Abstract
Branched-chain amino acid transaminases (BCATs) play a crucial role in the metabolism of leucine, isoleucine, and valine. They catalyze the last step of the synthesis and/or the initial step of the degradation of this class of amino acids. In Arabidopsis, seven putative BCAT genes are identified by their similarity to their counterparts from other organisms. We have now cloned the respective cDNA sequences of six of these genes. The deduced amino acid sequences show between 47.5% and 84.1% identity to each other and about 30% to the homologous enzymes from yeast (Saccharomyces cerevisiae) and mammals. In addition, many amino acids in crucial positions as determined by crystallographic analyses of BCATs from Escherichia coli and human (Homo sapiens) are conserved in the AtBCATs. Complementation of a yeast Δbat1/Δbat2 double knockout strain revealed that five AtBCATs can function as BCATs in vivo. Transient expression of BCAT:green fluorescent protein fusion proteins in tobacco (Nicotiana tabacum) protoplasts shows that three isoenzymes are imported into chloroplasts (AtBCAT-2, -3, and -5), whereas a single enzyme is directed into mitochondria (AtBCAT-1).
In plants, branched-chain amino acids are important compounds in many respects. Besides their function as building blocks of proteins, they play a pivotal role in the synthesis of a number of secondary products in plants (Lea and Ireland, 1999). In addition, the intermediates of branched-chain amino acid biosynthesis are also substrates for the synthesis of pantothenate and propionyl-CoA (Singh, 1999).
The pathways of branched-chain amino acid biosynthesis are well investigated in plants because several commercially important herbicides interfere with selected enzymes of these cascades (Wittenbach and Abell, 1999). In the branched-chain amino acid biosynthesis pathways, almost all enzymatic activities have been described and many genes and cDNAs have been characterized (for review, see Singh, 1999). The single exception is the final synthesis step, the conversion of respective 2-oxo acids to the branched-chain amino acids, which is catalyzed by branched-chain amino acid transaminases (BCATs). Very little is known about this final step besides a biochemical study, in which a respective activity from spinach (Spinacia oleracea) chloroplasts was enriched and enzyme activities for the synthesis of Val and for Leu and iso-Leu, respectively, could be differentiated (Hagelstein et al., 1997).
However, BCAT activity is not only involved in the biosynthesis, but also in the degradation of these three amino acids. First indications for the presence of branched-chain amino acid degradative pathways in plants have been reported in castor bean (Ricinus communis), where gluconeogenesis from Leu was reported (Stewart and Beevers, 1967) and in maize (Zea mays), where the respiration of Leu and its conversion to starch, sugars, or organic acids has been observed (Sodek and Wilson, 1973). The detection of branched-chain 2-oxo acids in peroxisomes of mung bean (Vigna radiata) further indicated the presence of branched-chain catabolic processes (Gerbling and Gerhardt, 1988, 1989).
The biochemical evidence for the involvement of the biotin-containing subunit of the 3-methylcrotonyl-CoA carboxylase in Leu degradation and the characterization of the respective cDNA provided evidence for the conversion of at least Leu in several plant species (Alban et al., 1993; Song et al., 1994; Wang et al., 1994; Weaver et al., 1995; Aubert et al., 1996). This enzyme is located in mitochondria, where the operation of Leu degradation is further confirmed by the detection of several other enzyme activities associated with Leu catabolism (Anderson et al., 1998). The unambiguous location of the isovaleryl-CoA dehydrogenase in mitochondria of Arabidopsis further substantiated the presence of a Leu degradation pathway in these organelles. Substrate specificity tests of this latter enzyme and the respective enzyme activity enriched from potato (Solanum tuberosum) also indicated activity in the conversion of isobutyryl-CoA, an intermediate of Val degradation in mammalian mitochondria (Faivre-Nitschke et al., 2001; Däschner et al., 2001). This prompted the speculation of whether Val also might be catabolized in mitochondria of Arabidopsis and potato, although such an activity has not been detected with the homologous enzyme from pea (Pisum sativum; Reinard et al., 2000).
Toward a better understanding of the branched-chain amino acid metabolism in higher plants, we have now characterized the complete cDNAs of six putative BCATs forming a gene family in Arabidopsis. Complementation analyses in a yeast (Saccharomyces cerevisiae) strain deficient in endogenous BCAT activities confirm the predicted BCAT function for five of these proteins. Subcellular localization studies reveal distinct intracellular compartmentation of the different proteins in the plant cell.
RESULTS
Identification and Cloning of BCATs in Arabidopsis (AtBCAT)
Several recent reports on genes and cDNAs encoding enzymes engaged in the degradation of the branched-chain amino acid Leu suggest the presence of a catabolic pathway in mitochondria (Anderson et al., 1998; Däschner et al., 1999, 2001). To identify the plant enzyme catalyzing the initial step of this degradation process, BCAT sequences from yeast (Kispal et al., 1996; accession no. P38891), human (Homo sapiens; Bledsoe et al., 1997; Davoodi et al., 1998; accession no. O15382), and Escherichia coli (Kuramitsu et al., 1985; Inoue et al., 1988; accession no. P00510) were used to screen the sequence data available in the public data bases by the tblastN algorithm (Altschul et al., 1990). This approach identified seven putative BCAT genes in Arabidopsis. After the completion of the Arabidopsis genome project, these genes were annotated on different bacteria artificial chromosome (BAC) clones as indicated in Table I. An additional putative gene annotated as BCAT like was found in later searches, but the predicted protein shows only limited similarity to the other AtBCATs and thus was not further analyzed. In addition to the genomic sequences, several EST sequences were identified, four of which potentially encode BCAT reading frames. Sequencing of these clones, which were kindly provided by the Arabidopsis Biological Resource Center (Ohio State University, Columbus), confirmed them to encode putative BCAT reading frames (Table I). In cDNA clone 97D16T7 (AtBCAT-1), however, four missing nucleotides, possibly lost in an aberrant splicing process, and in clone ATTS4608 (AtBCAT-4), a point mutation introducing a stop codon, disrupt the respective BCAT reading frames. Correct reading frames were identified in cDNA clones generated by reverse transcriptase (RT)-PCR from total Arabidopsis RNA.
Table I.
Survey of the putative BCAT genes identified in the Arabidopsis genome
| Gene | cDNA Accession No. | EST Clonea | BAC Clone | BAC Accession No. | Protein Identification | Chromosome |
|---|---|---|---|---|---|---|
| Atbcat-1 | AJ276123 | 97D16T7b | T27I1 | AC004122 | AAC34335 | 1 |
| Atbcat-2 | AJ271731 | 123A19T7c | T27I1 | AC004122 | AAC34333 | 1 |
| Atbcat-3 | AJ276124 | ATTS3314 | T16K5 | AL132965 | CAB66906 | 3 |
| Atbcat-4 | AJ271732 | ATTS4608d | MMB12 | AP000417 | BAB02558 | 3 |
| Atbcat-5 | AJ293804 | – | MPA24 | AB010075 | BAB10685 | 5 |
| Atbcat-6 | AJ312747 | – | F2J10 | AC015445 | AAF76437 | 1 |
| Atbcat-7 | – | – | F2J10 | AC015445 | AAF76438 | 1 |
| Atbcat like | – | – | T12H1 | AC009177 | AAF27025 | 3 |
The putative protein AAF27025 identified on BAC clone T12H1 is annotated as a BCAT-like protein, but shows only very limited sequence similarity to the other AtBCAT amino acid sequences.
Only expressed sequence tag (EST) clones used in this analysis are indicated.
Four nucleotides are missing in the AtBCAT-1 reading frame.
This cDNA contains unspliced intron 2, which shortens the N-terminal part of the AtBCAT-2 reading frame.
Contains a stop codon in the AtBCAT-4 reading frame, which is not present in the genomic sequence.
Of the other clones from the Arabidopsis Biological Resource Center, clone ATTS3314 (AtBCAT-3) encodes an apparently complete BCAT protein, but clone 123A19T7 (AtBCAT-2) encodes a BCAT reading frame most likely truncated at the N terminus. Inspection of genomic N-terminal sequences identified a putative intron in this cDNA clone, which shortens the BCAT reading frame at the N terminus. This clone was used to screen an Arabidopsis cDNA library and several fully spliced cDNA clones encoding N-terminally extended reading frames were identified. However, about 10% of the isolated cDNA clones still contained intron 2 with the shortened version of the reading frame. Potential alternative splicing was examined by RT-PCR analysis of total RNA. A single cDNA fragment corresponding in size to a completely spliced mRNA does not detect partially spliced mRNAs for AtBCAT-2 in the total steady-state RNA (data not shown). For AtBCAT-5 and -6, partial cDNA fragments were generated by RT-PCR and used to isolate complete cDNA clones from an Arabidopsis cDNA library.
No cDNA fragment could be amplified for AtBCAT-7 with gene-specific primers even in repeated experiments. In addition, no cDNA clone originating from this gene could be isolated in a hybridization with the AtBCAT-6-specific probe, which is 86% identical with the AtBCAT-7 nucleotide sequence. It remains unclear at present whether AtBCAT-7, which is located directly downstream of AtBCAT-6, is an actively transcribed gene.
Seven Members Form the BCAT Gene Family in Arabidopsis
The amino acid sequences deduced from the seven putative AtBCAT cDNAs share between 27.1% and 39.2% identical amino acids with their counterparts from non-plant organisms, between 48.4% and 76.0% with two putative BCAT sequences from potato cDNAs, and between 47.5% and 84.1% with each other (Table II). Weak amino acid sequence similarity (31.1%–37.9% in 87 to maximally 252 amino acids) is observed with part of a BCAT-like reading frame in the Arabidopsis genome. Alignment of the seven AtBCAT sequences shows extensive conservation across almost the complete reading frames. A Lys to which pyridoxalphosphate (PLP) is covalently linked in non-plant BCATs is conserved in all BCATs from Arabidopsis (Fig. 1, highlighted by a gray box). A Tyr (Y236 in AtBCAT-1, Y207 in HsBCATm, and Y164 in EcILVE) and a Glu (E268, E237, and E193) anchoring the PLP ring in the bottom of the active site cavity are also present in the plant sequences (Fig. 1, arrowheads 3 and 4). In addition, several other amino acids, with crucial functions revealed by crystallization of the respective proteins from E. coli (ILVE) and human (HsBCATm), are conserved in the Arabidopsis sequences. These include a Tyr (Y100, Y70, and Y31) and an Arg (R173, R143, and R97; Fig. 1, arrowheads 1 and 2; Okada et al., 1997; Yennawar et al., 2001). These residues are important for the interaction between the monomers via hydrogen bonding and seem to be essential for the formation of the putative substrate-binding pocket in HsBCATm (Yennawar et al., 2001). Substantial sequence variability characterizes the N termini (Fig. 1) of the AtBCATs, possibly encoding different targeting signals for the subcellular sorting of the individual enzymes.
Table II.
Comparison of the amino acid sequences deduced from the different BCAT cDNAs and genomic sequences by the BestFit algorithm of the GCG software package (Genetics Computer Group, Madison, WI)
| Organism
|
Arabidopsis
|
Potato
|
E.
coli
|
Yeast
|
Human
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | BCAT 2 | BCAT 3 | BCAT 4 | BCAT 5 | BCAT 6 | BCAT 7 | BCAT like | BCAT 1 | BCAT 2 | BCAT ilvE | BATm | BATc | BCAT m | BCAT c |
| BCAT-1 | 64.7 | 58.2 | 50.1 | 59.3 | 58.0 | 59.2 | 35.6* | 60.5 | 61.3 | 30.3 | 33.9 | 34.0 | 36.2 | 36.6 |
| BCAT-2 | – | 64.7 | 53.3 | 63.2 | 58.6 | 58.0 | 35.7* | 65.5 | 66.3 | 29.8 | 36.0 | 34.9 | 33.5 | 33.1 |
| BCAT-3 | – | – | 47.5 | 80.9 | 58.3 | 60.3 | 36.0* | 69.6 | 76.0 | 33.3 | 35.0 | 36.1 | 38.5 | 39.2 |
| BCAT-4 | – | – | – | 47.8 | 61.8 | 60.5 | 31.1* | 48.4 | 48.8 | 27.1 | 35.4 | 34.6 | 32.5 | 32.1 |
| BCAT-5 | – | – | – | – | 56.6 | 58.3 | 37.9* | 70.3 | 79.7 | 32.4 | 35.0 | 35.5 | 38.1 | 38.5 |
| BCAT-6 | – | – | – | – | – | 84.1 | 37.7* | 58.9 | 57.7 | 29.2 | 38.2 | 34.8 | 33.6 | 33.3 |
| BCAT-7 | – | – | – | – | – | – | 33.2* | 56.7 | 59.4 | 31.3 | 37.7 | 37.6 | 32.4 | 32.1 |
| BCAT-like | – | – | – | – | – | – | – | 37.0* | 37.8* | 36.3* | 33.8* | 37.3* | 29.0* | 28.0* |
The nos. represent the percentages of identical amino acids. BCAT amino acid sequences are available under the following accession nos.: potato, AAF07191 (StBCAT-1) and AAF07192 (StBCAT-2); E. coli, BAB38127; yeast, P38891 (mitochondrial) and P47176 (cytosolic); and human, O15382 (mitochondrial) and P54687 (cytosolic). Nos. marked by asterisks represent amino acids identical in partial sequences with lengths between 87 and 252 residues.
Figure 1.
Alignment of the amino acid sequences deduced from the seven AtBCAT cDNA sequences. Amino acid residues conserved in at least five AtBCATs are highlighted by black boxes. The conserved Lys suggested to bind pyridoxal phosphate is indicated by a gray box. Numbered arrowheads indicate important amino acids as determined by the crystallization of the respective enzymes from E. coli and human. Positions of the introns are given by gray vertical arrows.
AtBCATs Function as BCATs in Yeast
The comparatively high amino acid sequence similarity of the putative BCATs investigated here with their counterparts in other organisms strongly suggests an analogous function as BCATs. To experimentally confirm such an activity in vivo, the Arabidopsis reading frames were tested for their ability to suspend auxotrophy for branched-chain amino acids of a Δbat1/Δbat2 double knockout yeast strain (Kispal et al., 1996). This strain is not able to grow on minimal medium with Glc in the absence of a single, any combination of two, or all three branched-chain amino acids (data not shown). The transformation of the vector pRS425GPD without insert, which carries a leu2 marker, rescues the auxotrophy for Leu, but does not influence the dependency on an external supply of Val and iso-Leu. Of course, growth of the mutant can be rescued by the addition of all three branched-chain amino acids, although reduced growth rates compared with the wild-type cells may indicate an additional function of the BCATs in yeast (Kispal et al., 1996). We used this system with the aim to test the ability of AtBCATs to complement the auxotrophy for branched-chain amino acids.
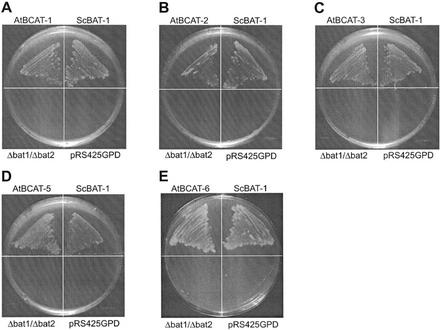
To this end, the respective full-length Arabidopsis cDNA sequences were cloned downstream of the yeast glycerol aldehyde-3-phosphate dehydrogenase promoter in vector pRS425GPD and transformed into Δbat1/Δbat2. Restored growth on minimal medium lacking the three branched-chain amino acids confirms the respective enzymatic activities for AtBCAT-1, -2, -3, -5, and -6 (Fig. 2). Growth parameters were comparable with the positive control in which ScBAT-1 was used for complementation. We assume that a mitochondrial AtBCAT will be transported to mitochondria also in yeast and there take over the function of the mitochondrial ScBAT1. The other complementing AtBCATs probably remain in the cytosol taking over the function of the yeast cytosolic enzyme. Because yeast cells lacking the mitochondrial enzyme but with an intact cytosolic protein can grow in the absence of branched-chain amino acids, a localization of the AtBCATs in the cytosol is sufficient to rescue the auxotrophy. No complementation was achieved with AtBCAT-4 (data not shown). To check whether an improper subcellular targeting might be responsible for the lack of complementation, the conserved region starting with amino acid 18 was C-terminally fused to the atp9 mitochondrial targeting sequence from Neurospora crassa (amino acids 1–59). However, even with this presequence, which correctly directs the conserved part of the homologous ScBAT-1 protein into mitochondria in yeast, no complementation is detectable (data not shown).
Figure 2.
Complementation analysis of the AtBCATs in the yeast double knockout mutant Δbat1/Δbat2. To examine the suggested BCAT function in vivo, the different AtBCAT cDNA sequences were cloned into pRS425GPD vectors and transformed into Δbat1/Δbat2. Restored growth on medium lacking Val, Leu, and iso-Leu indicates the competent BCAT activity of AtBCAT-1 (A), -2 (B), -3 (C), -5 (D), and -6 (E) in yeast. Complementation was also observed with ScBAT-1 as positive control. No growth is detected without transformation (Δbat1/Δbat2) or after transformation of the vector without insert (pRS425GPD).
A control plasmid carrying the Arabidopsis aspartate aminotransaminase 1 (AtAAT-1, data not shown; Schultz and Coruzzi, 1995) does not complement the auxotrophy, confirming that the presence of functional BCAT reading frames from Arabidopsis is responsible for complementation (Fig. 2).
Arabidopsis Encodes Three Plastid and One Mitochondrially Located BCAT
Previous analyses had indicated that branched-chain amino acids are synthesized in chloroplasts, whereas the degradation pathways are accommodated in mitochondria. The N-terminal extensions of AtBCAT-1, -2, -3, and -5 suggest targeting of these proteins to either of these organelles. Distinct intracellular localizations are predicted by the various sorting prediction programs (see “Materials and Methods”). Mitochondrial targeting of AtBCAT-1 and -2 is suggested by all programs used, whereas different subcellular destinations are predicted for AtBCAT-3 and -5. For AtBCAT-4 and -6, targeting to plastids or mitochondria is clearly excluded, but peroxisomal (AtBCAT-4) and cytosolic (AtBCAT-6) localizations are suggested by PSORT albeit with relatively low probabilities (0.59 and 0.65). To test the subcellular sorting of AtBCAT-1 to -6 in vivo, complete cDNAs (AtBCAT-3, -4, and -6) and fragments corresponding to N-terminal parts of different lengths were cloned in frame upstream of a plant-adapted GFP (smGFP4, Fig. 3A). The cauliflower mosaic virus 35S promoter drives expression of these fusion proteins after transient transformation into tobacco (Nicotiana tabacum) protoplasts.
Figure 3.
Subcellular localization of different AtBCATs. A, Constructs used for the transient transformation of tobacco protoplasts. cDNA fragments representing the 47 (AtBCAT-1), 126 (AtBCAT-2), 413 (AtBCAT-3), and 156 (AtBCAT-5) N-terminal amino acids were cloned in frame upstream of the green fluorescent protein (GFP) reading frame. Expression of the fusion proteins is under control of the cauliflower mosaic virus 35S promoter and the nopaline synthase terminator. B, Fluorescence images of the protoplasts taken with the following filter sets: GFP, D395x/440 DLCP/HQ510/50 and GG 475 LP; this set allows the specific detection of the GFP fluorescence. Fluorescein isothiocyanate (FITC), HQ 470/40/Q 495 LP/HQ 500 LP; this filter set allows the simultaneous detection of GFP and chlorophyll autofluorescence, and chloroplasts containing the GFP fusion protein will appear yellow. MitoTracker, HQ545/30/Q570 LP/HQ 610/75; this filter set is optimized for the specific visualization of the MitoTracker red dye. Rhodamine, BP530–585/FT600/LP615; this filter allows the detection of the chlorophyll autofluorescence. Space bars correspond to 10 μm. The GFP fluorescence of the AtBCAT-1:GFP fusion protein coincides with the pattern of Mitotracker red-stained mitochondria. The GFP fluorescences of AtBCAT-2, -3, and -5 fusion proteins are congruent with the autofluorescence of chlorophyll, as seen in the yellow FITC images, indicating their localization in chloroplasts.
Colocalization of the AtBCAT-1:GFP fluorescence with the MitoTracker Red fluorescence, a dye that is specifically enriched in mitochondria, identifies the AtBCAT-1:GFP fusion protein in mitochondria. This is consistent with the computer prediction and suggests a participation of this protein in the branched-chain amino acid degradation process (Fig. 3B, upper lane). In contrast, the GFP fluorescence patterns observed in protoplasts transformed with AtBCAT-2, -3, and -5:GFP constructs are congruent with the autofluorescence patterns of the chloroplasts, identifying these organelles as their subcellular residence (Fig. 3B, lower parts). The in vivo targeting of AtBCAT-2 contradicts the prediction of a mitochondrial localization of this protein.
No definitive localization is observed for the GFP fusion proteins with AtBCAT-4 and -6 moieties. For these, a weak and diffuse GFP fluorescence is seen that could possibly be interpreted as a cytosolic distribution of the respective fusion proteins (data not shown), which for would AtBCAT-6 be consistent with the prediction by PSORT.
DISCUSSION
Branched-Chain Amino Acid Transaminases in Arabidopsis
BCATs are PLP-dependent enzymes that occupy a special position among the aminotransferases. They are distantly related to all other aminotransferases and, based on profile and family profile analyses, are grouped with the bacterial d-amino acid aminotransferase and another bacterial enzyme in a separate branch called d-Ala (or d-amino acid) aminotransferase family (Jansonius, 1998; Mehta and Christen, 2000). Consistent with the observation that the mutual overall sequence identities between the majority of aminotransferases is too low for an alignment by standard analysis (Mehta et al., 1993), substantial sequence similarities of the AtBCATs are only found with BCATs from other organisms. This supports that the cDNAs analyzed here encode aminotransferases with branched-chain amino acids as preferred substrates. In addition, almost all amino acids identified in crucial positions within the three-dimensional structures of the homologous enzymes from E. coli and humans are conserved. This includes those residues that can recognize branched-chain amino acids in the E. coli enzyme; for example, tyrosines at positions 31 and 164 are present in the respective positions in the Arabidopsis proteins (positions 100 and 236) and human mitochondrial BCAT (70 and 207).
However, variation is observed concerning a Phe, which is found at position 37 in the E. coli and at position 75 in human BCAT. Although the same residue is found at the respective positions in AtBCAT-3, -5, and -6, a Tyr is present in AtBCAT-2 and -4 and an iso-Leu in AtBCAT-1 (I105). Similarly, a Val (V109 in E. coli) is only seen in AtBCAT-1 (position 185) and -6, but is replaced by a Leu in AtBCAT-2, -3, and -5 and a Ser in AtBCAT-4 (Okada et al., 1997; Yennawar et al., 2001). Nevertheless, the majority of these amino acids considered to be important for substrate binding are conserved in the AtBCATs. The complementation assays confirm the activity of AtBCAT-1, -2, -3, -5, and -6 toward branched-chain amino acids deduced from the protein similarity. The function of AtBCAT-4 remains unclear at present and an activity test with this protein will be necessary to determine its substrate specificity and function as aminotransferase.
AtBCATs Are Operative in Biosynthesis and/or Degradation Pathways
Analysis of the subcellular localization of the BCATs in Arabidopsis by the respective GFP fusion proteins revealed strong evidence for three plastidal and a single mitochondrial BCAT. The presence of BCATs in chloroplasts is consistent with previous analyses of branched-chain amino acid metabolism, which had indicated that the biosynthesis of these branched-chain amino acids is performed in chloroplasts. This was concluded from the cofractionation of all necessary enzyme activities with chloroplast fractions, including the two separable BCATs designated Val and Leu iso-Leu aminotransferases detected in spinach (Hagelstein et al., 1997).
In line with these chloroplast-located aminotransferases with different substrate specificities, the respective localization of three different proteins may suggest specialized enzymes that predominantly complete the synthesis of either Val, Leu, or iso-Leu, respectively. However, it is also possible that the different chloroplast BCATs are expressed in different plant tissues or developmental stages. Thus, substrate specificity tests as well as detailed expression studies are necessary to further elaborate the functions of the individual chloroplast BCATs. Such assays would also reveal whether these enzymes are responsible for the reversible formation of 2-aminobutyrate from 2-ketobutyrate, a key intermediate of the 3-hydroxybutyrate-co-3-hydroxyvalerate copolymer biosynthesis in transgenic plants. It was speculated that this reaction might be catalyzed by BCATs and might prevent an efficient biosynthesis of this biopolymer (Slater et al., 1999).
A BCAT activity has been suggested for mitochondria (Anderson et al., 1998). In this compartment, several enzyme activities necessary for Leu degradation have been detected and methylcrotonyl-CoA carboxylase and isovaleryl-CoA dehydrogenase have been localized here (Anderson et al., 1998; Däschner et al., 2001). In this cascade, AtBCAT-1 may be responsible for the initiation of mitochondrially located degradation of at least Leu. Detailed substrate specificity tests are again necessary to gather more information about the degradation of iso-Leu and Val, a breakdown pathway of the latter being suggested by activity of AtIVD toward respective degradation intermediates.
At present, the subcellular localization of AtBCAT-4 and -6 and their physiological role remain unclear. Import of these enzymes into mitochondria or chloroplasts is unlikely for two reasons. First, the comparatively short N termini do not show any characteristic feature of respective targeting peptides and consequently prediction programs do not suggest an organellar localization of these proteins. Second, the respective GFP fusion proteins are never observed to be associated with these organelles in repeated protoplast transformation experiments.
In some plant species, degradation of branched-chain amino acids is suggested in peroxisomes (Gerbling and Gerhardt, 1988, 1989). A low-probability prediction suggests peroxisomal targeting for AtBCAT-4, although none of the features of PTS-1 or PTS-2 targeting sequences is obvious. However, the fluorescence images do not support a localization of the respective fusion proteins in this compartment. Although the faint and diffuse distribution of the AtBCAT-4:GFP and AtBCAT-6:GFP fluorescence may suggest a cytosolic localization, the exact subcellular in vivo targeting of these polypeptides remains unclear and thus needs to be investigated in transgenic plants transformed with respective GFP fusion constructs.
Spatial and Regulatory Control of Synthetic and Degradative Pathways of Branched-Chain Amino Acids
Plants contain the capability for both the synthesis and degradation of Leu and possibly other branched-chain amino acids. These counteracting pathways are physically separated from each other in different subcellular compartments, suggesting that transporter systems for Leu and the other branched-chain amino acids may play a crucial role in the appropriate distribution. This compartmentalization requires a sensitive regulatory control of the relevant transporter systems and the various enzymes involved in branched-chain amino acid metabolism. The mitochondrial AtBCAT-1 initiates degradation and therefore may perform a central regulatory role in the branched-chain amino acid turnover. Though transaminases in plants are thought to be nonregulated enzymes (Lea and Ireland, 1999), a recent high-throughput investigation of gene expression in Arabidopsis revealed that expression of EST clone 123A19T7, the chloroplast-located AtBCAT-2, is enhanced upon dark adaption (Schaffer et al., 2001). This suggests that BCATs are regulated at the transcriptional level and thus may be checkpoints of branched-chain amino acid metabolism in plants.
MATERIALS AND METHODS
Arabidopsis BCAT cDNA Analysis
The cDNA of AtBCAT-1 was amplified with Klentaq polymerase (BD Biosciences, Palo Alto, CA) on total RNA after first strand synthesis primed by oligo(dT)-adapter primer DTXSC [5′-GACTCGAGTCGACATCGA-(dT)17] under conditions recommended for the polymerase used. In the first reaction, primers Atbcat1/2 (1–26) and XSC (5′-GACTCGAGTCGACATCGA) were used under the following conditions: 1 min at 94°C, 1 min at 46°C, and 2 min at 68°C. The obtained cDNAs were size fractionated on an agarose gel and fragments with sizes between 1.3 and 1.7 kb covering the expected size of about 1.5 kb were eluted from the gel and used as templates for a second PCR with primers Atbcat1/2 and Atbcat1/3 (1,368–1,346) with the following parameters: five cycles of 1 min at 94°C, 1 min at 58°C, and 2 min at 68°C followed by 30 cycles of 1 min at 94°C, 1 min at 56°C, and 2 min at 68°C. The resulting fragment of 1.4 kb was then directly cloned into pBluescript vectors (Stratagene, La Jolla, CA). Ligation and transformation into Escherichia coli was performed under standard conditions described elsewhere (Sambrook et al., 1989). The AtBCAT-4 cDNA (1.2 kb) was cloned by an analogous strategy after one round of PCR amplification with primers Atbcat4/5 (17–36) and Atbcat4/4 (1,204–1,180) and five cycles of 1 min at 94°C, 1 min at 56°C, and 1.5 min at 68°C, followed by 30 cycles of 1 min at 94°C, 1 min at 53°C, and 1.5 min at 68°C.
The cDNA sequence of AtBCAT-3 was determined on EST clone ATTS3314 (accession no. Z34554). For the isolation of complete cDNAs of AtBCAT-2, EST clone 123A19T7 was used as a probe to screen an Arabidopsis cDNA lambda ZAPII library (Stratagene) obtained from the Arabidopsis Biological Resource Center (Kieber et al., 1993). Plating, transfer, and hybridization were carried out following the manual for the lambda ZAPII system (Stratagene) and the recommendations given by the manufacturer of the Hybond N+ membranes (Amersham Biosciences, Uppsala). A cDNA clone for AtBCAT-5 was obtained by the same experimental procedure using a respective partial cDNA fragment as probe. This fragment was amplified from first strand cDNA synthesized on total RNA (see above) engaging a nested PCR strategy with primer pairs Atbcat5/3 (775–794), Atbcat5/2 (1,841–1,821), and Atbcat5/4 (831–850), Atbcat5/2. Screening of the Arabidopsis cDNA library was performed as described for AtBCAT-2.
Full-length AtBCAT-6 clones were isolated from the Arabidopsis cDNA library by hybridization with a cDNA fragment obtained in a PCR with primers bcat6/7.1 (231–255) and bcat6/7.2 (774–753) on first strand cDNA generated as indicated above.
All sequences are available in public databases under the following accession numbers: AJ276123 (AtBCAT-1), AJ271731 (AtBCAT-2), AJ276124 (AtBCAT-3), AJ271732 (AtBCAT-4), AJ293804 (AtBCAT-5), and AJ312747 (AtBCAT-6). Numbers describing the locations of the primers refer to the nucleotide numbering in the respective database entries.
Yeast (Saccharomyces cerevisiae) Complementation Analysis
For the complementation tests, AtBCAT-2, -3, -5, and -6 cDNAs were amplified with primer pairs Atbcat2GFP-H (34–63) and Atbcat2Y-R (1,284–1,258), Atbcat3GFP-H (85–108) and Atbcat3Y-R (1,405–1,381), Atbcat5Y-H (546–568) and Atbcat5Y-R (1,872–1,847), and Atbcat6GFP-H (5′-GCGGATCCTCCATAGATGGCT-3′) and Atbcat6.3 (1,113–1,093) on the respective cDNA clones as templates. These primers contain BamHI restriction sites attached at the 5′ ends to allow cloning into the respective site in vector pRS425GPD (Sikorski and Hieter, 1989). Analogous clones of AtBCAT-1 and -4 were generated by direct cloning of the BamHI/BamHI and BamHI/SalI full-length cDNA fragments into the vector. A plasmid carrying the ScBAT-1 coding region from amino acids 17 through 393 with the 5′-attached mitochondrial targeting sequence of the Neurospora crassa ATPase subunit 9 (amino acids 1–59) was used as positive control. To test the influence of subcellular targeting of AtBCAT-4 on complementation, a fragment coding the same N. crassa atp9 targeting signal was cloned upstream of the AtBCAT-4 reading frame, which codes for amino acids 18 through 354. As negative control, a cDNA fragment representing the complete Arabidopsis mitochondrial Asp amino transaminase 1 (AtAAT-1, accession no. U15026) was generated by RT-PCR with primers DTXSC 5′-GACTCGAGTCGACATCGA(dT)17 (first strand synthesis) and primer pair Ataat1-H (62–86)/Ataat1-R (1,605–1,579) with attached BamHI restriction sites. Cloning into pRS425GPD was done as above.
About 3 to 5 μg of each complementation clone and of the vector without insert were transformed into yeast strain Δbat1/Δbat2 (Kispal et al., 1996) and plated onto minimal medium (yeast nitrogen base without amino acids, 2% [w/v] Suc, 0.002% [w/v] adenine, His, and Lys). The plates were cultivated at 30°C and growth was monitored after 5 to 7 d.
Transient Expression of AtBCAT:GFP Fusion Proteins in Tobacco (Nicotiana tabacum) Protoplasts
For the transient expression of AtBCAT:smGFP4 fusion proteins, respective fragments were amplified on the various AtBCAT cDNA templates: AtBCAT-1, primers Atbcat1GFP-H (62–89) and Atbcat1GFP-R3 (236–209); AtBCAT-2, primers Atbcat2GFP-H and Atbcat2GFP-R (438–411); AtBCAT-3, primers Atbcat3GFP-H and Atbcat3GFP-R (1,340–1,318); AtBCAT-4, primers Atbcat4GFP-H (29–52) and Atbcat4GFP-R (322–299); AtBCAT-5, primers Atbcat5GFP-H (546–568) and Atbcat5GFP-R2 (1,047–1,022); and AtBCAT-6, primers Atbcat6GFP-H and Atbcat6GFP-R (1,085–1,065).
The amplified fragments correspond to the N-terminal parts of 47 (AtBCAT-1), 126 (AtBCAT-2), 413 (AtBCAT-3), and 156 (AtBCAT-5) amino acids. All primers used contain 5′-attached BamHI restriction sites, which were used for cloning into the respective site in vector psmGFP4 containing the smGFP reading frame (Davis and Vierstra, 1998). On average, 50 μg of DNA of the individual clones were transformed into about 500,000 tobacco protoplasts. Preparation and transformation of the protoplasts were carried out as described previously (Koop et al., 1996). MitoTracker Red staining was done according to a protocol given by the manufacturer (Molecular Probes, Eugene, OR).
Fluorescence Microscopy
Fluorescence of GFP, Mitotracker Red, and chlorophyll was visualized with an Axioplan I microscope (Carl Zeiss, Oberkochen, Germany) and the following filter sets: GFP, HQ D3295x/HQ510/50 and GG475LP; MitoTracker, HQ 545/30/HQ 610/75; rhodamine, BP530-585/FT600/LP615 (used for chlorophyll autofluorescence); and FITC, HQ 470/40/HQ 500 LP. All filters were purchased from AHF Analysentechnik (Tübingen, Germany). Images were taken with the Axiovision software.
Computer Analysis
Computer analyses were performed using the Blast algorithms at the National Center for Biotechnology Information server. The protein alignment was generated with the MegAlign program of the DNA Star sequence analysis package. The intron in the AtBCAT-2 cDNA clone 123A19T7 was identified using the Genscan Web Server at Massachusetts Institute of Technology (http://genes.mit.edu/GENSCAN.html) and the NatPlantGene Server at the Center for Biological Sequence Analysis, BioCentrum-DTU at the Technical University of Denmark (http://www.cbs.dtu.dk/services/NetPGene/). Predictions of subcellular targeting were done with PSORT (http://psort.nibb.ac.jp/), MiTop (http://www.mips.biochem. mpg.de/cgibin/proj/medgen/mitofilter), Predotar (http://www.inra.fr/Internet/Produits/Predotar/index.html), and ChloroP (http://www.cbs.dtu.dk/services/ChloroP/).
Miscellaneous Methods
Reverse transcription was carried out with Superscript II RT in a buffer supplied by the manufacturer as outlined in the manual (Life Technologies/Gibco-BRL, Cleveland). Sequence analyses of the clones carrying GFP fusion constructs and the negative test complementation clones were performed by cycle sequencing with the Thermo Sequenase fluorescent labeling kit or T7 DNA polymerase-based sequencing with the Cy5 Autoread sequencing kits with Cy5-dATP labeling mix (Amersham Biosciences). Sequencing fragments were detected and processed by an Alf Express sequencer. If not otherwise stated, PCR was carried out with 5 units of KlenTaq polymerase on about 100-ng DNA template with 35 cycles of 60 s at 94°C, 60 s at the melting temperature, and 90 s at 68°C.
ACKNOWLEDGMENTS
We are very grateful to Cornelia Prohl, Jana Gerber, and Roland Lill for the generous gift of the Δbat1/Δbat2 yeast strain. We thank Conny Guha and Bärbel Weber for excellent technical assistance and Axel Brennicke for his ongoing support. We also thank the Arabidopsis Biological Resource Center for the kind gift of cDNA clones 97D16T7, 123A19T7, ATTS3314, and ATTS4608.
Footnotes
This work was supported by the Fonds der Chemischen Industrie and by the Anfangsförderung der Universität Ulm. J.S. is a fellow of the Studienstiftung des Deutschen Volkes.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001602.
LITERATURE CITED
- Alban C, Baldet P, Axiotis S, Douce R. Purification and characterization of 3-methylcrotonyl-coenzyme A carboxylase from higher plant mitochondria. Plant Physiol. 1993;102:957–965. doi: 10.1104/pp.102.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson MD, Che P, Song J, Nikolau BJ, Wurtele ES. 3-Methylcrotonyl-coenzyme A carboxylase is a component of the mitochondrial leucine catabolic pathway in plants. Plant Physiol. 1998;118:1127–1138. doi: 10.1104/pp.118.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Alban C, Bligny R, Douce R. Induction of beta-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett. 1996;383:175–180. doi: 10.1016/0014-5793(96)00244-x. [DOI] [PubMed] [Google Scholar]
- Bledsoe RK, Dawson PA, Hutson SM. Cloning of the rat and human mitochondrial branched chain aminotransferases (BCATm) Biochim Biophys Acta. 1997;1339:9–13. doi: 10.1016/s0167-4838(97)00044-7. [DOI] [PubMed] [Google Scholar]
- Däschner K, Couee I, Binder S. The mitochondrial isovaleryl-coenzyme A dehydrogenase of Arabidopsisoxidizes intermediates of leucine and valine catabolism. Plant Physiol. 2001;126:601–612. doi: 10.1104/pp.126.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Däschner K, Thalheim C, Guha C, Brennicke A, Binder S. In plants a putative isovaleryl-CoA-dehydrogenase is located in mitochondria. Plant Mol Biol. 1999;39:1275–1282. doi: 10.1023/a:1006129220778. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- Davoodi J, Drown PM, Bledsoe RK, Wallin R, Reinhart GD, Hutson SM. Overexpression and characterization of the human mitochondrial and cytosolic branched-chain aminotransferases. J Biol Chem. 1998;273:4982–4989. doi: 10.1074/jbc.273.9.4982. [DOI] [PubMed] [Google Scholar]
- Faivre-Nitschke SE, Couee I, Vermel M, Grienenberger JM, Gualberto JM. Purification, characterization and cloning of isovaleryl-CoA dehydrogenase from higher plant mitochondria. Eur J Biochem. 2001;268:1332–1339. doi: 10.1046/j.1432-1327.2001.01999.x. [DOI] [PubMed] [Google Scholar]
- Gerbling H, Gerhardt B. Oxidative decarboxylation of branched-chain 2-oxo fatty acids by higher plant peroxisomes. Plant Physiol. 1988;88:13–15. doi: 10.1104/pp.88.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbling H, Gerhardt B. Peroxisomal degradation of branched-chain 2-oxo acids. Plant Physiol. 1989;91:1387–1392. doi: 10.1104/pp.91.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelstein P, Sieve B, Klein M, Jans H, Schultz G. Leucine synthesis in chloroplasts: Leucine/isoleucine aminotransferase and valine aminotransferase are different enzymes in spinach chloroplasts. J Plant Physiol. 1997;150:23–30. [Google Scholar]
- Inoue K, Kuramitsu S, Aki K, Watanabe Y, Takagi T, Nishigai M, Ikai A, Kagamiyama H. Branched-chain amino acid aminotransferase of Escherichia coli: overproduction and properties. J Biochem. 1988;104:777–784. doi: 10.1093/oxfordjournals.jbchem.a122549. [DOI] [PubMed] [Google Scholar]
- Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol. 1998;8:759–769. doi: 10.1016/s0959-440x(98)80096-1. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kispal G, Steiner H, Court DA, Rolinski B, Lill R. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J Biol Chem. 1996;271:24458–24464. doi: 10.1074/jbc.271.40.24458. [DOI] [PubMed] [Google Scholar]
- Koop HU, Steinmüller K, Wagner H, Rossler C, Eibl C, Sacher L. Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta. 1996;199:193–201. doi: 10.1007/BF00196559. [DOI] [PubMed] [Google Scholar]
- Kuramitsu S, Ogawa T, Ogawa H, Kagamiyama H. Branched-chain amino acid aminotransferase of Escherichia coli: nucleotide sequence of the ilvEgene and the deduced amino acid sequence. J Biochem. 1985;97:993–999. doi: 10.1093/oxfordjournals.jbchem.a135176. [DOI] [PubMed] [Google Scholar]
- Lea PJ, Ireland RJ. Nitrogen metabolism in higher plants. In: Singh BK, editor. Plant Amino Acids: Biochemistry and Biotechnology. New York: Marcel Dekker; 1999. pp. 1–47. [Google Scholar]
- Mehta PK, Christen P. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv Enzymol Relat Areas Mol Biol. 2000;74:129–184. doi: 10.1002/9780470123201.ch4. [DOI] [PubMed] [Google Scholar]
- Mehta PK, Hale TI, Christen P. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur J Biochem. 1993;214:549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x. [DOI] [PubMed] [Google Scholar]
- Okada K, Hirotsu K, Sato M, Hayashi H, Kagamiyama H. Three-dimensional structure of Escherichia colibranched-chain amino acid aminotransferase at 2.5 A resolution. J Biochem. 1997;121:637–641. doi: 10.1093/oxfordjournals.jbchem.a021633. [DOI] [PubMed] [Google Scholar]
- Reinard T, Janke V, Willard J, Buck F, Jacobsen HJ, Vockley J. Cloning of a gene for an acyl-CoA dehydrogenase from Pisum sativumL. and purification and characterization of its product as an isovaleryl-CoA dehydrogenase. J Biol Chem. 2000;275:33738–33743. doi: 10.1074/jbc.M004178200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Coruzzi GM. The aspartate aminotransferase gene family of Arabidopsisencodes isoenzymes localized to three distinct subcellular compartments. Plant J. 1995;7:61–75. doi: 10.1046/j.1365-313x.1995.07010061.x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK. Biosynthesis of valine, leucine, and isoleucine. In: Singh BK, editor. Plant Amino Acids: Biochemistry and Biotechnology. New York: Marcel Dekker; 1999. pp. 227–247. [Google Scholar]
- Slater S, Mitsky TA, Houmiel KL, Hao M, Reiser SE, Taylor NB, Tran M, Valentin HE, Rodriguez DJ, Stone DA et al. Metabolic engineering of Arabidopsis and Brassicafor poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production. Nat Biotechnol. 1999;17:1011–1016. doi: 10.1038/13711. [DOI] [PubMed] [Google Scholar]
- Sodek L, Wilson CM. Metabolism of lysine and leucine derived from storage protein during the germination of maize. Biochim Biophys Acta. 1973;304:353–362. doi: 10.1016/0304-4165(73)90253-5. [DOI] [PubMed] [Google Scholar]
- Song J, Wurtele ES, Nikolau BJ. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of 3-methylcrotonoyl-CoA carboxylase: identification of the biotin carboxylase and biotin-carrier domains. Proc Natl Acad Sci USA. 1994;91:5779–5783. doi: 10.1073/pnas.91.13.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CR, Beevers H. Gluconeogenesis from amino acids in germinating castor bean endosperm and its role in transport to the embryo. Plant Physiol. 1967;42:1587–1595. doi: 10.1104/pp.42.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wurtele ES, Keller G, McKean AL, Nikolau BJ. Molecular cloning of cDNAs and genes coding for beta-methylcrotonyl-CoA carboxylase of tomato. J Biol Chem. 1994;269:11760–11768. [PubMed] [Google Scholar]
- Weaver LM, Lebrun L, Franklin A, Huang L, Hoffman N, Wurtele ES, Nikolau BJ. Molecular cloning of the biotinylated subunit of 3-methylcrotonyl-coenzyme A carboxylase of Arabidopsis thaliana. Plant Physiol. 1995;107:1013–1014. doi: 10.1104/pp.107.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbach VA, Abell LM. Inhibitors of valine, leucine and isoleucine biosynthesis. In: Singh BK, editor. Plant Amino Acids: Biochemistry and Biotechnology. New York: Marcel Dekker; 1999. pp. 385–416. [Google Scholar]
- Yennawar N, Dunbar J, Conway M, Hutson S, Farber G. The structure of human mitochondrial branched-chain aminotransferase. Acta Crystallogr D Biol Crystallogr. 2001;57:506–515. doi: 10.1107/s0907444901001925. [DOI] [PubMed] [Google Scholar]