved His and Asp residues lost their ability to induce a cytokinin response in the protoplast system (Hwang and Sheen, 2001), suggesting that CKI1 is involved in cytokinin signaling. CKI1 is localized to the plasma membrane (Hwang and Sheen, 2001). Currently, we cannot exclude the possibility that overexpression of CKI1 provides higher kinase activities that nonspecifically activate unrelated signaling pathways. Another possibility is that CKI1 might be part of a cytokinin receptor complex or recognizes cytokinins in a manner distinct from that of CRE1/AHK4/WOL. Recent studies have shown that CKI1 is expressed in the ovule and endosperm, but not in the embryo, based on the expression patterns of a CKI1::GUS reporter construct in transgenic plants (T. Kakimoto, personal communication). Analyses of CKI1 mutants suggest that CKI1 is involved in female gametophyte development and homozygous cki1 mutant is lethal (Pischke et al., 2001). It remains to be shown whether cytokinin plays a role in reproductive organ development.
The function of AHK5 (CKI2) may also be related to cytokinin signaling. The dominant cki2 mutant was obtained in an activation-tagging screen, which also identified the dominant cki1 mutant (Kakimoto, 1996). A further analysis of AHK5 (CKI2) is necessary to elucidate its precise cellular functions.
THE ETHYLENE RECEPTOR FAMILY
Molecular and genetic studies have suggested that ethylene is perceived by a family of five receptors (ETR1, ETR2, EIN4, ERS1, and ERS2) in Arabidopsis (Chang et al., 1993; Hua et al., 1995, 1998; Sakai et al., 1998b; for review, see Bleecker and Kende, 2000; Chang and Stadler, 2001). The ethylene receptor family shows characteristic features of an N-terminal ethylene-binding transmembrane domain, a GAF protein-protein interaction domain, and a His protein kinase domain, but falls into two subfamilies based on amino acid sequence similarity: ETR1 and ERS1 with 68% identity and 80% similarity, and ETR2, EIN4, and ERS2 with 47% to 48% identity and 61% to 63% similarity (Fig. 4). ETR1 and ERS1 have all five typical His protein kinase motifs, but the other subfamily members lack most of the consensus motifs of a His protein kinase. Interestingly, ETR1, ETR2, and EIN4 carry a receiver domain, but not ERS1 and ERS2. Because these five proteins perform partially redundant functions, it is possible that they form complexes and carry out the phosphotransfer process (Hua and Meyerowitz, 1998; Hua et al., 1998; Urao et al., 2000). The analysis of etr1 ers1 double mutants will be useful to determine the role of His protein kinase activity in ethylene signaling.
It has been demonstrated that ethylene binds to ETR1 and ERS1 in yeast (Schaller and Bleecker, 1995; Rodriguez et al., 1999; Hall et al., 2000). It remains to be determined whether ethylene binds to ETR2, EIN4, and ERS2. Despite the genetic evidence that the receptors are negatively regulated by the ethylene signal (Hua and Meyerowitz, 1998), the biochemical mechanism of their action, such as modulation of His protein kinase activity by ethylene binding, is still unclear. The similarity of the ethylene receptors to sensor His protein kinases and the finding that ETR1 has His protein kinase activity in vitro (Gamble et al., 1998) suggest that ethylene signaling could be mediated by a two-component system. Currently, there is no evidence for the involvement of AHPs or ARRs in ethylene signaling. Instead, CTR1 (a RAF-like protein kinase), which may activate an MAPK cascade, has been proposed to be a direct target of ethylene receptor action (Kieber et al., 1993; Clark et al., 1998). However, it remains possible that additional ethylene signaling pathways mediated by two-component proteins occur in addition to the CTR1 signaling pathway (Lohrmann and Harter, 2002).
THE PHYTOCHROME FAMILY
Phytochromes are photoreceptors enabling plants to regulate growth and development in response to light signals (for review, see Genick and Chory, 2000; Smith, 2000). The cyanobacterial phytochrome Cph1 is a light-regulated His protein kinase and mediates phosphotransfer to the response regulator Rcp1 (Yeh et al., 1997). This discovery and sequence similarity of plant phytochromes to bacterial His protein kinases suggested that higher plant phytochromes might be His protein kinases and that light signaling in higher plants could use a light–regulated phosphotransfer mechanism. In Arabidopsis, there are five photoreceptor phytochromes: PHYA, PHYB, PHYC, PHYD, and PHYE (Sharrock and Quail, 1989; Clack et al., 1994). These phytochromes have two major structural domains (Fig. 2). The amino-terminal domain has a covalently attached linear tetrapyrrole chromophore for light absorption and photoreversibility. The carboxy terminus consists of two PAS domains and a domain related to the His protein kinase for signal transduction. Plant phytochromes are soluble proteins with structural features similar to those of sensor His protein kinases, with an N-terminal sensor and a C-terminal His protein kinase domain. However, none of the phytochromes contain the five conserved motifs essential for His protein kinase activity (Fig. 4). Analyses of phyB mutants have suggested that the His protein kinase-related domain is important for PHYB signaling, but removal of this domain does not eliminate PHYB activity (Krall and Reed, 2000). In addition to the direct interactions with NDPK2 or transcription factors (Smith, 2000), it has been proposed that plant phytochromes exert Ser/Thr, not His, kinase activity in response to light (McMichael and Lagarias, 1990). Oat (Avena sativa) phytochrome could be autophosphorylated on Ser/Thr residues in a light-dependent manner (Yeh and Lagarias, 1998). Using the yeast two-hybrid screen, a PHYA substrate PKS1 has been identified in Arabidopsis (Fankhauser et al., 1999). PKS1 encodes a basic soluble protein. These studies indicate that plant phytochromes have diverged from the ancestral His protein kinase as Ser/Thr kinases with a new activity.
HIS PHOSPHOTRANSFER PROTEINS
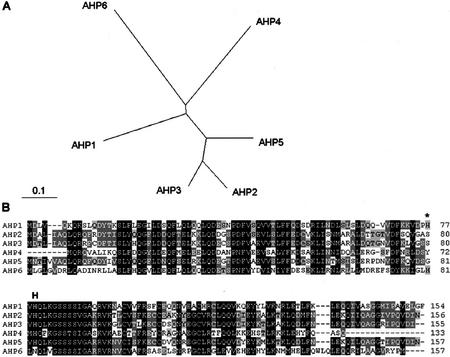
The functional importance of hybrid His protein kinases implicates the necessity of another player in the HIS-to-ASP phosphorelay to serve as an intermediate by acquiring and transferring phosphate to separate receiver proteins, ARRs. Five Arabidopsis genes, AHP1 through AHP5, encode putative intermediate proteins with a His phosphotransfer domain (Table II). All of them contain the highly conserved XHQXKGSSXS motif, which includes the His phosphorylation site (Fig. 5). Further extensive search of the Arabidopsis genome revealed a sixth gene (AHP6/APHP1) in which the His residue in the phosphorylation motif is replaced by an Asn. However, a potential His phosphorylation residue is found two amino acids away (Fig. 5). The amino acid sequences of AHP2 and AHP3 show 81% identity, suggesting possible functional redundancy of these genes. AHP2 has 45% identity with AHP1. Functional complementation analysis using a yeast ypd1 mutant demonstrated that AHP1, AHP2, and AHP3 could act as phosphorelay intermediates (Miyata et al., 1998; Suzuki et al., 1998). Yeast two-hybrid assays have been used to show that AHP1, AHP2, and AHP3 interact with ARR1. AHP2 also interacts with other B-type ARRs, ARR2, and ARR10, but not with A-type ARRs such as ARR3 and ARR4 (Suzuki et al., 2001b). Although phosphotransfer from AHP1 or AHP2 to ARR3 and 4 has been reported (Suzuki et al., 1998; Imamura et al., 1999), it is likely that AHPs preferentially interact with B-type ARRs. In vitro studies showed that AHP1 is able to acquire a phosphoryl group at the conserved His site (Suzuki et al., 1998). AHP proteins could interact with hybrid His protein kinases such as AtHK1, ETR1, CKI1, and CRE1 in the yeast and E. coli two-hybrid assays (Urao et al., 2000a; Suzuki et al., 2001a). Thus, AHPs could be phosphoryl-transfer intermediate proteins in multistep phosphorelay signal transduction. Analyses of AHP-green-fluorescent protein (GFP) fusions revealed that AHP1 and AHP2, but not AHP5, are translocated from the cytoplasm to the nucleus in a cytokinin-dependent manner (Hwang and Sheen, 2001). The results support the idea that AHPs form a physical link between the plasma membrane-localized sensor kinase and the nuclear response regulators in cytokinin signaling (Fig. 9). Ectopic expression of AHP2 caused hypersensitivity to cytokinin and inhibited root and hypocotyl elongation in the dark (Suzuki et al., 2002). However, the specific phosphorelay of AHPs from hybrid His protein kinases to ARRs has not been demonstrated. The action of AHPs does not seem to be rate limiting because overexpression of AHP proteins does not affect cytokinin-responsive reporter gene expression (Hwang and Sheen, 2001). Considering the larger number of sensors and response regulators, His protein kinases probably converge on and share AHP proteins to direct different signals on response regulators. The functional specificity of AHPs could be determined by other interacting proteins. Such functional interactions might be as important as the phosphorelay itself to transmit specific inputs from different signals and sensors to specific regulatory outputs.
Table II.
His-containing phosphotransfer proteins in Arabidopsis
| Namea | AGI IDb | BACc | Expressiond | AAe | Mutantf | Function | Refsg |
|---|---|---|---|---|---|---|---|
| AHP1 (AtHP3) | At3g21510 | AB019232 | 8 ESTs | 154 | 2G | Cytokinin | 1, 2 |
| AHP2 (AtHP1) | At3g29350 | AP001309 | Yes* | 156 | 1G/2S | Cytokinin | 1, 2 |
| AHP3 (AtHP2) | At5g39340 | AB009054 | Yes | 155 | 1S/1S | – | 1, 2 |
| AHP4 | At3g16360 | AB023046 | Yes | 127 | 1G/1S | – | 3 |
| AHP5 | At1g03430 | AC002560 | Yes | 157 | 2G | – | 3 |
| AHP6 (APHP1) | At1g80100 | AC009322.1 | – | 157 | 1G | – | 3 |
Alternative names are given in parentheses.
Systematic names given to genes by MIPS (http://mips.gsf.de/proj/thal/) from AGI sequencing project.
BAC accession nos. are for genomic DNA sequences deposited in NCBI database (http://www.ncbi.nlm.nih.gov/).
Gene expression based on northern analysis, cloned cDNA, or the presence of EST. The no. of ESTs in the MIPS database is indicated. An asterisk indicates that there is no EST in the database.
The no. of predicted amino acids.
The no. of Garlic (G) and SALK (S) T-DNA insertion lines available from TMRI/Syngenta (http://www.tmri.org) and Salk Institute Genomic Analysis laboratory (http://signal.salk.edu), respectively.
References: 1, Miyata et al. (1998); 2, Suzuki et al. (1998); 3, Suzuki et al. (2000).
Figure 5.
His-containing phosphotransfer proteins in Arabidopsis. A, Unrooted relationship tree of His-containing phosphotransfer proteins (AHPs). Programs used were Clustal X for alignment and TreeView for graphical output. The entire amino acid sequences were aligned. B, Alignment of deduced amino acid sequences of His phosphotransfer proteins in Arabidopsis. AHP1-5 contains the highly conserved XHQXKGSSXS motif, which includes the His phosphorylation site. The putative His phosphorylation residue of AHP6 is replaced by an Asn. H, Conserved His phosphorylation site; the asterisk marks a potential His phosphorylation residue of AHP6.
Figure 9.
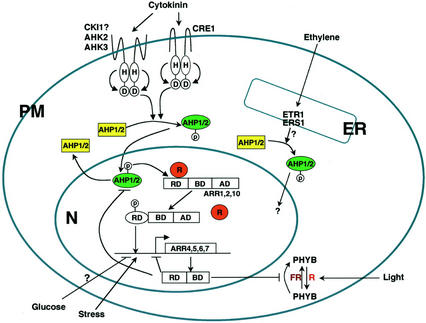
Model of the two-component signal transduction pathways in Arabidopsis. The cytokinin signal is perceived by multiple His protein kinases at the plasma membrane. Upon perception of the cytokinin signal, His protein kinases initiate a signaling cascade via the phosphorelay that results in the nuclear translocation of AHPs (Hwang and Sheen, 2001). Activated AHPs may interact with sequestered ARRs or ARR complexes, transfer the phosphate to the receiver domain of its cognate B-type ARR, releasing these activation-type ARRs from putative repressors in the nucleus. The dephosphorylated AHP shuttles back to the cytosol, where it can be rephosphorylated. The liberated ARRs bind to multiple cis elements in the promoter of target genes. The activation of the repressor-type ARRs as primary cytokinin response genes provides a negative feedback mechanism. In addition to the CTR1 signaling pathway, additional ethylene signaling pathways could be mediated by two-component components (Lohrmann and Harter, 2002). Red light and cytokinin signaling is converged at ARR4. ARR4 stabilizes the active form of PHYB by inhibiting dark reversion (Sweere et al., 2001). Stress and Glc may also modulate two-component signaling (Urao et al., 1998; F. Rolland and J. Sheen, unpublished data). RD, Response domain; BD, DNA binding domain; AD, transactivation domain; PM, plasma membrane; N, nucleus; R, putative repressor; FR, far-red light.
RESPONSE REGULATORS
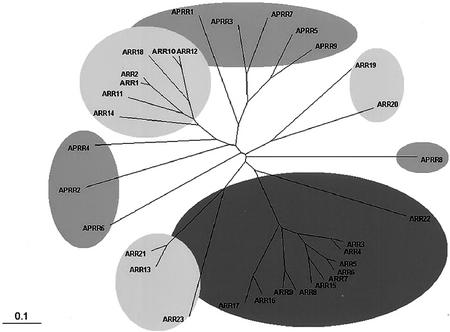
The completion of the Arabidopsis genome sequence has also revealed the existence of 32 genes encoding putative response regulators and related proteins that are not fused to the His protein kinase domain (Fig. 6; Table III; see also Schaller et al., 2002). Alignment of the predicted amino acid sequences shows that the receiver domains of Arabidopsis response regulators contain the conserved Asp and Lys residues, which are the hallmark in response regulators of prokaryotes and yeasts (Fig. 7). Based on the predicted protein domain structures and amino acid sequences, these response regulators fall into three distinct subfamilies: the A-type ARRs with only the receiver domain, the B-type ARRs with the receiver domain fused to the DNA-binding domain, and the pseudoresponse regulators with the atypical receiver domain (Figs. 2 and 7; Table III). Members of the APRR (Arabidopsis pseudoresponse regulator) family share significant sequence similarity with ARRs in the putative receiver domain, but they do not have conserved D-D-K motifs in the receiver domain (Fig. 2).
Figure 6.
Unrooted relationship tree of response regulators and response regulator-like proteins in Arabidopsis. The amino acid sequences of receiver domains of response regulators and response regulator-like proteins in Arabidopsis were aligned by the Clustal X program and the relationship tree was produced by the TreeView program. Response regulators and response regulator-like proteins in Arabidopsis are divided into three major groups: A type (11 genes), B type (12 genes), and pseudoresponse regulator (nine genes). We categorized proteins as response regulator-like (pseudoresponse regulators) if they did not have the conserved phosphate-accepting Asp within the receiver domain. A-, B-, and pseudo-type response regulators are in dark-gray, light-gray, and gray shade, respectively.
Table III.
Response regulators and response regulator-like proteins in Arabidopsis
| Namea | AGI IDb | BACc | Expressiond | AAe | Mutantf | Function | Refsg |
|---|---|---|---|---|---|---|---|
| A-type response regulator | |||||||
| ARR3 | At1g59940 | AC007258 | 7 ESTs | 231 | 2G/1S | Cytokinin | 1 |
| ARR4 (ATRR1 and IBC7) | At1g10470 | AC007067 | 12 ESTs | 259 | 4G/3S | Cytokinin/light | 1–3 |
| ARR5 (ATRR2 and IBC6) | At3g48100 | AL049658 | 7 ESTs | 184 | 2G | Cytokinin | 1–3 |
| ARR6 | At5g62920 | AB009053 | 8 ESTs | 186 | 2G/1S | Cytokinin | 1 |
| ARR7 | At1g19050 | ACC068602 | Yes* | 206 | 2G | Cytokinin | 1 |
| ARR8 (ATRR3) | At2g41310 | AC005662 | 1 EST | 225 | 4G | Cytokinin | 3, 4 |
| ARR9 (ATRR4) | At3g57040 | AL138655 | 1 EST | 234 | 3G/2S | Cytokinin | 3, 4 |
| ARR15 | At1g74890 | AC008263 | Yes | 206 | 2G | – | 5 |
| ARR16 | At2g40670 | AC007660 | – | 164 | 3G | – | 5 |
| ARR17 | At3g56380 | AL163972 | – | 122 | 3G | – | 5 |
| ARR22 | At3g04280 | AC016829 | – | – | 1G | – | – |
| B-type response regulator | |||||||
| ARR1 | At3g16857 | AP001308 | Yes | 690 | 5S | Cytokinin | 6 |
| ARR2 (ARP5) | At4g16110 | AL161543 | 1 EST | 644 | 1G/1S | Cytokinin | 6 |
| ARR10 (ARP4) | At4g31920 | AL021636 | 1 EST | 552 | 1G | Cytokinin | 4, 7 |
| ARR11 (ARP3) | At1g67710 | AC008113 | Yes | 521 | 4G/2S | – | 4, 7 |
| ARR12 | At2g25180 | AC018722 | 4 ESTs | 573 | 2G/1S | – | 4 |
| ARR13 | At2g27070 | AC005623 | – | 575 | 4G/1S | – | 4 |
| ARR14 | At2g01760 | AC006069 | 1 EST | 382 | 2G | – | 4 |
| ARR18 | At5g58080 | AB024029 | 2 ESTs | 632 | 6G | – | – |
| ARR19 | At1g49190 | AC016041 | – | 612 | 0 | – | – |
| ARR20 | At3g62670 | AL162651 | – | 426 | 3G | – | – |
| ARR21 | At5g07210 | AL163652 | – | 621 | 1S | – | – |
| ARR23 | At5g62120 | AB016880.1 | – | 145 | 1G/1S | – | – |
| Response regulator-like protein | |||||||
| APRR1 (TOC1) | At5g61380 | AB010073 | 10 ESTs | 618 | toc1 | Circadian | 8–11 |
| APRR2 | At4g18020 | AL021889 | 5 ESTs | 483 | 3G/3S | – | 8 |
| APRR3 | At5g60100 | AB019231 | – | 495 | 2G | Circadian | 12 |
| APRR4 | At5g49240 | AB016872 | – | 558 | 5G | – | |
| APRR5 | At5g24470 | AB025641 | 6 ESTs | 558 | 3S | Circadian | 12 |
| APRR6 | At1g68210 | AC016447 | – | – | 1G | – | |
| APRR7 | At5g02810 | AL162973 | 7 ESTs | 720 | 2G/2S | Circadian | 12 |
| APRR8 | At4g00760 | AF013294 | – | – | 1G/1S | – | |
| APRR9 (TL1) | At2g46790 | AC005310 | Yes | 468 | 1G/3S | Circadian | 9, 12 |
Alternative names are given in parentheses.
Systematic names given to genes by MIPS (http://mips.gsf.de/proj/thal/) from AGI sequencing project.
BAC accession nos. are for genomic DNA sequences deposited in NCBI database (http://www.ncbi.nlm.nih.gov/).
Gene expression is based on northern analysis, cloned cDNA, or the presence of EST. The no. of ESTs in the MIPS database is indicated. An asterisk indicates that there is no EST in the database.
The no. of predicted amino acids.
The no. of Garlic (G) and SALK (S) T-DNA insertion lines available from TMRI/Syngenta (http://www.tmri.org) and Salk Institute Genomic Analysis laboratory (http://signal.salk.edu), respectively, is presented. The toc1 mutant has a point mutation (Strayer et al. 2000).
References: 1, Imamura et al. (1998); 2, Brandstatter and Kieber (1998); 3, Urao et al. (1998); 4, Imamura et al. (1999); 5, D'Agostino et al. (2000); 6, Sakai et al. (1998a); 7, Lohrmann et al. (1999); 8, Makino et al. (2000); 9, Strayer et al. (2000); 10, Makino et al. (2002); 11, Matsushika et al. (2002); 12, Matsushika et al. (2000).
Figure 7.
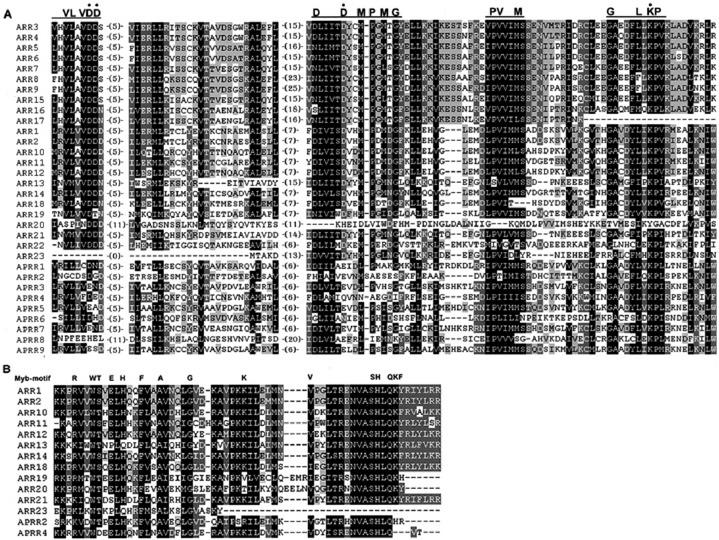
Alignment of deduced amino acids sequences of response regulators and response regulator-like proteins in Arabidopsis. A, The amino acid sequences of receiver domains of response regulators and response regulator-like proteins were aligned. The highly conserved amino acids are highlighted. The three conserved motifs are indicated above the alignments. The numbers indicate the amino acid gaps between the motifs. B, Alignment of putative DNA-binding B motifs of B-type response regulators and related proteins. The predicted amino acid sequences of the B motifs were aligned with the conserved Myb DNA-binding motif.
THE A-TYPE ARR FAMILY
The cDNA of seven members of the Arabidopsis A-type response regulators, ARR3 through ARR9, were first cloned by searching Arabidopsis expressed sequence tags (ESTs) based on homologies to the sequences of cyanobacteria response regulators (Imamura et al., 1998, 1999). Their domain structures are similar to the CheY E. coli response regulator involved in chemotaxis (Imamura et al., 1998). Further Arabidopsis EST and genomic database searches revealed that there are three additional A-type ARR family members, ARR15, ARR16, ARR17, and ARR22 (Fig. 6; Table III). The A-type ARRs are mainly composed of a receiver domain and short N- and C-terminal extensions without any typical output domain (Fig. 2). The amino acid sequences of receiver domains show 50% to 93% identity among A-type response regulators, but are very distinct from those of the B-type ARRs, displaying less than 30% amino acid identity in the receiver domain (Fig. 7A). Interestingly, five pairs of highly homologous ARRs (Fig. 6) are located on the duplicated regions in chromosome 1 (ARR3 and ARR4, ARR7 and ARR15), chromosomes 3 and 5 (ARR5 and ARR6), and chromosomes 2 and 3 (ARR8 and ARR9, ARR16 and ARR17) of the Arabidopsis genome (Fig. 8; AGI, 2000; Vision et al., 2000). These ARRs are likely to have redundant functions. ARR22 is more similar to the receiver domains of the hybrid kinases than to the other response regulators (Schaller et al., 2002). The expression of ARR3 to ARR9 was induced very rapidly and specifically by cytokinin (Brandstatter and Kieber, 1998; Taniguchi et al., 1998; Kiba et al., 1999), indicating that these A-type ARR genes are primary cytokinin response targets and possibly involved in cytokinin signaling. The expression patterns of an ARR5::GUS reporter construct revealed the presumed endogenous cytokinin action sites in transgenic plants, such as the shoot and root meristems. However, treatment with exogenous cytokinin activated ARR5::GUS expression ubiquitously (D'Agostino et al., 2000). Thus, the cytokinin signaling pathway may exist in most cell types (Hwang and Sheen, 2001). Nitrate application also activated ARR3 through ARR9 expression (Taniguchi et al., 1998; Kiba et al., 1999), presumably due to the elevation of cytokinin levels by nitrate (Takei et al., 2001). Expression of ARR4 and ARR5 was also found to be sensitive to environmental stress such as drought, salt, and low temperature (Urao et al., 1998), suggesting a molecular link between stress and cytokinin signaling.
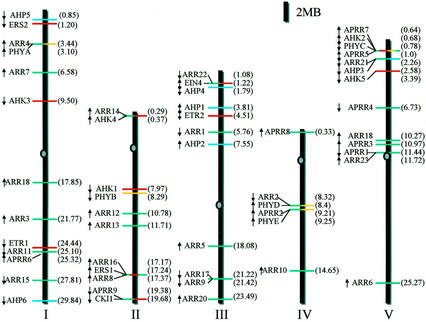
Figure 8.
Locations of putative two-component regulators on the Arabidopsis chromosomes. Ovals on the chromosomes represent the centromeres. The arrows show the direction of transcription. The numbers in parentheses indicate the position of the first exon of each two-component gene from one end of the chromosome in megabase (Mb). The bars represent AHKs and ethylene receptors (red), phytochromes (yellow), ARRs (green), and AHPs (blue).
In a protoplast transient expression system, ARR4, ARR5, ARR6, and ARR7 acted as negative regulators to repress the expression of the cytokinin-responsive ARR6 promoter (Hwang and Sheen, 2001), providing a negative feedback mechanism in cytokinin signaling. The distinct target genes of ARRs and relative degree of the negative regulation may direct specific signaling mechanisms to mediate particular cytokinin responses in different cell types during plant growth and development. GFP fusions of ARR5, ARR6, and ARR7 are exclusively localized in the nucleus and this localization is not affected by cytokinin treatment, whereas ARR4-GFP is localized both in the cytoplasm and nucleus (Hwang and Sheen, 2001; Sweere et al., 2001; I. Hwang and J. Sheen, unpublished data). Interestingly, recent studies have found that ARR4 interacts with the N terminus of PHYB and stabilizes its active form by inhibiting the dark reversion (Sweere et al., 2001). Because PHYB does not have His protein kinase activity, it is unlikely that ARR4 is a direct target of PHYB. This result raises the possibility that ARR4 could serve as a signaling module at which cytokinin and light signal transduction pathways converge to integrate information from these two signals (Hwang and Sheen, 2001; Lohrmann and Harter, 2002). These studies support the idea that response regulators could be key components in plant signal transduction at which different signaling pathways interact and integrate. Currently, the physiological roles of most A-type ARRs in plant development remain mostly unknown (Table III). Further functional analyses are required to elucidate their precise role in plant signal transduction.
THE B-TYPE ARR FAMILY
There are 12 putative B-type ARRs in the Arabidopsis genome (Figs. 2 and 7; Table III). The amino acid sequences of the receiver domains have 30% to 72% identity among B-type ARRs, with the exception of ARR23. Although ARR23 contains the conserved phosphoryl-accepting Asp, it might be a pseudogene because its predicted coding region has only 145 amino acids and a partial B motif (Fig. 7; Table III; Schaller et al., 2002). In addition to the N-terminal receiver domain, B-type ARRs have a large C-terminal region (Fig. 2). This C terminus contains an 80-amino acid stretch (B motif) that distantly resembles the DNA-binding domain of the c-Myb proto-oncogene protein and a glutamine-rich domain (Lohrmann et al., 1999, 2001; Sakai et al., 2000). The conserved B motif of the B-type response regulators suggests a role as transcription factors. The B motifs of ARR1 and ARR2 bound synthetic oligonucleotides with a 5′-(A/T) GAT(A/T)-3′ core, and their C-terminal halves functioned as a transactivation domain when fused to the GAL4 DNA-binding domain in bombarded plant cells (Sakai et al., 2000). ARR2 could also bind specifically to a conserved promoter of the plant nCI genes (the nuclear genes for mitochondrial respiratory complex I; Lohrmann et al., 2001). It was also shown that the C-terminal domain of ARR11 could activate transcription when fused to the GAL4 DNA-binding domain (Lohrmann et al., 1999). The presence of a potential nuclear localization signal VRK(R/K) R in the C-terminal regions of ARR1 and ARR2 is another indication that these B-type ARRs are transcriptional factors. Transient expression of the GFP fusions of ARR1, ARR2, ARR10, and ARR11 showed their nuclear localization in Arabidopsis protoplasts (Hwang and Sheen, 2001; I. Hwang and J. Sheen, unpublished data). Deletion of the receiver and the transactivation region or mutation of the conserved Asp in the receiver domain did not affect nuclear localization of ARR2 (Hwang and Sheen, 2001).
Emerging evidence supports the model that the B-type ARRs control cytokinin-inducible A-type ARR gene expression (Hwang and Sheen, 2001; Sakai et al., 2001) and are key players in cytokinin signaling. For instance, ARR1 overexpression in transgenic Arabidopsis plants caused hypersensitivity to cytokinin in shoot regeneration, inhibition of root elongation, and induction of several A-type ARR genes expression, whereas an arr1 mutant appeared to be partially resistant to cytokinin (Sakai et al., 2001). The lack of overt phenotypes in the arr1 mutant could be due to functional redundancy provided by other B-type ARRs. In Arabidopsis protoplasts, overexpression of several B-type ARRs such as ARR1, ARR2, and ARR10 activated the promoter of the cytokinin-responsive gene, ARR6 (encoding an A-type ARR), in the absence of cytokinin. Cytokinin treatment further induced ARR6 promoter activity (Hwang and Sheen, 2001). The function of ARR2 in cytokinin signaling was further confirmed in transgenic Arabidopsis plants where overexpression of ARR2 mimicked cytokinin effects and promoted cell proliferation and shoot formation in tissue cultures in the absence of exogenous cytokinin (Hwang and Sheen, 2001). Using the established protoplast system and transgenic assays, the function of other members of the B-type ARR family in cytokinin signaling can now be tested. However, to elucidate the precise functions of all ARR genes, examining their expression patterns in plants is critical. The expression of ARR1, ARR2, ARR10, and ARR11 are generally detectable in all tissues, but they do display distinct spatial expression patterns (Urao et al., 1998; Lohrmann et al., 1999). ARR1 and ARR2 are predominantly expressed in roots (Sakai et al., 1998a), in which the expression of ARR10 and ARR11 is hardly detectable (Lohrmann et al., 1999). Thus, different members of the B-type ARR family could be responsible for cytokinin actions in distinct cell types or act in other two-component signaling pathways. In contrast to A-type ARRs, the expression of B-type ARRs is not affected by either cytokinins or nitrate or other plant hormones (Imamura et al., 1999; Kiba et al., 1999; Lohrmann et al., 1999).
In a plant transient assay, the truncation of N-terminal receiver domain of ARR1 led to an increase in the transcription activation function of ARR1, and in transgenic plants, the truncated ARR1 increased the expression of A-type ARR genes (Sakai et al., 2000, 2001). It was proposed that cytokinin-dependent phosphorylation of ARR1 could relieve the inhibition imposed by the receiver domain. However, overexpression of the wild-type ARR1 and ARR2 in protoplasts or transgenic plants is sufficient to mimic cytokinin responses in the absence of exogenous cytokinin (Hwang and Sheen, 2001; Sakai et al., 2001). In addition, the mutation of the presumed phospho-accepting Asp residue in ARR2 did not alter its transcription activity based on the ARR6 promoter assay (Hwang and Sheen, 2001). An alternative explanation for the importance of the two-component phosphorelay could be that the cytokinin-dependent phosphotransfer to the receiver domain activates ARR proteins by liberating them from endogenous repressors (Fig. 9). Phosphorylation of the Asp residue in the receiver domain of ARRs may not directly regulate nuclear localization, DNA binding, or intrinsic transcription activities.
THE ARR-LIKE FAMILY
Extensive analyses of the Arabidopsis genome sequence revealed the presence of nine genes encoding ARR-like proteins, called pseudoresponse regulators (APRR1–APRR9; Table III; Makino et al., 2000). The receiver domains of APRRs show 30% to 36% identity with the receiver domain of ARR1. However, the three invariant amino acid residues (D-D-K) in typical bacteria and yeast RRs are not conserved (Figs. 2 and 7A). The phosphate-accepting Asp (D) of the receiver-like domain is replaced by Glu (E), Asn (N), or Gln (Q). However, despite significant sequence divergence, many APRRs have Asp residues in the conserved motifs (Fig. 7A) and could still act as the final outputs of two-component phosphorelay in plants.
Some APRR proteins contain a distinctive C-terminal motif identified within the CO (CONSTANS) transcription factors (Makino et al., 2000; Strayer et al., 2000). The C motif (or CCT motif) is rich in basic amino acids (Arg and Lys) and contains a putative nuclear localization signal (Makino et al., 2000; Strayer et al., 2000). The function of APRR1/TOC1 is best understood from the analysis of the toc1 (timing of CAB expression) mutant with shortened periods of circadian rhythms (Strayer et al., 2000). The APRR1/TOC1 protein is localized in the nucleus and regulates photoperiodic control of flowering. The expression of APRR1/TOC1 itself is also subject to circadian rhythm (Putterill et al., 1995; Kobayashi et al., 1999; Makino et al., 2000; Strayer et al., 2000). Interestingly, the APRR2 and APRR4 proteins contain a DNA-binding B motif at their C terminus (Fig. 7B). Analyses of APRR1 and APRR2 in vitro showed that their receiver-like domains did not contain phospho-accepting activity (Makino et al., 2000). Expression of several APRR genes is controlled by circadian rhythm with a coordinated sequential expression of APRR9, APRR7, APRR5, APRR3, and APRR1/TOC1 after dawn in a 24-h photoperiod (Matsushika et al., 2000, 2002; Makino et al., 2001, 2002). These rhythmic events of APRR transcription are proposed to be a basis of the presumed Arabidopsis circadian clock, but their physiological roles in plant development is still unclear.
CONCLUSION AND PERSPECTIVES
The Arabidopsis genome has 54 genes distributed among five chromosomes that encode putative two-component elements and related proteins (Fig. 8). Recent progress has provided compelling evidence for the involvement of a two-component circuitry in Arabidopsis cytokinin signaling (Hwang and Sheen, 2001; Inoue et al., 2001; Sakai et al., 2001; Suzuki et al., 2001a; Haberer and Kieber, 2002; Lohrmann and Harter, 2002). However, the physiological functions of most two-component regulators in plant signal transduction remain to be determined (Urao et al., 2000b; Lohrmann and Harter, 2002; Schaller et al., 2002). By analogy to the yeast osmosensing pathway, it has been proposed that the ethylene receptors and a putative osmosensor act as sensor His protein kinases and transmit signals through an MAPK cascade in Arabidopsis (Clark et al., 1998; Urao et al., 1999). The importance of His protein kinase activities and specific MAPK cascades will need to be demonstrated for both signaling pathways in plant cells.
Apparently, plants also adopted the prokaryotic two-component signaling pathways without a link to eukaryotic MAPK cascades. A multistep two-component phosphorelay system with distinct AHK, AHP, and ARR proteins plays a central role in cytokinin signal transduction (Fig. 9). This Arabidopsis cytokinin signaling pathway consists of four steps: (a) Distinct plasma membrane His protein kinases appear to initiate a phosphorelay cascade upon cytokinin perception; (b) the signals initiated at these His protein kinases converge on AHP proteins that serve as phosphorelay carriers between the cytokinin receptors and the downstream nuclear responses; (c) nuclear AHP translocation enables the activation of B-type ARR proteins, which in turn activate the transcription of A-type ARRs; and (d) the transcriptional activation of A-type ARRs provides a negative feedback mechanism in controlling the transient induction of primary cytokinin responsive genes (Hwang and Sheen, 2001; Inoue et al., 2001; Sakai et al., 2001; Suzuki et al., 2001a). The B-type ARRs appear to be the central rate-limiting regulators in cytokinin signaling, manifested by their abilities to mimic a broad spectrum of cytokinin actions when overexpressed in transgenic Arabidopsis plants and tissues. The B-type ARRs may interact with other tissue-specific proteins to mediate different cytokinin responses. The A-type ARRs may act as negative regulators in cytokinin signaling by competing with B-type ARRs for AHP bindings. Transiently expressed A-type ARRs may also interact with other effectors to mediate yet unknown, secondary responses to cytokinin (Haberer and Kieber, 2002). The proposed model provides a framework for dissecting the molecular mechanisms underlying cytokinin actions in various plant cytokinin responses (Fig. 9). Further studies will be required to elucidate the details of cytokinin perception and protein-protein interactions that are essential in cytokinin signaling in different cell types.
The use of E. coli and yeast mutant complementation has provided a rapid functional assay for plant two component proteins. The yeast two-hybrid assay has also shown direct interactions between many Arabidopsis two-component molecules (Imamura et al., 1998; Lohrmann et al., 2001; Suzuki et al., 1998, 2001a, 2001b). However, it is well known that two-component signaling elements are promiscuous and can interact with elements from non-physiological two-component systems (Stock et al., 1989, 2000; Schaller et al., 2002). The physiological functions and specificity of the interactions among two-component regulators could be further clarified in plant cells. In addition, detailed expression analyses of all two-component elements at the cellular level and the systematic isolation and characterization of knockout mutants will help to define their precise roles in plants. With the exception of AtHK1 and ARR19, Arabidopsis T-DNA insertion mutant lines of 52 two-component genes are now available (Tables I–III). The potential functional redundancy of two-component elements may explain the difficulty in isolating cytokinin signaling mutants and recessive ethylene receptor mutants by classical genetic screens. Thus, combining multiple loss-of-function mutations (Hua and Meyerowitz, 1998) will be necessary to reveal the physiological functions of two-component signal transduction pathways in planta.
Two-component systems may play important roles in the plant-signaling network that connects cytokinin, ethylene, light, stress, and Glc signals. Phenotypic analysis of a Glc insensitive mutant gin2 has revealed a new molecular link between Glc and cytokinin signaling through the regulation of ARR gene expression (Rolland et al., 2002; F. Rolland and J. Sheen, unpublished data). Cytokinin and red light signaling are found to converge at ARR4 (Sweere et al., 2001). Stress signaling may also utilize two-component elements (Urao et al., 1998). The challenge will be to understand the molecular and biochemical mechanisms underlying different plant signaling pathways that employ two-component elements. The identification of target genes of response regulators will facilitate the dissection of the complex network of plant signal transduction.
ACKNOWLEDGMENTS
We thank Filip Rolland for critical reading of the manuscript; Tastuo Kakimoto, Kazuo Shinozaki, and Klaus Harter for sharing unpublished results; and Eric Schaller for sharing his review.
Footnotes
This work was supported by the National Science Foundation (grant nos. IBN9723610 and DBI007692) and by the National Institutes of Health (grant no. GM60493).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005504.
LITERATURE CITED
- AGI. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell. 1998;10:1009–1019. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chang C, Stadler R. Ethylene hormone receptor action in Arabidopsis. Bioessays. 2001;23:619–627. doi: 10.1002/bies.1087. [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124:1706–1717. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elich TD, Chory J. Phytochrome: If it looks and smells like a histidine kinase, is it a histidine kinase? Cell. 1997;91:713–716. doi: 10.1016/s0092-8674(00)80458-4. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genick UK, Chory J. Red light sensing in plants. Curr Biol. 2000;10:R651–654. doi: 10.1016/s0960-9822(00)00694-1. [DOI] [PubMed] [Google Scholar]
- Haberer G, Kieber JJ. Cytokinins. New insights into a classic phytohormone. Plant Physiol. 2002;128:354–362. doi: 10.1104/pp.010773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Findell JL, Schaller GE, Sisler EC, Bleecker AB. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 2000;123:1449–1458. doi: 10.1104/pp.123.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Lamparter T. Prokaryotes and phytochrome. The connection to chromophores and signaling. Plant Physiol. 1999;121:1059–1068. doi: 10.1104/pp.121.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999;40:733–742. doi: 10.1093/oxfordjournals.pcp.a029600. [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- Kiba T, Taniguchi M, Imamura A, Ueguchi C, Mizuno T, Sugiyama T. Differential expression of genes for response regulators in response to cytokinins and nitrate in Arabidopsis thaliana. Plant Cell Physiol. 1999;40:767–771. doi: 10.1093/oxfordjournals.pcp.a029604. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Krall L, Reed JW. The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc Natl Acad Sci USA. 2000;97:8169–8174. doi: 10.1073/pnas.140520097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Buchholz G, Keitel C, Sweere U, Kircher S, Baurle I, Kulda J, Schafer E, Harter K. Differential expression and nuclear localization of response regulator-like proteins from Arabidopsis thaliana. Plant Biol. 1999;1:495–505. [Google Scholar]
- Lohrmann J, Harter K. Plant two-component signaling systems and the role of response regulators. Plant Physiol. 2002;128:363–369. doi: 10.1104/pp.010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Baurle I, Keitel C, Kozma-Bognar L, Brennicke A, Schafer E, Kudla J, Harter K. The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol Genet Genomics. 2001;265:2–13. doi: 10.1007/s004380000400. [DOI] [PubMed] [Google Scholar]
- Mahonen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T. Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:791–803. doi: 10.1093/pcp/41.6.791. [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Oda Y, Mizuno T. Light response of the circadian waves of the APRR1/TOC1 quintet: When does the quintet start singing rhythmically in Arabidopsis? Plant Cell Physiol. 2001;42:334–339. doi: 10.1093/pcp/pce036. [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002;43:58–69. doi: 10.1093/pcp/pcf005. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: II. Characterization with CCA1-overexpressing plants. Plant Cell Physiol. 2002;43:118–122. doi: 10.1093/pcp/pcf006. [DOI] [PubMed] [Google Scholar]
- McMichael RW, Jr, Lagarias JC. Phosphopeptide mapping of Avena phytochrome phosphorylated by protein kinases in vitro. Biochemistry. 1990;29:3872–3878. doi: 10.1021/bi00468a011. [DOI] [PubMed] [Google Scholar]
- Pischke MS, Fernandez DE, Sussman MR (2001) Characterizing the role of the CKI1 histidine kinase in Arabidopsis thaliana (abstract no. 285). 12th International Conference on Arabidopsis Research
- Miyata S, Urao T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of genes for two-component phosphorelay mediators with a single HPt domain in Arabidopsis thaliana. FEBS Lett. 1998;437:11–14. doi: 10.1016/s0014-5793(98)01188-0. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Sakai H, Aoyama T, Oka A. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 2000;24:703–711. doi: 10.1046/j.1365-313x.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Bono H, Oka A. Two-component response regulator from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol. 1998a;39:1232–1239. doi: 10.1093/oxfordjournals.pcp.a029325. [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science. 2001;294:1519–1521. doi: 10.1126/science.1065201. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998b;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Taniguchi M, Sugiyama T. His-Asp phosphorelay signaling: a communication avenue between plants and their environment. Plant Mol Biol. 2000;42:273–278. doi: 10.1023/a:1006334926388. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Mathews D, Gribskov M, Walker JC. Two-component signaling elements and histidyl-to-aspartyl phosphorelays. In: Somerville C, Meyerowitz E, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. (in press) [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Smith H. Phytochromes and light signal perception by plants: an emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stock JB, Ninfa AJ, Stock AM. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Imamura A, Ueguchi C, Mizuno T. Histidine-containing phosphotransfer (HPt) signal transducers implicated in His-to-Asp phosphorelay in Arabidopsis. Plant Cell Physiol. 1998;39:1258–1268. doi: 10.1093/oxfordjournals.pcp.a029329. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Ishikawa K, Yamashino T, Mizuno T. An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: overexpression of AHP2 in plants results in hypersensitiveness to cytokinin. Plant Cell Physiol. 2002;43:123–129. doi: 10.1093/pcp/pcf007. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. The Arabidopsis sensor his-kinase, ahk4, can respond to cytokinins. Plant Cell Physiol. 2001a;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Sakurai K, Ueguchi C, Mizuno T. Two types of putative nuclear factors that physically interact with Histidine-containing phosphotransfer (Hpt) domains, signaling mediators in His-to-Asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol. 2001b;42:37–45. doi: 10.1093/pcp/pce011. [DOI] [PubMed] [Google Scholar]
- Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, Kudla J, Nagy F, Schafer E, Harter K. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science. 2001;294:1108–1111. doi: 10.1126/science.1065022. [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 2001;42:85–93. doi: 10.1093/pcp/pce009. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T. Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 1998;429:259–262. doi: 10.1016/s0014-5793(98)00611-5. [DOI] [PubMed] [Google Scholar]
- Thomason P, Kay R. Eukaryotic signal transduction via histidine-aspartate phosphorelay. J Cell Sci. 2000;113:3141–3150. doi: 10.1242/jcs.113.18.3141. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C, Koizumi H, Suzuki T, Mizuno T. Novel family of sensor Histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:231–235. doi: 10.1093/pcp/pce015. [DOI] [PubMed] [Google Scholar]
- Urao T, Miyata S, Yamaguchi-Shinozaki K, Shinozaki K. Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 2000a;478:227–232. doi: 10.1016/s0014-5793(00)01860-3. [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Yamaguchi-Shinozaki K, Shinozaki K. Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 1998;427:175–178. doi: 10.1016/s0014-5793(98)00418-9. [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Shinozaki K. Plant histidine kinases: an emerging picture of two-component signal transduction in hormone and environmental responses. Sci Signal Transduction Knowledge Environment (STKE) 2001;109:RE18. doi: 10.1126/stke.2001.109.re18. [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Shinozaki K. Two-component systems in plant signal transduction. Trends Plant Sci. 2000b;5:67–74. doi: 10.1016/s1360-1385(99)01542-3. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Yamashino T, Mizuno T. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001;42:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]